Abstract
The filamentous bacterium Streptomyces coelicolor differentiates by forming specialized, spore-bearing aerial hyphae that grow into the air. Using microarrays, we identified genes that are down-regulated in a mutant unable to erect aerial hyphae. Through this route, we identified a previously unknown layer of aerial mycelium surface proteins (the “chaplins”). The chaplins share a hydrophobic domain of ∼40 residues (the “chaplin domain”), and all have a secretion signal. The five short chaplins (ChpD,E,F,G,H) have one chaplin domain, whereas the three long chaplins (ChpA,B,C) have two chaplin domains and a C-terminal “sorting signal” that targets them for covalent attachment to the cell wall by sortase enzyme. Expression of the two chaplin genes examined (chpE, chpH) depended on aerial hyphae formation but not sporulation, and egfp fusions showed their expression localized to aerial structures. Mass spectrometry of cell wall extracts confirmed that the short chaplins localized to the cell surface. Deletion of chaplin genes caused severe delays in aerial hyphae formation, a phenotype rescued by exogenous application of chaplin proteins. These observations implicate the chaplins in aerial mycelium formation, and suggest that coating of the envelope by the chaplins is required for aerial hyphae to grow out of the aqueous environment of the substrate mycelium into the air.
Keywords: Streptomyces, differentiation, hydrophobic protein, cell-surface protein, sortase, microarray
The environmental variability of terrestrial ecosystems requires that organisms living in the soil must develop ways of adapting to all conditions they may encounter. Two such groups of organisms, the filamentous fungi and the filamentous bacteria, have independently adopted similar strategies for growth and propagation in their terrestrial environments. They both form a substrate or feeding mycelium when growing in a hydrophilic environment, and respond to stress (nutritional, physical, chemical, or biological) or encounters with hydrophobic environments by erecting a hydrophobic aerial structure in which spores or fruiting bodies will form. This response to changing environments implies an ability to modulate the surface layers of the mycelial filaments in accordance with the environmental conditions.
Filamentous fungi, when confronted by hydrophobic conditions, secrete highly surface-active proteins known as hydrophobins. Hydrophobins are small (∼100-amino-acid) proteins that are produced during the formation of aerial hyphae and fruiting bodies. They show little overall sequence conservation, but share a characteristic hydropathy profile and have eight conserved cysteine residues that are involved in disulphide bond formation (Wösten and de Vocht 2000). They self-assemble at hydrophilic–hydrophobic interfaces (e.g., at air–water interfaces, or water–hydrophobic solid surfaces) to form an amphipathic protein film, made up of interwoven pairs of rodlets (Wösten et al. 1993). Hydrophobins serve multiple functions, acting as surfactants to mediate the escape of hyphae from aqueous environments, conferring hydrophobicity to aerial structures, and attaching hyphae to hydrophobic surfaces (for review, see Wessels 1997).
In the filamentous bacterium Streptomyces coelicolor, early studies identified an outer hydrophobic layer termed the fibrous sheath that is present around aerial hyphae but absent from vegetative hyphae (Hopwood and Glauert 1961; Wildermuth et al. 1971). Three proteins/polypeptides, RdlA, RdlB, and SapB, have been shown to perform functions analogous to those of the fungal hydrophobins, and may contribute to the formation of this fibrous sheath. The rodlin proteins, RdlA and RdlB, have an amphipathic profile, and assemble to form a basketwork of paired rodlets on the surface of aerial hyphae and spores, very similar in appearance to that of the fungal hydrophobins (Claessen et al. 2002). They are expressed during aerial hyphae formation and seem to be involved in adhesion to hydrophobic surfaces. However, loss of the rodlin proteins does not affect the growth of aerial hyphae, or the overall hydrophobicity of the aerial structures (Claessen et al. 2002).
On rich media such as R2YE, erection of the aerial mycelium is mediated, at least in part, by a modified 18-amino-acid hydrophobic peptide called SapB (spore-associated protein B; Guijarro et al. 1988). However, on other media such as minimal medium containing mannitol as the sole carbon source, S. coelicolor raises an aerial mycelium without production of SapB (Willey et al. 1991), implying that there may be at least two pathways by which aerial hyphae can form. Like fungal hydrophobins, SapB acts as a surfactant, coating the air–water interface and reducing the surface tension to allow the growth of the aerial mycelium (Tillotson et al. 1998). It is not, however, associated with the aerial structures and, like the rodlins, is not thought to contribute to the hydrophobicity of the aerial mycelium and the spores (Wösten and Willey 2000). The gene for SapB has remained elusive, and although various lines of evidence had suggested that it may be nonribosomally encoded (Willey et al. 1991, 1993), no such candidate for this has emerged from the S. coelicolor genome sequence (Bentley et al. 2002). The expression of SapB depends on a class of genes termed the bld genes, which are required for aerial hyphae formation (Kelemen and Buttner 1998). Studies of the known bld genes have revealed that, for the most part, they specify regulatory proteins: bldB (Pope et al. 1998), bldC (A.C. Hunt, pers. comm.), bldD (Elliot et al. 1998, 2001), and bldM (Molle and Buttner 2000) encode known or putative DNA-binding proteins; bldN encodes an extracytoplasmic function (ECF) sigma factor (Bibb et al. 2000; Bibb and Buttner 2003), and bldG encodes an anti-sigma factor antagonist (Bignell et al. 2000). The expression of SapB is impaired in all bld mutant strains examined to date (Willey et al. 1991).
The findings that loss of the rodlin proteins does not affect the growth of aerial hyphae or the overall hydrophobicity of the aerial mycelium (Claessen et al. 2002), and that SapB is only required for aerial mycelium formation on a subset of media (Willey et al. 1991) imply that other, as yet uncharacterized proteins must be involved in creating the hydrophobic surface of aerial hyphae. Using DNA microarray technology, we have identified a set of genes whose expression depends genetically on the developmental sigma factor gene, bldN. Through this route, we have gone on to characterize a family of secreted, hydrophobic proteins that form a previously unknown layer of the aerial surface structures, and which are important for the growth of aerial hyphae.
Results
Comparison of global transcription in the bldN null mutant J2177 and its bldN+ congenic parent J1915
To examine the effect of bldN on gene expression during S. coelicolor development, we compared the global transcription profiles of J1915 (bldN+) and J2177 (bldN) using DNA microarrays. Strains were cultivated on R2YE solid medium, and mycelium was harvested for RNA purification at six time points. For J1915, these time points corresponded to early and late vegetative growth, early and late aerial hyphae formation, and early and late sporulation. As the inocula for J1915 and J2177 were different in nature (spores vs. mycelium), the inoculum densities were adjusted to give similar biomass accumulation for both during vegetative growth. J2177 was then harvested at the same times as J1915. bldN is not expressed at the earliest stages of vegetative growth (Bibb et al. 2000), so the first RNA samples from J1915 and J2177 were used as the references (green) to which all subsequent samples (red) were compared for their respective time courses. The microarray experiments were carried out in duplicate.
There are 7825 annotated protein-coding genes in the S. coelicolor genome (Bentley et al. 2002), and 7071 of these were represented on the DNA microarrays used in these experiments (90.4%). Seventeen genes were identified whose expression levels were induced greater than twofold in more than three time points (i.e., the time during which σBldN would be active) in J1915 but not in the bldN mutant, J2177 (Fig. 1A). For five of these genes, these results were confirmed by S1 nuclease protection experiments (Fig. 1B).
Figure 1.
(A) Expression profiles of genes whose expression differs in a bldN mutant strain and its congenic parent, J1915. Profiles indicate the extent of gene expression in each sample relative to the reference (the first RNA sample/time point taken), with red indicating an increase in transcript abundance and green indicating a decrease, relative to the reference. Black represents unchanged expression levels, and gray indicates the absence of data. (B) S1 nuclease protection assays examining the expression of selected genes from 1A, to confirm dependence of expression on bldN. hrdB was used as a control for RNA integrity and RNA loading levels. The RNA used for these experiments was the same as was used for the microarray analysis shown in A.
Identification of the chaplin family of cell surface-associated proteins
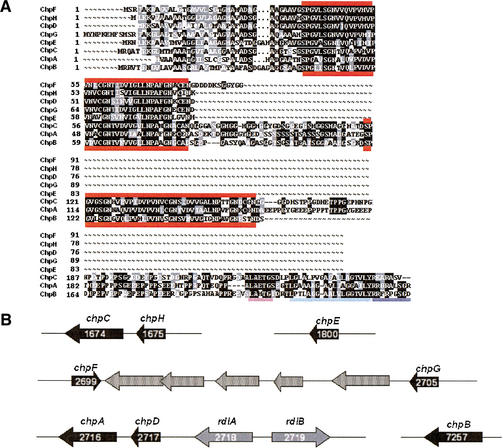
Six of the 17 genes whose expression appeared to be downregulated in the bldN mutant encoded secreted proteins. One of these, SCO2718, was recently characterized and named rdlA (Claessen et al. 2002). It encodes one of the two rodlin proteins on the surface of aerial hyphae and spores, and its expression appears to require the formation of aerial hyphae (Claessen et al. 2002). Another of the genes that specifies a secreted protein, SCO4002, encodes a product unlike any annotated protein in the databases. Three of the secreted proteins were found to resemble each other (SCO1800, SCO1675, and SCO1674) and to be part of a larger family, with five other members in S. coelicolor (SCO2699, SCO2705, SCO2716, SCO2717, and SCO7257). Database analysis showed that members of this family of proteins are present in other actinomycetes [e.g., Streptomyces verticillus (Sanchez et al. 2001), and Thermobifida fusca (NCBI Microbial Genomes Annotation Project)], but have not so far been identified in any organism outside of this order. For reasons described below and in the accompanying manuscript (Claessen et al. 2003), this family of secreted proteins has been designated the chaplin family, for coelicolor hydrophobic aerial proteins, with gene designations chpA–H (Fig. 2A).
Figure 2.
(A) Alignment of the eight chaplin proteins. The thick red lines mark the chaplin domains. The requirements for covalent cell wall attachment are underlined at the C-terminal end of ChpA–C: the LAXTG sortase recognition motif is underlined in pink, the hydrophobic region following is underlined in light blue, and the positively charged tail is underlined in dark blue. (B) Organization of the chaplin genes. Chaplin genes are shown as black arrows, rodlin genes are shown as gray arrows, and unrelated intervening genes are shown as stippled arrows. Within the rodlin and chaplin gene arrows are the corresponding SCO numbers in white, indicating the position of the gene in the chromosome (genes are numbered 1–7825).
We have termed the region of similarity shared by the members of the chaplin family the chaplin domain (Fig. 2A). In S. coelicolor, three of the chaplins (ChpA–C; the long chaplins) are ∼210–230 amino acids in length (after cleavage of the leader peptide), and contain two chaplin domains separated by ∼35 amino acids. The other five chaplin proteins (ChpD–H; the short chaplins) are only ∼50–60 amino acids in length and contain a single chaplin domain. The chaplin domain itself is very hydrophobic, containing ∼60%–65% hydrophobic amino acids. There is no obvious pattern to the distribution of the hydrophobic residues, but there are three absolutely conserved GN motifs, spaced 12–13 residues apart, which may contribute to the structure of these domains. Secondary structure analysis predicts the chaplin domains to be rich in β-sheets, with the GN residues falling at the turns separating the individual β-sheets. There are also two highly conserved cysteine residues in all chaplin domains except for ChpE and the second (C-terminal) chaplin domain of ChpB.
The genetic organization of the chaplin genes is also striking (Fig. 2B). The genes encoding two of the three long chaplins are found next to those for a short chaplin: chpH is directly upstream of chpC, and chpD is immediately upstream of chpA. chpD and chpA are also immediately adjacent to rdlA and rdlB, which encode the two rodlin proteins (Claessen et al. 2002).
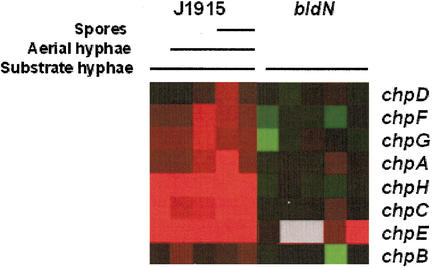
To determine whether the expression of all chp genes was affected by the bldN mutation, the microarray data were reanalyzed, looking specifically at chp expression. Expression of each was greater in the bldN+ strain than in the bldN mutant strain (Fig. 3). This difference was most striking for chpH,C and chpE, which were identified in the original screen, and were up-regulated at all times examined. chpD,F,G, and chpA all appeared to be temporally regulated, with their expression increasing in the wild type during late aerial hyphae formation and early sporulation, suggesting that they might function during the later stages of development, whereas the expression of chpB appeared to be unchanged between the wild-type and mutant strains. To confirm the apparent dependence of these genes on bldN, S1 nuclease protection assays were carried out for each of the chaplin genes. There was either very little expression, or a complete absence of expression, of the eight genes in the bldN mutant strain.
Figure 3.
Expression profiles of the chaplin genes in the bldN mutant strain and its congenic parent, J1915. The color representation is as described in the legend for Figure 1A.
Potential cell wall localization of the long chaplins
The three long chaplins (ChpA–C) are potential targets for the cell wall sorting enzyme, sortase (Mazmanian et al. 1999). S. coelicolor is predicted to have seven sortase enzymes (Pallen et al. 2001) and up to 36 potential substrates, including the three long chaplins (A. Hesketh, pers. comm.). Sortases are transpeptidases that recognize a conserved C-terminal motif, typically consisting of an LPXTG signature sequence, followed by a string of hydrophobic amino acids, and a positively charged tail (Navarre and Schneewind 1994; Mazmanian et al. 1999). They cleave between the threonine and glycine residues of the LPXTG motif, and mediate covalent attachment of the C-terminal end of the target protein to the peptidoglycan crossbridge, which, in Streptomyces, is a single glycine. ChpA, ChpB, and ChpC have an LAXTG motif, followed by a hydrophobic region of 22–23 amino acids and a 7–9-amino-acid tail with at least three lysine or arginine residues (Fig. 2A). The region between the second chaplin domain and the likely sortase-recognition sequence is very proline-rich, and secondary structure analysis predicts it to form a long coil. This is likely to represent a peptidoglycan-spanning domain, which is typically rich in proline residues and repeating sequences (Fischetti 2000).
Spatial localization of chpH and chpE expression
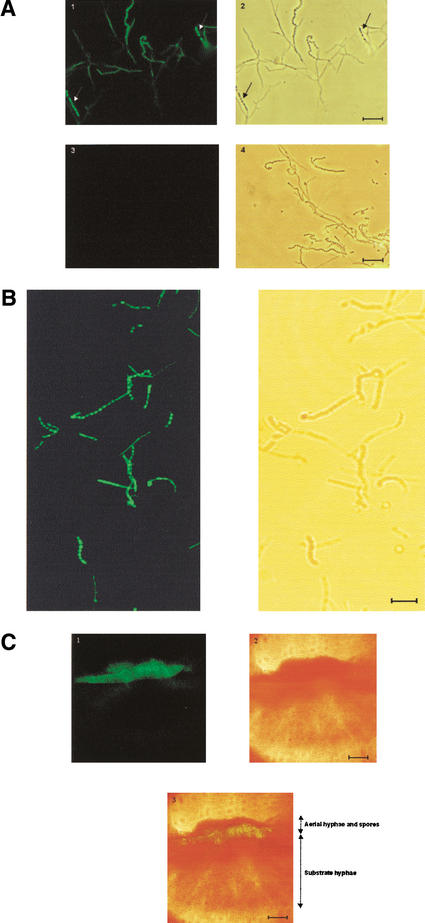
The temporal correlation of chp gene transcription with the formation of aerial hyphae in surface-grown cultures, and the dependence of this expression on bldN, prompted us to examine the spatial location of chpE and chpH transcription within differentiating colonies. The promoter region of each gene was inserted upstream from the enhanced green fluorescent protein (egfp) gene in pIJ8660 (Sun et al. 1999). Analysis by fluorescence light microscopy (Fig. 4A) and confocal microscopy (Fig. 4B) revealed that there was expression of chpH in some hyphal filaments, but that chpH expression was strongest in the spores. A similar pattern of expression was also seen for chpE (data not shown). A cross-section of a colony containing the chpH–egfp transcriptional fusion construct also revealed chpH expression to occur primarily in the aerial mycelium (Fig. 4C).
Figure 4.
Localization of chpH promoter activity using the egfp reporter gene. (A) Light microscope images of M600 carrying a chpH–egfp transcriptional fusion, or egfp alone. (Left) Fluorescent images. (Right) Phase contrast images. Panels 1 and 2 show images of M600 containing the chpH–egfp transcriptional fusion, and panels 3 and 4 show M600 containing promoterless egfp. Arrows in panels 1 and 2 indicate spore chains that show the greatest fluorescence. (B) Confocal microscope images of M600 containing the chpH–egfp transcriptional fusion. The strain containing egfp alone was used to adjust the fluorescence, removing any visible autofluorescence before examining the chpH–egfp-containing strain. (C) Confocal microscope image of a cross-section of a colony (M600 carrying a chpH–egfp transcriptional fusion). Panel 1 shows the fluorescent image, panel 2 shows the phase contrast image, and panel 3 shows an overlay of panel 1 on 2, illustrating the localization of fluorescence to the aerial structures. Bars: A,B, 10 μm; C, 100 μm.
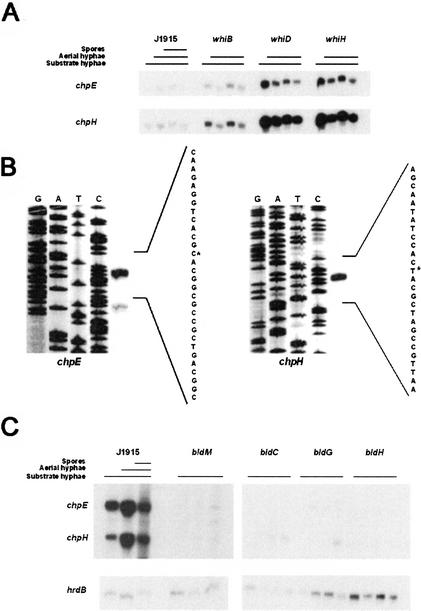
Given the abundant expression of chpE and chpH observed in spores, we were interested to determine whether sporulation was required for this expression. We conducted S1 nuclease protection experiments using RNA harvested from representative sporulation-deficient mutants (whiB, whiD, and whiH), and found that both chpE and chpH were expressed in all three whi mutant strains (Fig. 5A). We also introduced the chpH–egfp transcriptional fusion into whiH and whiD mutant strains, and found substantial chpH expression in the aerial hyphae (data not shown). Sporulation is therefore not required for the expression of these chaplins.
Figure 5.
(A) S1 nuclease protection assays of chpE and chpH expression in J1915 and whiB, whiD, and whiH mutant strains during growth on solid medium (R2YE). (B) High-resolution S1 nuclease mapping of the 5′ end of the chpE (left) and chpH (right) transcripts in S. coelicolor J1915. The most likely transcription start points are indicated by asterisks. Lanes G, A, T, and C are a dideoxy sequencing ladder generated using the same oligonucleotide as was used to make the S1 mapping probe. (C) S1 nuclease protection assays of chpE and chpH transcription in J1915 and bldM, and bldC, bldG, and bldH mutant strains during growth on solid medium (R2YE). hrdB served as a positive control.
Expression of the chaplins in bld mutants
High-resolution S1 nuclease mapping was conducted to determine the transcription start sites for chpE and chpH. A single start site was detected for each: 43 nt upstream from the translation start site of chpE and 44 nt upstream of the translation start site of chpH (Fig. 5B). Examination of the sequence upstream from the transcription start sites for these two genes, and generally upstream of the remaining chaplin genes, did not reveal any sequences resembling a σBldN target promoter (Bibb et al. 2000), suggesting that the transcriptional dependence on bldN was indirect.
σBldN directs transcription from the major promoter of another developmental regulatory gene, bldM (Bibb et al. 2000), so it was possible that the effects of the bldN mutation on chp expression were caused by the absence of BldM. S1 nuclease protection assays were conducted to compare the expression of chpH and chpE in the bldM null mutant strain J2151 and its congenic bldM+ parent strain, J1915. The expression of chpH and chpE was drastically reduced in the bldM mutant compared to J1915 (Fig. 5C), raising the possibility of regulation by BldM, although the dependence of these genes on bldM could also be indirect. To determine whether these genes were expressed in any bld mutant strains, S1 nuclease mapping experiments were conducted using RNA isolated from bldC, bldG, and bldH strains. Compared to wild-type levels, the expression of chpH and chpE in each of the bld mutant strains examined was significantly decreased (Fig. 5C). This suggests that although there is some expression of these chaplin genes in the absence of differentiation, wild-type levels of expression are associated with the formation of aerial hyphae.
Disruption of the chp genes affects aerial hyphae formation and sporulation
To address the function of the chaplin proteins, a number of gene disruptions were created. chpH was replaced with an apramycin resistance cassette. Knockout of chpH conferred no obvious phenotype: colonies appeared wild type when grown on media containing different carbon sources, coverslip impressions examined by light microscope revealed no sporulation defects, and scanning electron microscopy (SEM) revealed that the majority of spores appeared wild-type, with a few abnormal spore protrusions and spore sizes (data not shown).
We also knocked out the three genes (chpA,B,C) encoding the sortase-targeted chaplins. Loss of chpA (in a chpA,D double knockout), or chpB, or chpC (in a chpC,H double knockout) caused no discernible change in phenotype. However, combinations of these knockouts did affect colony differentiation. Loss of chpA and chpC (in a chpA,C,D,H quadruple knockout) delayed aerial hyphae formation and sporulation (Fig. 6A,B), and a more severe phenotype was seen when an additional deletion removed the last remaining sortase-targeted chaplin gene, chpB, to generate a chpA,B,C,D,H quintuple mutant, with a greater delay in aerial hyphae formation, and an almost complete lack of sporulation (Fig. 6A,B). When these strains did finally produce aerial mycelium, the surface of the colonies was hydrophobic. The delay in aerial hyphae formation of the quadruple mutant could be complemented by the reintroduction of a single long chaplin gene (chpC) together with genes for the two short chaplins (chpD and chpH; data not shown).
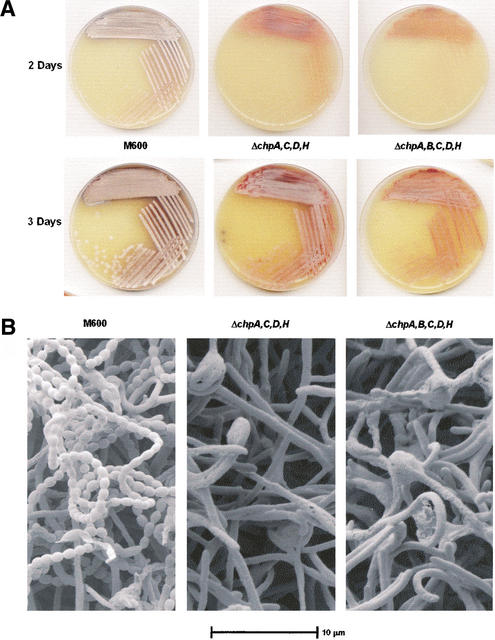
Figure 6.
(A) Delayed aerial hyphae formation of ΔchpA,C,D,H and ΔchpA,B,C,D,H strains, compared with the wild-type strain, M600, when grown on MS agar. (B) Scanning electron microscope images of M600, ΔchpA,C,D,H and ΔchpA,B,C,D,H after growth for 4 d on MS agar.
Localization of the short chaplin proteins on the colony surface
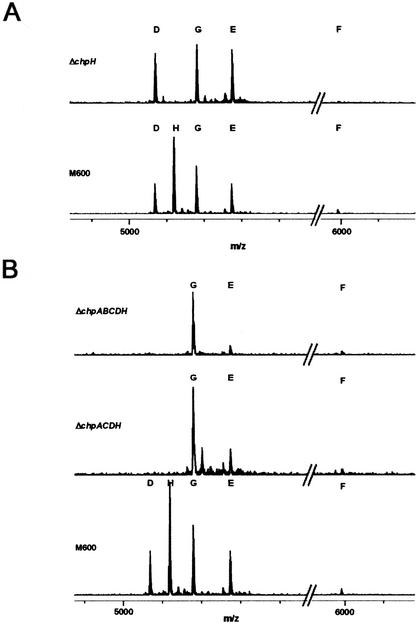
To examine the potential cell wall localization of the chaplin proteins, surface-grown S. coelicolor cultures were subjected to extraction procedures previously used to isolate the rodlin proteins from S. coelicolor and the hydrophobin SC3 from the filamentous fungus Schizophyllum commune (Wösten et al. 1993; Claessen et al. 2002). Cell wall fractions were extracted with boiling SDS, and the SDS-insoluble matter was extracted with trifluoroacetic acid (TFA). The TFA-soluble fraction was then analyzed by MALDI-TOF mass spectrometry, and the profile of the wild-type M600 strain was compared with those of various mutants. As shown in Figure 7A, five peaks, corresponding to the five short chaplins, were evident in the M600 sample, whereas for the ΔchpH strain there were only four peaks, due to the loss of the peak corresponding to ChpH. This demonstrated that the short chaplin proteins were indeed localized to the cell envelope, and showed that assembly of the short chaplins on the cell surface could occur in the absence of other individual chaplin proteins. The chaplin protein profiles of the quadruple (chpA,C,D,H) and quintuple (chpA,B,C,D,H) mutants, lacking two or all three of the sortase-targeted chaplins, respectively, were also examined, and, in both cases, the remaining three short chaplins (ChpE,F,G) were detected (Fig. 7B), suggesting that the cell wall-attached chaplins were not absolutely required for polymerization of the short chaplins on the cell surface.
Figure 7.
MALDI-TOF mass spectra of the short chaplins in M600 and ΔchpH (A), and ΔchpA,C,D,H and ΔchpA,B,C,D,H (B). The X-axis shows the mass (m)/charge (z), where z = 1.
The masses of the short chaplin proteins were considered to be accurate within 0.5 Da; however, for four of the five chaplin proteins, the calculated mass differed from the predicted monoisotopic masses by 2 Da (Table 1; as the MALDI-TOF procedure adds a proton to the observed molecular weight). This could correspond to the loss of two hydrogens through disulphide bond formation between the two conserved cysteine residues. This explanation is supported by the observation that the mass of ChpE, which lacks the cysteine residues, did not show this 2-Da difference.
Table 1.
Molecular weight of the chaplins: Predicted versus observed using MALDI-TOF mass spectrometry
| Protein | Predicted monoisotopic mass (Da) | Observed mass (Da) |
|---|---|---|
| ChpD | 5067.52 | 5066.49 |
| ChpH | 5117.65 | 5116.61 |
| ChpG | 5178.51 | 5177.48 |
| ChpE | 5270.74 | 5271.65 |
| ChpF | 5989.85 | 5988.63 |
Extracellular rescue of the chp mutants by purified chaplins
To determine whether the exogenous application of chaplin proteins was capable of rescuing the defect in aerial hyphae formation of the chp quintuple mutant (lacking the three sortase-targeted chaplins, chpA,B, and chpC, and two of the short chaplins, chpD and chpH), samples were applied to the surface of the quintuple mutant after 24 h of growth on MS media. TFA-extracted material, resuspended in either water or 2% TFA, was found to restore aerial hyphae formation within 4 h of application, whereas the application of water, 2% TFA, or 2% SDS (which had been used to wash the cell walls prior to TFA extraction) had no effect on aerial hyphae formation (Fig. 8). The TFA-extracted material used was purified from wild-type S. coelicolor and would therefore have contained the rodlin proteins, RdlA and RdlB, in addition to the chaplins. However, in the accompanying report in this issue, Claessen et al. (2003) obtained comparable results using TFA-extracted material derived from an rdlAB double mutant, showing that the extracellular rescue of aerial hyphae formation in chp mutants does not require the rodlin proteins.
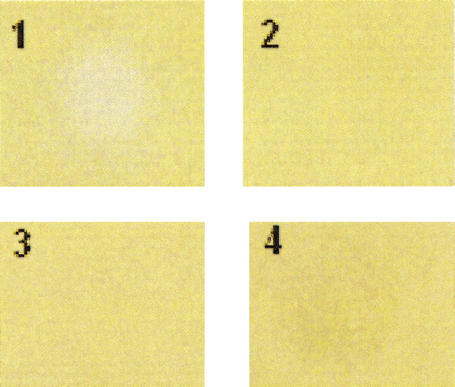
Figure 8.
Extracellular rescue of aerial hyphae formation of the ΔchpA,B,C,D,H mutant using cell-wall extract containing the short chaplins. The panels show application of short chaplin-containing extract (1), water (2), 2% SDS (3), and 2% TFA (4).
SapB is not derived from the chaplins
A partial N-terminal sequence of SapB (TXGXR) and its complete amino acid content have been determined (Willey et al. 1991), but the sapB gene has not been identified. Accordingly, we examined the possibility that the 18-amino-acid SapB peptide might be derived by processing of one of the chaplin proteins, but we found that none of the chaplin sequences was compatible with this hypothesis. All eight of the chaplin proteins lacked the TXGXR motif, and indeed, did not contain a single arginine residue outside of the signal peptide or cell wall-anchored region; nor did they contain the correct complement of amino acids predicted to make up SapB.
Discussion
A model of chaplin assembly and function
We have identified a novel family of secreted proteins, termed the chaplins, required for efficient formation of aerial mycelium in S. coelicolor. There are eight members of this family, all sharing a highly conserved, hydrophobic “chaplin” domain. Five of these proteins (ChpD–H) have a single chaplin domain, whereas the remaining three (ChpA–C) have two such domains, and are likely to be covalently attached to the cell wall by the sortase enzyme.
Based on the work presented here, we propose the following speculative model of chaplin assembly and function. Chaplins are expressed primarily during the formation of aerial hyphae and sporulation, and are secreted via the Sec pathway. The three long chaplins (ChpA–C) become covalently attached to the glycine crossbridge of the peptidoglycan through the action of one or more sortases. The five short chaplins (ChpD–H) are exported to the surface of the hyphae, where they polymerize through the interaction between chaplin domains, to form a hydrophobic layer. Because the long chaplins are sortase targets, heteropolymerization of the short and long chaplins serves to anchor the hydrophobic layer to the cell wall peptidoglycan. The hydrophobic chaplin layer could then function either alone, or in conjunction with the surfactant SapB on certain media, to permit hydrophobic aerial hyphae to break surface tension and grow into the air, where they differentiate to form chains of exospores.
The anchoring of the hydrophobic chaplin layer to the cell surface cannot, however, occur solely through the heteropolymerization of the short chaplins with the long, cell wall-anchored chaplins, because the remaining three short chaplins (ChpE,F,G) were still present in cell wall fractions derived from the quintuple (chpA,B, C,D,H) mutant lacking all three cell wall-bound chaplins (ChpA,B,C), as demonstrated by MALDI-TOF mass spectrometry. It is therefore possible that the short chaplins could interact with other components of the cell wall, such as carbohydrates. The observation that the quadruple and quintuple chaplin mutant strains were severely delayed in the formation of an aerial mycelium suggests that the remaining three chaplin proteins cannot fully substitute for the loss of the other five. This could be because of functional specialization of the different chaplins; for example, the proposed role for the long chaplins in the covalent attachment of the hydrophobic chaplin layer to the cell wall. Alternatively, a simple “critical concentration” of chaplin proteins may be required for polymerization, and this concentration may take much longer to reach in a strain lacking chpA,B,C,D,H; this is supported by the observation that exogenous application of a solution containing all of the short chaplin proteins could restore aerial hyphae formation.
Comparisons with hydrophobic cell-surface proteins in other bacteria
Although the chaplins bear no sequence similarity to any proteins found outside of the actinomycetes, the unusual combination of sortase-targeted proteins and homologous shorter secreted proteins is also characteristic of the internalin family of Listeria monocytogenes. The internalins are a family of leucine-rich repeat proteins, the most studied of which are surface proteins involved in the induction of phagocytosis by non-phagocytic cell types, thereby promoting internalization of the bacteria into mammalian host cells (Gaillard et al. 1991). Like the chaplins, where there are two chaplin domains in the long proteins and only one in the short proteins, the number of leucine-rich repeats in the long, cell wall-anchored internalins is greater than in the two short internalins, InlB and InlC, which lack a C-terminal sortase recognition motif (Gaillard et al. 1991; Dramsi et al. 1997). Of these two short internalins, InlB is associated with the lipoteichoic acid of the bacterial cell wall, although a considerable amount can also be found in a free secreted form (Jonquieres et al. 1999), whereas InlC is found primarily in the cytosolic compartment of infected host cells (Engelbrecht et al. 1996). In contrast, all the internalins from the animal pathogen Listeria ivanovii are of the short variety lacking the cell wall anchor, and there is no indication that they are cell wall-associated (Engelbrecht et al. 1998).
A striking analogy can also be drawn between the chaplins and the proteins that make up the curli fibrils in Escherichia coli (Olsén et al. 1989). Curli are highly aggregated surface fibrils implicated in binding to human proteins (e.g., see Sjöbring et al. 1994; Ben Nasr et al. 1996), in colonization of inert surfaces (Vidal et al. 1998), and in biofilm formation (Austin et al. 1998). The curli fibrils are expressed under conditions of low temperature, low nutrient condition, low osmolarity, and during stationary phase (Prigent-Combaret et al. 2000), and are assembled by the aggregation/precipitation of secreted, soluble CsgA monomers (Olsén et al. 1993). CsgB, a CsgA homolog that makes up a minor component of the curli fibrils themselves (Arnqvist et al. 1994), is believed to act primarily as a nucleator protein for the polymerization of CsgA (Hammar et al. 1996), and may be anchored to the membrane by the lipoprotein CsgG (Loferer et al. 1997). The ChpA, B, and C proteins may therefore play an anchoring/nucleation role analogous to that of CsgG and CsgB. Interestingly, like the chaplins, the CsgA and CsgB curli proteins have regularly spaced GN motifs and are predicted to be rich in β-sheets (Hammar et al. 1996).
The curli proteins also share many features with the hydrophobins of the filamentous fungi. Both of these classes of proteins can be considered “adhesins”, binding to particular host proteins in the case of the curli proteins, and to hydrophobic surfaces in the case of the hydrophobins. In addition, they both self-assemble to form filaments that bind thioflavin T and Congo red, behavior characteristic of so-called amyloid filaments (de Vocht et al. 2000; Wösten 2001; Chapman et al. 2002), like those associated with bovine spongiform encephalopathy (BSE) and Alzheimer's disease. In the accompanying report, Claessen et al. (2003) show that the chaplin proteins are also capable of self-assembling to form amyloid-like filaments.
Roles of the chaplins, the rodlins, and SapB
The growth of aerial hyphae and the physical properties conferred on these hyphae depend on at least three groups of proteins and polypeptides: the chaplins, the rodlins, and SapB. Together, they appear to do in S. coelicolor what is accomplished by a single family of proteins, the hydrophobins, in the filamentous fungi. The hydrophobins, like SapB, aid in the escape of hyphae from aqueous environments. Like the rodlins, the hydrophobins are involved in attaching hyphae to hydrophobic surfaces and providing the surface of aerial structures with a basketwork of paired rodlets. And like the chaplins, the hydrophobins are required for growth of the aerial hyphae out of the aqueous environment of the vegetative mycelium and into the air.
We assume that hyphal hydrophobicity is a prerequisite for aerial hyphae formation. When the chpA,B, C,D,H strain did finally produce an aerial mycelium, the surface of the colonies was hydrophobic. Perhaps this is because the remaining three chaplins (ChpE,F,G) are still present on the hyphal surface (as seen by mass spectrometry), and the additional loss of these proteins would eliminate aerial mycelium formation. The chpA,B,C,D,H strain also has the capacity to synthesize the amphipathic rodlin proteins, RdlA and RdlB. Deletion of the rodlin genes alone does not affect aerial hyphae formation in any detectable way, nor does it have any impact on the hydrophobic nature of the aerial hyphae and spores (Claessen et al. 2002). However, the chaplins and rodlins might have some functional redundancy, and deletion of both classes of genes might be required to completely eliminate aerial hyphae formation, hyphal hydrophobicity, and hyphal attachment to hydrophobic surfaces.
Sortase enzymes in S. coelicolor
S. coelicolor has seven potential sortase genes, two of which lie adjacent to each other in the chromosome and encode products highly similar to SrtA of Staphylococcus aureus, whereas the remaining five encode proteins more divergent in sequence (Pallen et al. 2001). A second sortase, SrtB, was recently identified in S. aureus (Mazmanian et al. 2002), but it is not closely related to any of the five divergent sortase homologs in S. coelicolor. In contrast to SrtA, which recognizes an LPXTG sorting signal, SrtB recognizes the pentapeptide sequence NPQTN (Mazmanian et al. 2002), showing that different sortase enzymes have their own subset of targets. In many cases, the genes specifying the proposed target substrates are adjacent to the gene for a particular sortase enzyme (Pallen et al. 2001). None of the three long chaplin genes is next to a sortase gene. However, based on their LAXTG recognition motif, which is very similar to the LPXTG recognition motif of SrtA (Navarre and Schneewind 1994), it seems most likely that one or both of the SrtA homologs in S. coelicolor would be responsible for the cell wall anchoring of the long chaplin proteins.
Materials and methods
Strains and growth conditions
The S. coelicolor strains used in these studies are summarized in Table 2. They were cultivated on solid R2YE, MS (mannitol plus soya flour), and DNA medium (Kieser et al. 2000). For DNA extraction, they were grown in TSB (trypticase soy broth), and for protoplast preparation, they were grown in YEME medium supplemented with 34% sucrose, MgCl2 to 5 mM and glycine to 0.5%. Protoplast manipulations and transformations were carried out as described by Kieser et al. (2000). Conjugation from E. coli required passage of plasmids through the dam dcm hsdS E. coli strain ET12567 (MacNeil et al. 1992) as described by Ryding et al. (1999) in order to bypass the methyl-specific restriction system of S. coelicolor. E. coli DH5α (Hanahan 1983) was used for plasmid construction and routine subcloning.
Table 2.
Streptomyces coelicolor strains used in this study
| Streptomyces strain names | Genotype | Parent strain | Source or reference |
|---|---|---|---|
| J1915 | ΔglkA119 SCP1− SCP2− | M145 | Kelemen et al. 1996 |
| M600 | SCP1− SCP2− | Chakraburtty and Bibb 1997 | |
| J2177 (bldN) | ΔglkA119 bldN::hyg SCP1− SCP2− | J1915 | Bibb et al. 2000 |
| J2151 (bldM) | ΔglkA119 bldM::hyg SCP1− SCP2− | J1915 | Molle and Buttner 2000 |
| J2152 (whiD) | ΔglkA119 whiD::hyg SCP1− SCP2− | J1915 | Molle et al. 2000 |
| J2210 (whiH) | whiH::hyg SCP1− SCP2− | M145 | Ryding et al. 1998 |
| J2402 (whiB) | whiB::hyg SCP1− SCP2− | M145 | Flärdh et al. 1999 |
| WC103 (bldG) | bldG103 hisA1 uraA1 strA1 Pg1− SCP1− SCP2− | J1501 | Champness 1988 |
| WC109 (bldH) | bldH109 hisA1 uraA1 strA1 Pg1− SCP1− SCP2− | J1501 | Champness 1988 |
| J2161 (bldC) | ΔbldC::aac(3)IV SCP1− SCP2− | M600 | A. Hunt, unpubl. |
| J3140 (chpH) | ΔchpH::aac(3)IV SCP1− SCP2− | M600 | This work |
| J3141 (chpC,H) | ΔchpCH::aac(3)IV SCP1− SCP2− | M600 | This work |
| J3142 (chpA,D) | ΔchpAD::aac(3)IV SCP1− SCP2− | M600 | This work |
| J3143 (chpA,C,D,H) | ΔchpAD ΔchpCH::aadA SCP1− SCP2− | M600 | This work |
| J3144 (chpB) | ΔchpB::aac(3)IV SCP1− SCP2− | M600 | This work |
| J3145 (chpA,B,C,D,H) | ΔchpAD ΔchpCH::aadA ΔchpB::aac(3)IV SCP1− SCP2− | M600 | This work |
RNA isolation, microarray analysis, and S1 nuclease mapping
For RNA isolation, S. coelicolor strains were grown on cellophane discs on R2YE medium, and RNA was extracted as described by Kelemen et al. (1996) for J1915 and bldN, and using the modified Kirby mix procedure as described by Elliot et al. (2001) for all other RNA preparations. Two additional phenol/chloroform and chloroform extractions were included before the final precipitation step for all RNA. Quantitation of RNA was carried out by UV spectroscopy and was checked by agarose gel electrophoresis.
Generation of cDNA for microarray analysis was done as described by Huang et al. (2001). The first RNA samples for J1915 and bldN were used as the references to which subsequent samples were compared, and were labeled with Cy-3 (green), whereas the later samples were labeled with Cy-5 (red). Fifteen micrograms of total RNA was denatured together with 5 μg of 72% G-C content hexamers (total volume of 13 μL) at 75°C for 15 min before snap cooling on ice. Three microliters of Cy3- or Cy5-dCTP (Amersham Pharmacia Biotech) were added to each reaction, together with a 14-μL cocktail containing Superscript II buffer (6 μL of a 5× solution), DTT (3 μL of a 0.1 M solution), dNTPs (3 μL of a mixture of 4 mM dATP, 4 mM dTTP, 10 mM dGTP, and 0.2 mM dCTP), and Superscript II (2 μL of 200U/μL) to give a total volume of 30 μL. The reactions were incubated for 10 min at room temperature before transfer to 42°C for 2–3 h. The reactions were stopped, and the RNA degraded, by adding 1 μL of 1 M NaOH and heating to 65°C for 10 min before being neutralized by the addition of 1 μL of 1 M HCl. The labeled cDNA was purified using Microcon-10 filters (Amicon); then, 500 μL of TE was placed in each Microcon filter, and the reverse transcription reactions were added to this before spinning for 20 min at 13,000 rpm. This step was repeated and centrifugation continued until ∼6 μL remained. The purified samples were then recovered by inverting the filters into fresh tubes and spinning for 1 min.
The Cy3- and Cy5-labeled cDNA samples were mixed together. SSC, SDS, and polyA were added as described (DeRisi et al. 1997). The mixture was heated at 100°C for 2 min and applied to a microarray. The microarray was hybridized at 65°C for 10–12 h. The microarray was washed as described (DeRisi et al. 1997), and scanned using a GenePix 4000B scanner (Axon Instruments). Microarray data were analyzed using the software programs Cluster and Treeview (Eisen et al. 1998).
S1 nuclease mapping was carried out in order to verify the microarray results, to identify the transcription start sites for chpH and chpE, and to determine gene expression of chpH and chpE in different mutant strains. For each S1 nuclease reaction, 30–40 μg of RNA was hybridized to a γ-32P end-labeled probe at 45°C for 4–15 h following denaturation at 65°C for 10–15 min. S1 nuclease (Roche) digestions were performed as described by Kieser et al. (2000), and the reaction products were separated on 6% sequencing gels. Probes having a unique end label were generated by 5′ end labeling of the primer internal to the coding sequence (Table 3) using [γ-32P] ATP and T4 polynucleotide kinase (Cambio), and using this labeled primer together with an upstream, unlabeled primer (Table 3) in PCR reactions. DNA fragments of 200–900 bp were generated. When examining the expression of chpE and chpH in the various bld mutant strains, the probes for chpE, chpH, and hrdB (positive control for RNA integrity and loading) were added together to each RNA sample. Sequencing ladders were generated as described in Elliot et al. (2001), using the internal oligonucleotide as primer for the cycle sequencing reactions.
Table 3.
Oligonucleotides used in this study
| Sequence 5′ to 3′ | |
|---|---|
| Gene disruption primers | |
| SC5H1.35c Forward (chpB) | ACTGCCCGCGACCATGTCTGGAAGGCAGGGAGCCTCATGATTCCGGGGATCCGTCGACC |
| SC5H.135c Reverse (chpB) | CCGCCCTGACGGCGCGGGTGCCCCCGGTCGACGCGGTCATGTAGGCTGGAGCTGCTTC |
| SCC46.02c Forward (chpA,D) | CGTCATTCAGGCATGACCGCAGAGAAGGGAAAGTTCATGATTCCGGGGATCCGTCGACC |
| SCC46.01c Reverse (chpA,D) | GGACCGGCCGGGGGCGTCGGGGTGGCGGGGGTGGGGTCATGTAGGCTGGAGCTGCTTC |
| SCI52.16c Reverse (chpC,H) | GGTGCGGAGCCCGCTGCGTGTACCGGTGCGCCCGCCTCATGTAGGCTGGAGCTGCTTC |
| SCI52.17c Forward (chpC,H) | CAATTTTGGGATCGCGTTTGCCAACTAGGAGGAACAATGATTCCGGGGATCCGTCGACC |
| SCI52.17c Reverse (chpH) | CGGCGGAATCAGATCCTCACGGGGCGAGGCACAACGTCATGTAGGCTGGAGCTGCTTC |
| S1 nuclease mapping primers | |
| I52.17c up (chpH) | CGGAGTGGACGAGCGGGTGC |
| I52.17c down IchpH) | GGAAAGGACGCCGGGCGAGTGC |
| SCI5.08c up (chpE) | GCGTGCCCTCGTCCTCTCCG |
| SCI5.08c down (chpE) | CCAGGTTGCCCGAGGCGACG |
| SCC46.03c up (SCO2718) | GTCGTCGCCGATCGCCATCG |
| SCC46.03c down (SCO2718) | CCCGTTGTCGTCCCCGATGG |
| SCD63 up (SCO4772) | GGTGACCGGCCCCTTTCGCG |
| SCD63 down (SCO4772) | GGACGACACCCGCAGTCGGTACG |
| 2SC10A7 up (SCO4002) | GGGAAGAAGGGAAGCAAAGGG |
| 2SC10A7 down (SCO4002) | GCTGCGTGGTGGACGAGTTGC |
| Complementation primers | |
| I52.16c end (chpC) | CGAGTACGGACACTGGGAG |
| I52.17c up (chpH) | see S1 nuclease mapping primers |
| C46 Nhe up | GTCGGCTAGCGCGGCCGGGGCGGGTGTT |
| C46 Nhe down | GTCGGCTAGCGAACCAGCAGTGCGCGGAC |
Transcriptional fusions with egfp
Transcriptional fusions between promoter fragments (corresponding to the S1 probes) of chpE and chpH and the enhanced green fluorescent protein (egfp) gene were constructed to visualize the location of gene expression. The 419-bp and 334-bp promoter fragments of chpE and chpH, respectively, were amplified using Turbo Pfu (Promega), and were cloned into the egfp-containing pIJ8660 vector (Sun et al. 1999) digested with EcoRV. PCR was carried out using I5 down and I52.17c down, together with the universal primer to confirm the presence of insert, and the correct insert orientation. Following conjugation of the plasmids into S. coelicolor, along with pIJ8660 without insert, spore stocks were prepared. Glass coverslips sterilized in ethanol were inserted at an ∼60° angle into MS medium, and 5 μL of each spore preparation was used to inoculate along the underside of the coverslip, at the agar interface. Fluorescence microscopy was then carried out as described by Sun et al. (1999) after growth for 48 h, 72 h, or 96 h. Examination of EGFP expression in the colony cross-section required growth of the colony for 6 d on minimal medium supplemented with mannitol.
Construction of the chaplin mutants
Chaplin mutants were created by replacing the entire coding sequence for individual genes, or pairs of genes in the cases of chpC and chpH or chpA and chpD, with the apramycin-resistance cassette (aac(3)IV) or the cassette conferring resistance to spectinomycin and streptomycin (aadA) using the PCR-targeted method of Gust et al. (2003). This method involves replacement of S. coelicolor genes carried on cosmids in E. coli with a selectable marker generated by PCR using primers with a 39-nt gene-specific extension. The selectable marker cassette includes not only the antibiotic resistance gene, but also oriT, which allows the direct transfer of the mutagenized cosmid into S. coelicolor by conjugation (Gust et al. 2003). Cosmids I52 (chpH and chpC), C46 (chpD and chpA), and 5H1 (chpB) were introduced into E. coli BW25113 (Datsenko and Wanner 2000) by electroporation, and the appropriate genes were disrupted by electroporation of the oriT/aac(3)IV or oriT/aadA cassette that had been amplified using oligonucleotides containing gene-specific extensions (Table 3). The resulting cosmid was introduced into E. coli ET12567 before conjugation into S. coelicolor M600. Null mutations in each gene, or gene pair, were generated by a double crossover of the mutagenized cosmid with the chromosome to give an apramycin-resistant, or spectinomycin/streptomycin-resistant, and kanamycin-sensitive phenotype. The mutations were confirmed by PCR and Southern blot hybridization. Individual null mutants (chpH, chpA, and chpD, and chpC and chpH) were all created by gene replacement with the apramycin-resistance cassette. The fourfold (chpH, chpC, chpD, and chpA) and fivefold (chpH, chpC, chpD, chpA, and chpB) mutants were, however, created in a stepwise manner involving multiple cassettes. chpD and chpA were initially replaced with the apramycin-resistance cassette. This cassette was then removed using a second targeting event. Protoplasts of the mutant were generated, and were transformed using the mutagenized cosmid that had had the oriT and apramycin-resistance cassette removed [the disruption cassette is flanked by FRT sites (for FLP recognition targets), which allow the removal of intervening sequences in the presence of the FLP recombinase in E. coli (Datsenko and Wanner 2000)]. chpH and chpC were then replaced with the spectinomycin/streptomycin-resistance cassette as described above, and chpB was replaced with the apramycin-resistance cassette.
Complementation of the quadruple mutant (with the apramycin cassette removed) was achieved by introducing chpC and chpH [amplified from M600 chromosomal DNA using I52.16c end and I52.17c up primers (see Table 3) and Pfu Turbo DNA polymerase] into the EcoRV site of the conjugative plasmid pSET152 (Bierman et al. 1992), and chpD (amplified in the same way as above, using SCC46.01c Nhe and SCC46.02c Nhe as primers) into the unique NheI site of the chpC/H-containing pSET152 clone. The sequences of chpC, chpH and chpD were confirmed by sequencing, and the construct was introduced into the fourfold mutant after passage through E. coli ET12567. pSET152 alone was also introduced into the fourfold mutant as a negative control.
Purification and analysis of small secreted chaplin proteins
Wild-type and chaplin mutant strains were grown on cellophane discs on the surface of MS medium. After 4 d, the resulting biomass was harvested, and 3 mL of HEPES buffer (0.1 M) was added. The suspension was sonicated on ice for 3 × 15 sec on half-power before spinning at 7500 rpm for 10 min. The supernatant was removed and the remaining pellet was resuspended in 3 mL 2% SDS before heating to 100°C for 10 min. The suspension was then spun again, the supernatant was removed, and the insoluble pellet was again subjected to boiling for 10 min in 3 mL 2% SDS. The suspension was transferred to 2-mL Eppendorf tubes and spun at 13,000 rpm. The supernatant was discarded, and the pellet was frozen in liquid nitrogen and dried in a Speedy Vac for ∼1 h. The dried pellet was resuspended in ∼2–4 mL trifluoroacetic acid (TFA); the soluble fraction was taken, and was dried down using a stream of air. The dried material was then resuspended in ∼2%–4% TFA, or water. The TFA soluble extract was mixed in a 1:1 ratio with 10 mg/mL sinapinic acid in 50% acetonitrile containing 0.1% TFA. The mixture was then spotted onto a thin layer of sinapinic acid that had been applied to a MALDI-TOF sample template. The sample was dried, and a thin layer of sinapinic acid was applied again. The resulting crystals were washed twice using 0.1% TFA before being analyzed by MALDI-TOF mass spectrometry (Bruker Reflex III). Samples were calibrated using a mixture of oxidized bovine insulin (stock solution of 100 pmole/μL was diluted to 10 pmole/μL in 0.1% TFA) and a standard peptide mixture used for mass fingerprinting (at 1 pmole/μL), containing 20 pmole of insulin and 8 pmole of the peptide mixture.
Acknowledgments
We thank Mike Naldrett for performing the MALDI-TOF mass spectroscopy, Kim Findlay for taking the scanning electron micrographs, Jongho Sun for his help with the GFP work, Alison Hunt and Dagmara Jakimowicz for the use of their RNA, members of the Kao lab for all their assistance, Andrew Hesketh for helpful insights and discussion, and David Hopwood and Keith Chater for their comments on the manuscript. This work was funded by a postdoctoral fellowship from the Alberta Heritage Foundation for Medical Research (to M.A.E.), and by a grant-inaid to the John Innes Centre from the BBSRC.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.264403.
References
- Arnqvist A., Olsén, A., and Normark, S. 1994. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by σ70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 13: 1021–1032. [DOI] [PubMed] [Google Scholar]
- Austin J.W., Sanders, G., Kay, W.W., and Collinson, S.K. 1998. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 162: 295–301. [DOI] [PubMed] [Google Scholar]
- Ben Nasr A., Olsén, A., Sjöbring, U., Müller-Esterl, W., and Björck, L. 1996. Assembly of human contact phase proteins and release of bradykinin at the surface of curli-expressing Escherichia coli. Mol. Microbiol. 20: 927–935. [DOI] [PubMed] [Google Scholar]
- Bentley S.D., Chater, K.F., Cerdeño-Tarraga, A.M., Challis, G.L., Thomson, N.R., James, K.D., Harris, D.E., Quail, M.A., Kieser, H., Harper, D., et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417: 141–147. [DOI] [PubMed] [Google Scholar]
- Bibb M.J. and Buttner, M.J. 2003. The Streptomyces coelicolor developmental transcription factor σBldN is synthesized as a pro-protein. J. Bacteriol. 185: 2338–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M.J., Molle, V., and Buttner, M.J. 2000. σBldN, an extracytoplasmic function RNA polymerase σ factor required for aerial mycelium formation in Streptomyces coelicolor A3(2). J. Bacteriol. 182: 4606–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierman M., Logan, R., O'Brien, K., Seno, E.T., Rao, R.N., and Schoner, B.E. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116: 43–49. [DOI] [PubMed] [Google Scholar]
- Bignell D.R., Warawa, J.L., Strap, J.L., Chater, K.F., and Leskiw, B.K. 2000. Study of the bldG locus suggests that an anti-anti-σ factor and an anti-σ factor may be involved in Streptomyces coelicolor antibiotic production and sporulation. Microbiology 146: 2161–2173. [DOI] [PubMed] [Google Scholar]
- Chakraburtty R. and Bibb, M. 1997. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 179: 5854–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champness W.C. 1988. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J. Bacteriol. 170: 1168–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M.R., Robinson, L.S., Pinkner, J.S., Roth, R., Heuser, J., Hammar, M., Normark, S., and Hultgren, S.J. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295: 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D., Wösten, H.A., van Keulen, G., Faber, O.G., Alves, A.M., Meijer, W.G., and Dijkhuizen, L. 2002. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol. Microbiol. 44: 1483–1492. [DOI] [PubMed] [Google Scholar]
- Claessen D., Rink, R., de Jong, W., Siebring, J., de Vreugd, P., Boersma, F.G.H., Dijkhuizen, L., and Wösten, H.A.B. 2003. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Datsenko K.A. and Wanner, B.L. 2000. One step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vocht M.L., Reviakine, I., Wösten, H.A., Brisson, A., Wessels, J.G., and Robillard, G.T. 2000. Structural and functional role of the disulfide bridges in the hydrophobin SC3. J. Biol. Chem. 275: 28428–28432. [DOI] [PubMed] [Google Scholar]
- DeRisi J.L., Iyer, V.R., and Brown, P.O. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686. [DOI] [PubMed] [Google Scholar]
- Dramsi S., Dehoux, P., Lebrun, M., Goossens, P.L., and Cossart, P. 1997. Identification of four new members of the internalin multigene family of Listeria monocytogenes EGD. Infect. Immun. 65: 1615–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown, P.O., and Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 95: 14863–14968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot M., Damji, F., Passantino, R., Chater, K.F., and Leskiw, B. 1998. The bldD gene of Streptomyces coelicolor A3(2): A regulatory gene involved in morphogenesis and antibiotic production. J. Bacteriol. 180: 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot M.A., Bibb, M.J., Buttner, M.J., and Leskiw, B.K. 2001. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol. Microbiol. 40: 257–269. [DOI] [PubMed] [Google Scholar]
- Engelbrecht F., Chun, S.-K., Ochs, C., Hess, J., Lottspeich, F., Goebel, W., and Sokolovic, Z. 1996. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 21: 823–837. [DOI] [PubMed] [Google Scholar]
- Engelbrecht F., Domínguez-Bernal, G., Hess, J., Dickneite, C., Greiffenberg, L., Lampidis, R., Raffelsbauer, D., Daniels, J.J.D., Kreft, J., Kaufmann, S.H.E., et al. 1998. A novel PrfA-regulated chromosomal locus, which is specific for Listeria ivanovii, encodes two small, secreted internalins and contributes to virulence in mice. Mol. Microbiol. 30: 405–417. [DOI] [PubMed] [Google Scholar]
- Fischetti V.A. 2000. Surface proteins of Gram-positive bacteria. In Gram-positive pathogens (eds. V.A. Fischetti, R.P. Novick, J.J. Ferretti, D.A. Portnoy, and J.I. Rood), pp. 11–24. ASM Press, Washington, DC.
- Flärdh K., Findlay, K.C., and Chater, K.F. 1999. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2). Microbiology 145: 2229–2243. [DOI] [PubMed] [Google Scholar]
- Gaillard J.L., Berche, P., Frebel, C., Gouin, E., and Cossart, P. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65: 1127–1141. [DOI] [PubMed] [Google Scholar]
- Guijarro J., Santamaria, R., Schauer, A., and Losick, R. 1988. Promoter determining the timing and spatial localization of transcription of a cloned Streptomyces coelicolor gene encoding a spore-associated polypeptide. J. Bacteriol. 170: 1895–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust B., Challis, G.L., Fowler, K., Kieser, T., and Chater, K.F. 2003. Gene replacement by PCR targeting in Streptomyces and its use to identify a protein domain involved in the biosynthesis of the sesquiterpene odour geosmin. Proc. Natl. Acad. Sci. 100: 1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammar M., Bian, Z., and Normark, S. 1996. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl. Acad. Sci. 93: 6562–6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166: 557–580. [DOI] [PubMed] [Google Scholar]
- Hopwood D.A. and Glauert, A.M. 1961. Electron microscope observations on the surface structures of Streptomyces violaceoruber. J. Gen. Microbiol. 26: 325–330. [DOI] [PubMed] [Google Scholar]
- Huang J., Lih, C.J., Pan, K.H., and Cohen, S.N. 2001. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes & Dev. 15: 3183–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonquieres R., Bierne, H., Fielder, F., Gounon, P., and Cossart, P. 1999. Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: A novel mechanism of protein association at the surface of gram-positive bacteria. Mol. Microbiol. 34: 902–914. [DOI] [PubMed] [Google Scholar]
- Kelemen G.H. and Buttner, M.J. 1998. Initiation of aerial mycelium formation in Streptomyces. Curr. Opin. Microbiol. 1: 656–662. [DOI] [PubMed] [Google Scholar]
- Kelemen G.H., Brown, G.L., Kormanec, J., Potûcková, L., Chater, K.F., and Buttner, M.J. 1996. The positions of the σ-factor genes, whiG and sigF, in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2). Mol. Microbiol. 21: 593–603. [DOI] [PubMed] [Google Scholar]
- Kieser T., Bibb, M.J., Buttner, M.J., Chater, K.F., and Hopwood, D.A. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, UK.
- Loferer H., Hammar, M., and Normark, S. 1997. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol. Microbiol. 26: 11–23. [DOI] [PubMed] [Google Scholar]
- MacNeil D.J., Gewain, K.M., Ruby, C.L., Dezeny, G., Gibbons, P.H., and MacNeil, T. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111: 61–68. [DOI] [PubMed] [Google Scholar]
- Mazmanian S.K., Liu, G., Ton-That, H., and Schneewind, O. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285: 760–763. [DOI] [PubMed] [Google Scholar]
- Mazmanian S.K., Ton-That, H., Su, K., and Schneewind, O. 2002. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. 99: 2293–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V. and Buttner, M.J. 2000. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol. Microbiol. 36: 1265–1278. [DOI] [PubMed] [Google Scholar]
- Molle V., Palframan, W.J., Findlay, K.C., and Buttner, M.J. 2000. WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 182: 1286–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre W.W. and Schneewind, O. 1994. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in Gram-positive bacteria. Mol. Microbiol. 14: 115–121. [DOI] [PubMed] [Google Scholar]
- Olsén A., Jonsson, A., and Normark, S. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338: 652–655. [DOI] [PubMed] [Google Scholar]
- Olsén A., Arnqvist, A., Hammar, M., Sukupolvi, S., and Normark, S. 1993. The RpoS σ factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol. Microbiol. 7: 523–536. [DOI] [PubMed] [Google Scholar]
- Pallen M.J., Lam, A.C., Antonio, M., and Dunbar, K. 2001. An embarrassment of sortases—A richness of substrates? Trends Microbiol. 9: 97–102. [DOI] [PubMed] [Google Scholar]
- Pope M.K., Green, B., and Westpheling, J. 1998. The bldB gene encodes a small protein required for morphogenesis, antibiotic production, and catabolite control in Streptomyces coelicolor. J. Bacteriol. 180: 1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret C., Prensier, G., Le Thi, T.T., Vidao, O., Lejeune, P., and Dorel, C. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: Role of flagella, curli and colanic acid. Environ. Microbiol. 2: 450–464. [DOI] [PubMed] [Google Scholar]
- Ryding N.J., Kelemen, G.H., Whatling, C.A., Flärdh, K., Buttner, M.J., and Chater, K.F. 1998. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 29: 343–357. [DOI] [PubMed] [Google Scholar]
- Ryding N.J., Bibb, M.J., Molle, V., Findlay, K.C., Chater, K.F., and Buttner, M.J. 1999. New sporulation loci in Streptomyces coelicolor A3(2). J. Bacteriol. 181: 5419–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C., Du, L., Edwards, D.J., Toney, M.D., and Shen, B. 2001. Cloning and characterization of a phosphopantetheinyl transferase from Streptomyces verticillus ATCC15003, the producer of the hybrid peptide-polyketide antitumor drug bleomycin. Chem. Biol. 8: 725–738. [DOI] [PubMed] [Google Scholar]
- Sjöbring U., Pohl, G., and Olsén, A. 1994. Plasminogen, absorbed by Escherichia coli expressing curli or by Salmonella enteritidis expressing thin aggregative fimbriae, can be activated by simultaneously captured tissue-type plasminogen activator (t-PA). Mol. Microbiol. 14: 443–452. [DOI] [PubMed] [Google Scholar]
- Sun J., Kelemen, G.H., Fernández-Abalos, J.M., and Bibb, M.J. 1999. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145: 2221–2227. [DOI] [PubMed] [Google Scholar]
- Tillotson R.D., Wösten, H.A., Richter, M., and Willey, J.M. 1998. A surface active protein involved in aerial hyphae formation in the filamentous fungus Schizophillum commune restores the capacity of a bald mutant of the filamentous bacterium Streptomyces coelicolor to erect aerial structures. Mol. Microbiol. 30: 595–602. [DOI] [PubMed] [Google Scholar]
- Vidal O., Longin, R., Prigent-Combaret, C., Dorel, C., Hooreman, M., and Lejeune, P. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: Involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180: 2442–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels J.G. 1997. Hydrophobins: Proteins that change the nature of the fungal surface. Adv. Microb. Physiol. 38: 1–45. [DOI] [PubMed] [Google Scholar]
- Wildermuth H., Wehrli, E., and Horne, R.W. 1971. The surface structure of spores and aerial mycelium in Streptomyces coelicolor. J. Ultrastruct. Res. 35: 168–180. [DOI] [PubMed] [Google Scholar]
- Willey J., Santamaria, R., Guijarro, J., Geistlich, M., and Losick, R. 1991. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell 65: 641–650. [DOI] [PubMed] [Google Scholar]
- Willey J., Schwedock, J., and Losick, R. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes & Dev. 7: 895–903. [DOI] [PubMed] [Google Scholar]
- Wösten H.A.B. 2001. Hydrophobins: Multipurpose proteins. Ann. Rev. Microbiol. 55: 625–646. [DOI] [PubMed] [Google Scholar]
- Wösten H.A. and de Vocht, M.L. 2000. Hydrophobins, the fungal coat unraveled. Biochim. Biophys. Acta. 18: 79–86. [DOI] [PubMed] [Google Scholar]
- Wösten H.A. and Willey, J.M. 2000. Surface-active proteins enable microbial aerial hyphae to grow into the air. Microbiology 146: 767–773. [DOI] [PubMed] [Google Scholar]
- Wösten H., De Vries, O., and Wessels, J. 1993. Interfacial self-assembly of a fungal hydrophobin into a hydrophobic rodlet layer. Plant Cell 5: 1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]