Abstract
The integrity of chromosomes during cell division is ensured by both trans-acting factors and cis-acting chromosomal sites. Failure of either these chromosome integrity determinants (CIDs) can cause chromosomes to be broken and subsequently misrepaired to form gross chromosomal rearrangements (GCRs). We developed a simple and rapid assay for GCRs, exploiting yeast artificial chromosomes (YACs) in Saccharomyces cerevisiae. We used this assay to screen a genome-wide pool of mutants for elevated rates of GCR. The analyses of these mutants define new CIDs (Orc3p, Orc5p, and Ycs4p) and new pathways required for chromosome integrity in DNA replication elongation (Dpb11p), DNA replication initiation (Orc3p and Orc5p), and mitotic condensation (Ycs4p). We show that the chromosome integrity function of Orc5p is associated with its ATP-binding motif and is distinct from its function in controlling the efficiency of initiation of DNA replication. Finally, we used our YAC assay to assess the interplay of trans and cis factors in chromosome integrity. Increasing the number of origins on a YAC suppresses GCR formation in our dpb11 mutant but enhances it in our orc mutants. This result provides potential insights into the counterbalancing selective pressures necessary for the evolution of origin density on chromosomes.
Keywords: Chromosome integrity, gross chromosomal rearrangements, origin recognition complex, genomic stability, mitotic chromosome condensation
Proper cell division requires that replicated chromosomes not only segregate from one another but also segregate intact. Indeed, during a normal cell division, the integrity of chromosomes is maintained with remarkable fidelity. This fidelity is achieved through the action of trans-acting factors and specific cis-acting chromosomal sites. Failures in these chromosome integrity determinants (CIDs) can lead to gross chromosomal rearrangements (GCRs), including terminal deletions, translocations, and interstitial deletions. GCRs have been implicated in cancer progression and genome evolution.
Although very important insights into chromosome integrity could arise from a genome-wide screen for mutations that enhance GCR formation, such a screen has not been reported previously. GCRs rearrange or delete large regions of chromosomes that are likely to perturb essential functions. Thus cells with a GCR are rarely viable and can be detected only by very sensitive assays (Chen and Kolodner 1999; Tennyson et al. 2002). These assays are constrained by the requirement to analyze a very large number of cells. Because of this constraint, GCRs have only been assessed in mutants defective for functions already suspected to be important in the prevention of GCRs. This candidate approach has identified CIDs as a large number of DNA repair proteins as well as a few DNA replication factors and cell cycle regulators (Kolodner et al. 2002). The functions of these proteins support the model that GCRs often occur by the misrepair of a double-strand break (DSB). However, it is likely that this candidate gene approach has discovered only a subset of CID genes. Therefore, the identification of additional chromosome integrity determinants would be greatly facilitated by a simple and rapid assay for GCRs that would allow genome-wide mutant screens.
Although the focus of the candidate approach on DNA repair has provided significant insight into the role of DSB misrepair in GCR formation, very little is known about the cellular defects that cause DSBs in the first place. A few studies have shown that DSBs can result from stalled replication forks or unreplicated DNA in mitosis (Michel 2000; Rothstein et al. 2000; Pflumm and Botchan 2001). Thus, a rich source of DSBs may be labile DNA structures generated as a consequence of perturbations in factors necessary for essential DNA metabolism like DNA replication or condensation. To recover mutations in these factors, they must be hypomorphs, reducing the activity of the factor sufficiently to cause occasional DSBs and GCRs, but leaving sufficient activity to ensure viability of the organism. Finding such special alleles further emphasizes the importance of a simple and rapid assay for GCRs.
Here we use yeast artificial chromosomes (YACs) to develop an assay in budding yeast for GCRs. The sensitivity and ease of this assay allowed us to screen a large collection of hypomorphs in essential genes. The genes identified by these hypomorphs demonstrate that this YAC assay is a powerful tool to identify new gene products/processes that lead to GCR. In addition, we use this assay to assess the interplay between our mutants and cis factors of chromosome integrity, providing important clues to fundamental aspects of chromosome structure.
Results
YAC assay for GCRs
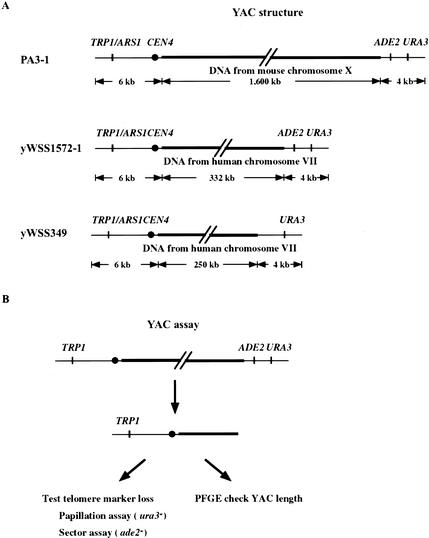
To perform a genome-wide screen for mutations that potentially cause DSBs, we needed a simple, sensitive, and rapid method for detecting GCRs. For this purpose, we developed a genetic assay for GCRs, by exploiting the assets of three YACs, PA3-1, yWSS1572-1, and yWSS349 (Fig. 1A; Table 1). These YACs are extremely telocentric with a short arm of 6 kb and long arms between 250 and 1600 kb (Burke et al. 1987; Kuhn and Ludwig 1994). Loss of the two telomere markers, URA3 and ADE2, is likely to occur by a GCR that simultaneously eliminates both genes and is easily detected by a colony color or papillation assay (Fig. 1B). Finally, these YACs have a similar size range as endogenous chromosomes but are dispensable for cell growth. As a result, DSBs that lead to very large GCRs are tolerated. Thus, the size, sequence content, and genetic markers on the YAC are well suited for use in a genetic screen for GCRs.
Figure 1.
YAC assay for GCRs. (A) Structure of the YACs (PA3-1, yWSS1572-1, and yWSS349; Heard et al. 1994; van Brabant et al. 1999, 2001). The human insert sequences of yWSS1572-1 and yWSS349 are from nonoverlapping regions of human Chromosome VII. (B) Diagram of genetic and physical assays for GCRs. We identified cells that retained the YAC (TRP+) but lost the telomere markers (ura- and ade-) by the papillation and sector assays, respectively. YACs with telomere marker loss were analyzed by PFGE to assess the presence and size of GCRs (see Materials and Methods).
Table 1.
Yeast strains
| Strain | Genotype |
|---|---|
| YPH925 | MATx ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 cyh2 kar1Δ15 |
| DH1003/102 | MATa leu2 trp1 ura3-52 can1 ade2::KAN/y WSS1572-1 |
| DH2004-1/101 | MATa leu2 trp1 ura3-52 can 1 ade2::KAN/PA3-1 |
| DH3701-1 | MATx trp1 ura3-52 his3Δ200 ade2 can1 bar1 |
| DH3701-2 | MATa trp1 ura3-52 his3Δ200 ade2 can1 bar1 |
| DH3701-2/103 | MATa trp1 ura3-52 his3Δ200 ade2 can1 bar1/y WSS349 |
| DH3701-2/104 | MATa trp1 ura3-52 his3Δ200 ade2 can1 bar1/yWSS349-5oriΔ |
| DH3702-1 | MATx ura3-52 ade2 trp1 lys2-801 can1 bar1 |
| DH3702-2 | MATa ura3-52 ade2 trp1 lys2-801 can1 bar1 |
| DH3702-2/201 | MATa ura3-52 ade2 trp1 lys2-801 can1 bar1/PDK243 |
| DH3702-2/202 | MATa ura3-52 ade2 trp1 lys2-801 can1 bar1/PDK368-7 |
| DH3702-2/103 | MATa trp1 ura3 lys2-801 ade2 can1 bar1/yWSS349 |
| DH3702-2/104 | MATa trp1 ura3-52 lys2-801 ade2 can1 bar1/yWSS349-5oriΔ |
| DH3710 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 bar1 can1 dpb11-27 |
| DH3710/201 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 bar1 can1 dpb11-27/PDK243 |
| DH3710/202 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 bar1 can1 dpb11-27/PDK368-7 |
| DH3710/101 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 can1 bar1 dpb11-27/PA3-1 |
| DH3710/102 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 can1 bar1 dpb11-27/yWSS1572-1 |
| DH3710/103 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 bar1 can1 dpb11-27/yWSS349 |
| DH3710/104 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 bar1 can1 dpb11-27/yWSS349-5oriΔ |
| DH3711 | MATa ura3-52 ade2 trp1 lys2-801 leu2 can1 dpb11-27 |
| DH3711/101 | MATa ura3-52 ade2 trp1 lys2-801 leu2 can1 dpb11-27/PA3-1 |
| DH3711/102 | MATa ura3-52 ade2 trp1 lys2-801 leu2 can1 dpb11-27/yWSS1572-1 |
| DH3705 | MATa ura3-52 ade2 trp1 lys2-801 leu2 can1mre11::KAN |
| DH3705/101 | MATa ura3-52 ade2 trp1 lys2-801 leu2 can1mre11::KAN/PA3-1 |
| DH3705/102 | MATa ura3-52 ade2 trp1 lys2-801 leu2 can1 mre11::KAN/yWSS1572-1 |
| DH3720 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 can1 orc3-70 |
| DH3720/102 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 can1 orc3-70/yWSS1572-1 |
| DH3720/103 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 can1 orc3-70/yWSS349 |
| DH3720/104 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 can1 orc3-70/yWSS349-5oriΔ |
| DH3720/201 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 can1 orc3-70/PDK243 |
| DH3720/202 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 can1 orc3-70/PDK368-7 |
| DH3721 | MATa ura3-52 ade2 trp1 lys2-801 leu2 bar1 can1 orc3-70 |
| DH3721/102 | MATa ura3-52 ade2 trp1 lys2-801 leu2 bar1 can1 orc3-70/yWSS1572-1 |
| DH3721/103 | MATa ura3-52 ade2 trp1 lys2-801 leu2 bar1 can1 orc3-70/yWSS349 |
| DH3721/104 | MATa ura3-52 ade2 trp1 lys2-801 leu2 bar1 can1 orc3-70/yWSS349-5oriΔ |
| DH3730 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 can1 orc5-70 |
| DH3730/102 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 can1 orc5-70/yWSS1572-1 |
| DH3730/103 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 can1 orc5-70/yWSS349 |
| DH3730/104 | MATa ura3-52 ade2 trp1 his3Δ200 leu2 can1 orc5-70/yWSS349-5oriΔ |
| DH3731 | MATa ura3-52 ade2 trp1 lys2-801 leu2 bar1 can1 orc5-70 |
| DH3731/201 | MATa ura3-52 ade2 trp1 lys2-801 leu2 bar1 can1 orc5-70/PDK243 |
| DH3731/202 | MATa ura3-52 ade2 trp1 lys2-801 leu2 bar1 can1 orc5-70/PDK368-7 |
| DH3731/102 | MATa ura3-52 ade2 trp1 lys2-801 leu2 bar1 can1 orc5-70/yWSS1572-1 |
| DH3731/103 | MATa ura3-52 ade2 trp1 lys2-801 leu2 bar1 can1 orc5-70/yWSS349 |
| DH3731/104 | MATa ura3-52 ade2 trp1 lys2-801 leu2 bar1 can1 orc5-70/yWSS349-5oriΔ |
| W303-1 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3, 112 can1-100n |
| W303-1/103 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3, 112 can1-100n/yWSS349 |
| W303-1/104 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3, 112 can1-100n/yWSS349-5oriΔ |
| W303-1/201 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3, 112 can1-100n/pDK243 |
| W303-1/202 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3, 112 can1-100n/pDK368-7 |
| YB0057 | MATa ade2-1 ura3-1 his3-11, 15 trp1-1 leu2-3, 112 can1-100 orc5-1 |
| YB0057/103 | MATa ade2-1 ura3-1 his3-11, 15 trp1-1 leu2-3, 112 can1-100 orc5-1/yWSS349 |
| YB0057/104 | MATa ade2-1 ura3-1 his3-11, 15 trp1-1 leu2-3, 112 can1-100 orc5-1/yWSS349-5oriΔ |
| YB0057/201 | MATa ade2-1 ura3-1 his3-11, 15 trp1-1 leu2-3, 112 can1-100 orc5-1/pDK243 |
| YB0057/202 | MATa ade2-1 ura3-1 his3-11, 15 trp1-1 leu2-3, 112 can1-100 orc5-1/pDK368-7 |
| DH3706 | MATa trp1 ura3-52 ade2 leu2 cdc9-1 can1 |
| DH3706/103 | MATa trp1 ura3-52 ade2 leu2 cdc9-1 can1/yWSS349 |
| DH3706/104 | MATa trp1 ura3-52 ade2 leu2 cdc9-1 can1/yWSS349-5oriΔ |
| DH3707 | MATa trp1 ura3-52 ade2 leu2 his3 cdc17-1 can1 |
| DH3707/103 | MATa trp1 ura3-52 ade2 leu2 his3 cdc17-1 can1/yWSS349 |
| DH3707/104 | MATa trp1 ura3-52 ade2 leu2 his3 cdc17-1 can1/yWSS349-5oriΔ |
YPH925 is in an Sc288 background, W303-1 and YB0057 strains are in a W303 background, and the rest of the strains are in an A364A background.
To assess the usefulness of these YACs for assaying GCRs, we began by examining telomere marker loss in wild-type cells. We grew cells in media lacking uracil to select for the telomere marker URA3 and then measured the percentage of total YAC-bearing cells (TRP+) that fail to grow in the absence of uracil. This percentage is proportional to the rate of loss of URA3 function from the YAC (see Materials and Methods). Greater than 95% of the URA3-defective YACs had also lost ADE2 function. Because these YACs simultaneously had lost both telomere markers, they were excellent candidates for GCRs. The formation of GCRs was confirmed by pulse field gel electrophoresis (PFGE; data not shown; see below). The percentage of URA3 marker loss was 10-2 for PA3-1 and 10-4 for both yWSS1572-1 and yWSS349. This telomere marker loss is 6–8 orders of magnitude higher than the spontaneous loss of URA3 and CAN1 from the end of Chromosome V (this study; Chen and Kolodner 1999). This difference is likely to reflect both the increased sensitivity of the YAC to detect GCRs and the increased propensity of the YAC to undergo GCRs (see Discussion).
Given the high spontaneous rate of GCRs on these YACs, we asked whether the rate of GCRs would respond to two different conditions known to enhance their formation. First, cells were irradiated with γ-rays (Fig. 2A). Concurrent loss of both telomere marker functions was elevated in a dose-dependent fashion, consistent with the induction of GCRs. For the same dose, telomere marker inactivation was observed more frequently for the large YAC than the small YAC as expected given its larger target size and the propensity of γ irradiation to induce random DSBs.
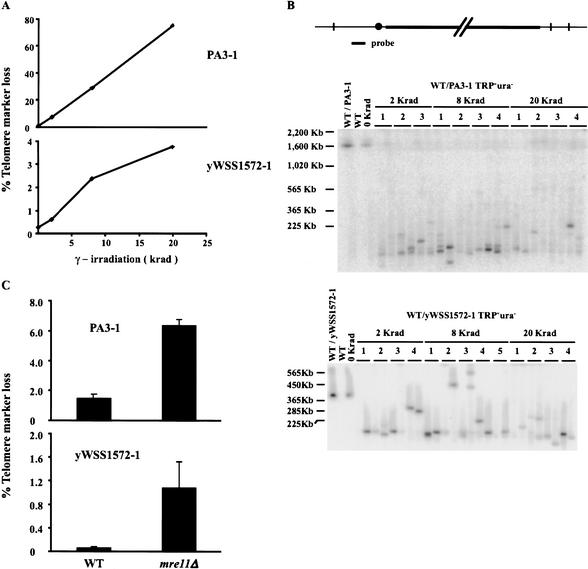
Figure 2.
(A) Cells that harbored the YACs, PA3-1 and yWSS1572-1, were subjected to different doses of γ irradiation and then assayed for telomere marker loss (see Materials and Methods). (B) DNA was prepared from TRP+ ura- cells identified in A and subjected to PFGE and Southern analysis. The YAC-specific probe is a region of pBR322 that lies on the short arm between the TRP1 gene and the centromere. (C) PA3-1 and yWSS1572-1 were transferred into an mre11Δ mutant and the isogenic wild type and tested for telomere marker loss (as described in A).
YACs lacking functional telomere markers were analyzed by PFGE (Fig. 2B). Almost all YACs migrated more rapidly than expected if they had suffered GCRs, validating our genetic assay. In the case of the large YAC, these GCRs were very large (Fig. 2B, top panel). The bias for large deletions is consistent with the induction of random DSBs by γ irradiation followed by significant resection of the YAC prior to de novo telomere addition (Slijepcevic and Bryant 1998). Another observation also supported DNA metabolism of the YACs after initial formation of the DSBs. Multiple-sized YACs were found in a culture derived from a single irradiated cell (Fig. 2B). In some cases, one of the YACs within a culture migrated more slowly than the starting YAC (Fig. 2B, bottom panel, 8 krad isolate 3). The generation of multiple-sized YACs, including extra long ones, from a single irradiated cell is consistent with breakage–bridge–fusion–bridge cycles (McClintock 1939).
Like γ irradiation, inactivation of Mre11p is also known to elevate the rate of GCRs (Chen and Kolodner 1999). When MRE11 was deleted in strains with the YACs, telomere marker loss was elevated ∼4-fold for PA3-1 and ∼20-fold for yWSS1572-1 relative to wild type (Fig. 2C). In mre11Δ cells, the absolute rate of YAC GCRs was 105-fold greater than the rate observed for GCRs at the end of Chromosome V (this study; Chen and Kolodner 1999). The size of these YACs, as assayed by PFGE, showed a pattern of GCRs very similar to those observed in irradiated cells (data not shown). The detection of GCRs in both mre11Δ and γ-irradiated cells demonstrated that these YACs could be used as a sensitive reporter in a genome-wide screen for mutants with enhanced rates of GCRs. These mutants are defective in chromosome integrity determinants and are defined as having a cid- phenotype.
Genome-wide screen for chromosome integrity determinants identifies DNA replication and condensation factors
Mutations can cause a cid- phenotype by enhancing the misrepair of DSBs or inducing more DSBs. The generation of DSBs is likely to occur by mutations that perturb essential DNA metabolism like DNA replication, sister chromatid cohesion, or condensation. To isolate this class of cid- mutants, we mutagenized strains harboring our YACs to generate a collection of random temperature-sensitive lethal (ts) mutants (Fig. 3A). This collection served three purposes. First, ts mutations will lie in essential genes and therefore be enriched for mutations in essential DNA metabolism. Second, at the permissive temperature for growth, many of these mutants will be hypomorphic and therefore have the potential to stimulate DSBs. Third, the inviability of the mutants at the nonpermissive temperature facilitates the easy cloning and identification of the genes responsible for the cid- phenotype at the permissive temperature.
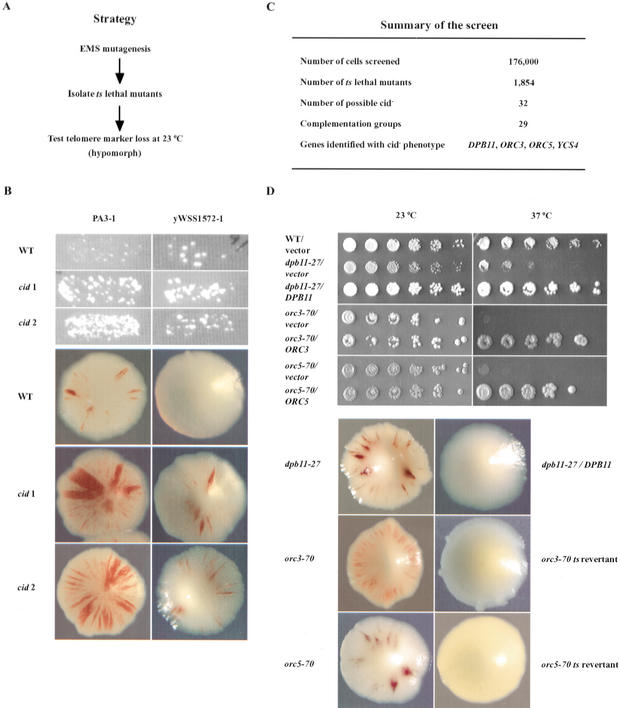
Figure 3.
Genome-wide screen for chromosome integrity determinant (cid-) mutants. (A) Steps in the screen for cid- mutants. (B) Candidate cid- mutants identified after mutagenesis of a strain with PA3-1. Wild-type and two candidate cid- mutants were tested for telomere marker loss by the papillation (top three panels) and sector (bottom three panels) assays. (C) Summary of the screen. (D) Complementation test. (Top panel) Plasmids harboring DPB11, ORC3, or ORC5 suppress the temperature-sensitive growth defect of three cid- mutants henceforth referred to as dpb11-27, orc3-70, and orc5-70, respectively. Fivefold serial dilutions of cultures were spotted onto selective medium (SC-LEU to maintain the ARS/CEN plasmids), and incubated at 23°C or 37°C. (Bottom panel) Linkage of ts growth and cid- phenotype. Cultures of cid- mutants were reverted to temperature resistance by integration of the wild-type gene (dpb11-27) or by spontaneous reversion (orc3-70, orc5-70). Revertants were then tested for telomere marker loss by the sectoring assay.
Strains with either a large (PA3-1) or small (yWSS1572-1) YAC were used to generate a collection of 1854 mutants that were temperature sensitive for growth. Only 32 of the mutants exhibited elevated GCRs at the permissive temperature as measured by the YAC assays (Fig. 3B). Ninety percent of the mutations fell into distinct complementation groups represented by a single allele (Fig. 3C). This indicated that the screen was not saturated, as expected given the small size of the ts collection (1800) relative to the number of essential yeast genes (2000). For most mutants (78%), the increased telomere marker loss was observed with both the sector assay and papillation assay. Mutants were backcrossed, following the ts phenotype, and the YACs in the original mutagenized strains were replaced with a new copy of the unmutagenized YAC (see Materials and Methods). For 12 of the backcrossed ts mutants with the fresh YAC, the GCR phenotype was still observed, indicating that it was caused by a defect in trans and linked to the ts mutation.
To identify the mutated gene in these promising cid- mutants, each mutant was tested for complementation against a subset of cohesin and condensin mutants and/or transformed with a genomic library and selected for growth at the nonpermissive temperature. For three of the cid- mutants in different complementation groups, complementing clones were obtained. The ends of the clones were sequenced, and genes within the clones were identified through the Saccharomyces Genome Database. The clones included genes that encode the replication proteins Dpb11p, Orc3p, or Orc5p (Fig. 3C). A plasmid with the isolated gene was shown to complement the ts lethality of the relevant cid- mutant (Fig. 3D, top panels). Furthermore, the ts and GCR phenotypes were linked by coreversion of these two phenotypes (Fig. 3D, bottom panels). Two other cid- mutants fell into a single complementation group, and both failed to complement the ycs4-2 mutation (data not shown).
Ycs4p is a component of the condensin complex required for mitotic condensation (Freeman et al. 2000). Dpb11p has been implicated in initiation, polymerase switching, Okazaki fragment maturation, and the S-phase checkpoint (Araki et al. 1995; Kamimura et al. 1998; Reid et al. 1999; Wang and Elledge 1999; Masumoto et al. 2000). Orc3p and Orc5p act in a complex with the Orc1, Orc2, Orc4, and Orc6 proteins to bind DNA and recruit proteins for the initiation of eukaryotic DNA replication (Bell 2002). Therefore all five of these CID mutations (henceforth named dpb11-27, orc3-70, orc5-70, ycs4-6, and ycs4-8) elevated GCRs by altering components of established DNA metabolic functions, suggesting that these defects may directly cause DSBs.
The mutations in the ORC genes were sequenced. In both cases a single base-pair substitution was found. In orc5-70, Gly 42 was changed to Arg 42. This changes a conserved amino acid of the Walker A motif (Gly–Lys–Thr), implicated in ATP binding (Haber and Walker 1991; Loo et al. 1995). In orc3-70, Ser 335 was changed to Ala 335. The orc3-70 allele maps to the region of this protein necessary for interaction with Orc5p (Lee and Bell 1997; Dhar et al. 2001). These results suggest that the cid- phenotype of orc3-70 and orc5-70 may result from compromising a single function of ORC that requires both Orc3p and Orc5p.
As an initial characterization of the cid- phenotype of our mutants, we began by examining their ability to induce GCRs of different YACs (Fig. 4A). The dpb11-27 and the orc mutations caused telomere marker loss of the small YAC at rates very similar to mre11Δ (cf. Figs. 4A and 2C). YACs that had lost telomere markers were examined by PFGE (Fig. 4B). These YACs migrated more rapidly than the original YAC, indicating they had suffered GCRs. The dpb11-27 mutation also caused telomere marker loss of the large YAC at rates very similar to mre11Δ (cf. Figs. 4A and 2C). We were unable to transfer the large YAC into either of the orc strains, presumably because the YACs are too unstable to be propagated in these mutants. Thus, the cid- phenotype of dpb11-27 and the orc mutants is not confined to a single YAC but rather is observed for YACs of distinct size and sequence. In contrast, both ycs4 alleles stimulate telomere marker loss of the large YAC but not the small YAC (data not shown), perhaps reflecting a more stringent requirement for condensin and condensation on longer chromosomes.
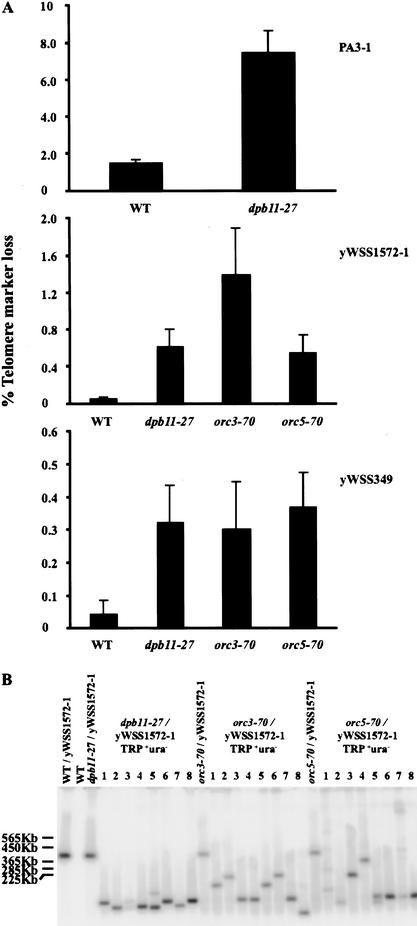
Figure 4.
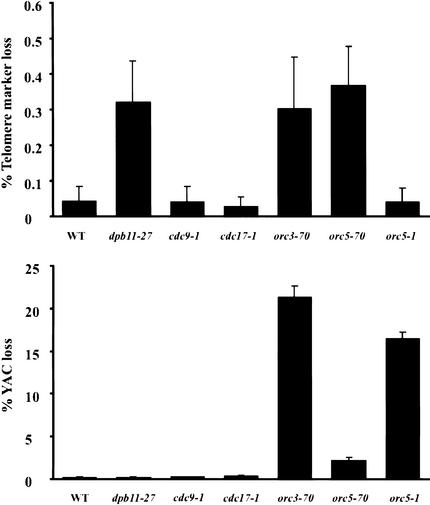
Quantitative analysis of the cid- phenotype of dpb11-27, orc3-70, and orc5-70 mutants. (A) Quantitation of telomere marker loss for wild-type and cid- strains with three different YACs, PA3-1, yWSS1572-1, and yWSS349 (see Materials and Methods). (B) Random TRP+ ura- clones with yWSS1572-1 were analyzed by PFGE as described in Figure 2B. Note all three mutants gave a similar size distribution for GCRs of the YAC. Although the GCRs in the dpb11-27 strain are all large here, this distribution is an artifact of a small sample size (data not shown).
Chromosome integrity defines novelfunctions for Dpb11p and Orc5p
The prevalence of DNA replication factors from the previous candidate approach and our genome-wide approach raised the question whether any DNA replication defect might cause elevated GCRs. To address this question, we compared the rates of GCR and total YAC loss in our mutants with mutants defective in DNA polymerase α (cdc17-1), DNA ligase (cdc9-1), and ORC (orc5-1). The orc5-1 allele has an established defect in the initiation of DNA replication (Liang et al. 1995; Loo et al. 1995). Cells that lost the entire YAC could be identified because they became simultaneously trp- and ura-, indicating loss of genetic markers on both YAC arms.
Only the alleles from our screen increase the rate of GCRs (Fig. 5, top panel). The failure to observe a cid- phenotype for at least cdc9-1 and orc5-1 cannot be attributed to a failure to perturb ligase or ORC function at 23°C. Under these growth conditions, the cdc9-1 mutant divides significantly more slowly than wild type (data not shown), and the orc5-1 mutant exhibits elevated total YAC loss (Fig. 5, bottom panel) and reduced DNA initiation (Fig. 6C). Thus, the presence of a cid- phenotype in our mutants but not cdc9-1 and orc5-1 demonstrates that telomere marker loss of the YAC is caused by only a subset of DNA replication defects. In addition, dpb11-27 induces only telomere marker loss, whereas orc5-70 and orc3-70 induce both telomere marker loss and total YAC loss. These phenotypic differences suggest that our cid- mutations may cause GCRs by different mechanisms.
Figure 5.
Telomere marker loss and total YAC loss of the yWSS349 in DNA replication mutants. Telomere marker loss and total YAC loss were quantitated for wild-type and mutant strains with the yWSS349 grown at 23°C (see Materials and Methods). The reported values are the average of three to six independent trials. Error bars indicate the standard error of the mean.
Figure 6.
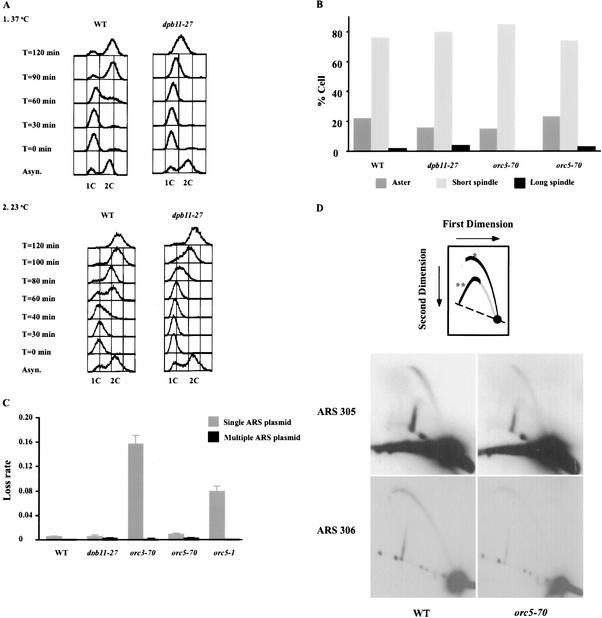
S-phase progression, S-phase checkpoint, and DNA replication initiation in cid- mutants. (A) Flow cytometry analysis of wild-type (WT) and dpb11-27 cells. Wild-type and dpb11-27 cells were grown to log phase at 23°C in YPD and arrested in G1 by α-factor. Cells were released from α-factor arrest into fresh YPD at 23°C (panel 2) or 37°C (panel 1). At the indicated times, aliquots were withdrawn and treated with RNase, stained with propidium iodide, then subject to FACScan analysis. 1C and 2C indicate the DNA content of G1 and G2/M cells, respectively. (B) Competency of S-phase checkpoint in dpb11-27, orc3-70, and orc5-70 mutants. When budding yeast are challenged with hydroxyurea, the S-phase checkpoint is triggered, and cells arrest with a short spindle. Defects in the S-phase checkpoint lead to spindle elongation. To assess the S-phase checkpoint in our cid- mutants, cells were arrested in G1 (as described in A) and released into 0.2 M hydroxyurea at 23°C or 37°C. After 90 and 120 min, aliquots of cells were withdrawn and fixed. Microtubule morphology was visualized by indirect immunofluorescence (see Materials and Methods) and scored as asters, short spindles (<2 μm) or long spindles (>2 μm). Identical results were obtained for 90 and 120 min. Only 120 min at 23°C is shown. (C) Plasmid loss assay for DNA replication initiation defects in cid- mutants. Mutants with defects in DNA replication initiation but not elongation show elevated mitotic loss rates for plasmids with a single ARS (pDK243), which is suppressed when the plasmid has multiple ARSs (pDK368-7; Hogan and Koshland 1992). To test our cid- mutants, we introduced these plasmids into our cid- strains and assayed plasmid loss at 23°C (see Materials and Methods). Error bars indicate standard errors of the mean obtained from four trials. (D) Neutral/neutral 2D gels at ARS305 and ARS306 for wild-type and orc5-70 cells. Cells grown at 23°C in YPD were divided into two portions: One portion was harvested, and the other portion was synchronized by α-factor then released into fresh medium. S-phase cells were collected at 1-min intervals. Total DNA was isolated, digested with EcoRV or PstI, run on N/N 2D gels, and blotted to membranes. The membranes were hybridized with probes to ARS305 and ARS306 (see Materials and Methods). *, bubbles; **, simple Ys.
To elucidate these potential different mechanisms, we characterized S-phase progression, the S-phase checkpoint, and replication initiation in our mutants (see Materials and Methods). S-phase progression is severely impaired in the dpb11-27 mutant at 37°C (Fig. 6A, panel 1) and delayed by 30 min at 23°C (Fig. 6A, panel 2), the temperature at which the cid- phenotype was observed. When challenged with hydroxyurea, the dpb11-27 mutant exhibited S-phase arrest comparable to wild-type cells at 23°C (Fig. 6B) or 37°C (data not shown). In addition, dpb11-27 mutant and wild-type cells show very similar sensitivity to UV or γ irradiation (data not shown). Taken together, these results suggest that the S-phase and other DNA-related checkpoints are intact in the dpb11-27 mutant. Analysis of DNA replication initiation using a plasmid loss assay revealed no defect in the dpb11-27 mutant (Fig. 6C). Because the dpb11-27 allele exhibits a defect in S-phase progression but not in the S-phase checkpoint or initiation of DNA replication, we suggest that this mutant specifically compromises the DNA elongation function of Dpb11p, and this elongation defect causes elevated GCRs. A previous study had implicated the S-phase checkpoint activity of Dpb11p in chromosome integrity (Myung et al. 2001b). Therefore, our studies expand the role of Dpb11p in chromosome integrity by defining a new function for its DNA elongation activity in chromosome integrity.
Like dpb11-27, the orc mutants were also analyzed for DNA replication defects. No defects in S-phase progression (data not shown) or the S-phase checkpoint were observed at either permissive (Fig. 6B) or nonpermissive temperatures (data not shown). These mutants are also indistinguishable from wild type in their sensitivity to UV and γ irradiation (data not shown). Thus, GCRs in these orc mutants are not likely to be the consequence of defects in elongation or checkpoints. The analysis of initiation by the plasmid loss assay showed that the orc5-1 mutant exhibited an initiation defect as expected from previous studies, but orc5-70 had none (Fig. 6C). Because the orc5-70 result was unexpected, initiation of DNA replication in this mutant was examined at two yeast origins by 2D gels (Fig. 6D). Quantitation of these gels reveals no change in the ratio of bubbles to simple Ys in the mutant compared with wild-type cells, corroborating the absence of a defect in initiation at 23°C. Therefore, from our analyses of chromosome integrity and DNA replication initiation in the orc mutants (Figs. 5, 6C), the orc5-1 has an initiation defect and elevated total YAC loss but no detectable increase in GCRs. In contrast, orc5-70 has no detectable initiation defect but shows elevated formation of GCRs. These opposite phenotypes suggest that chromosome integrity involves a novel function of Orc5p, defined by orc5-70, which is distinct from its initiation function defined by orc5-1. The orc3-70 has phenotypes of both orc5-1 and orc5-70 including an initiation defect, elevated YAC loss, and elevated formation of GCRs. This suggests that orc3-70 compromises both ORC activities defined by orc5-1 and orc5-70.
GCRs can be enhanced or suppressed by increasing origin number
The cid- phenotype of our DNA replication mutants might result from a combination of partial inactivation of the gene products and their interaction with cis factors. Indeed, reducing the number of origins on a YAC enhances interstitial deletions in rad9 checkpoint mutants (van Brabant et al. 2001). Similarly reducing the number of origins might alter the cid- phenotype of our dpb11 and orc mutants. Phenotypic interactions between origins and ORC are expected because of their biochemical interaction. In addition, GCRs of the YAC are elevated to very similar levels in dpb11-27 and mre11Δ mutants (Figs. 4A, 2C), whereas GCRs at the end of Chromosome V are 60-fold higher in the mre11Δ mutant than the dpb11-27 mutant (data not shown). This relative change in cid- phenotype for dpb11-27 between the YAC and Chromosome V suggests that the CID function of Dpb11p is influenced by a cis factor, perhaps origins. Therefore, we examined the effect of decreasing origin number on YAC stability in the dpb11-27 mutant as well as our orc mutants.
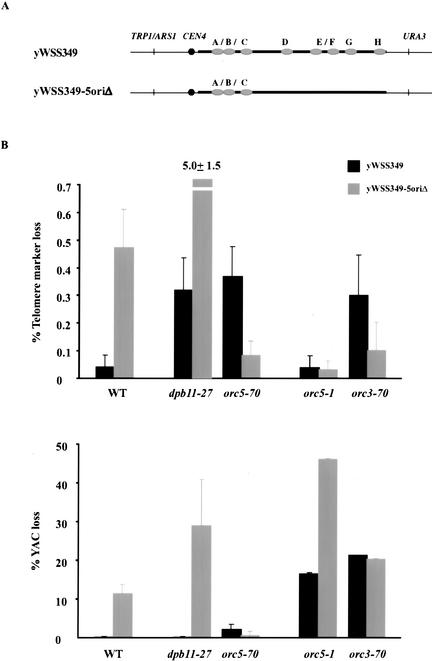
For these studies, we used the YAC yWSS349 (wild-type YAC) and its derivative, yWSS349-5oriΔ (5oriΔ YAC), in which five of the eight origins were deleted (Fig. 7A; van Brabant et al. 1999, 2001). These two YACs were introduced into our cid- mutants as well as orc5-1, cdc17-1, and cdc9-1, and the telomere marker loss and total YAC loss were monitored. For cdc17-1 and cdc9-1, no changes in either telomere marker loss or total YAC loss were observed (data not shown).
Figure 7.
Telomere marker loss and total YAC loss of yWSS349 and yWSS349-5oriΔ in dpb11-27, orc5-70, orc5-1, and orc3-70 mutants. (A) Schematic diagram of yWSS349 and yWSS349-5oriΔ. The origins (A–H) on yWSS349 were mapped, and five were deleted to give rise to yWSS349-5oriΔ. (B) Telomere marker loss (top panel) and total YAC loss (bottom panel) were quantitated for wild-type and mutant strains with the YAC yWSS349 and yWSS349-5oriΔ grown at 23°C. The numbers reported are the average of six independent trials. Error bars indicate standard error of the mean. Values are presented for both yWSS349-5oriΔ (gray) and yWSS349 (black).
However, in dpb11-27 and orc5-70 mutants, deleting origins has opposite effects on telomere marker loss and total YAC loss (Fig. 7B). In the dpb11-27 mutant, GCRs and total YAC loss increase dramatically for the 5oriΔ YAC compared with the wild-type YAC. This increase is significantly greater than expected from the rates of GCRs and YAC loss if the effects of dpb11-27 and deleting origins were additive. We conclude that the presence of more active origins on the wild-type YAC reduces the rate of GCRs in dpb11-27. In contrast, in the orc5-70 mutant, both telomere marker loss and total YAC loss decrease dramatically for the 5oriΔ YAC compared with the wild-type YAC. Thus, more origins on the wild-type YAC must enhance GCRs and total YAC loss in the orc5-70 mutant. Therefore, increasing origin number has opposite effects on YAC stability in dpb11-27 and orc5-70 mutants.
The consequence of deleting origins from the YAC is also very different in the orc5-70 and orc5-1 mutants (Fig. 7B). In the orc5-1 mutant, the rate of total YAC loss but not GCRs increases dramatically for the 5oriΔ YAC compared with the wild-type YAC. The total YAC loss increases to as high as 46% per cell division. Therefore, more origins on the wild-type YAC dramatically stabilize it in orc5-1 mutants but destabilize it in orc5-70 mutants (see above). This observation provides another phenotypic difference between these two alleles, further supporting the idea that these alleles compromise distinct functions of ORC. Furthermore, the notion that both of these functions might be compromised in the orc3-70 mutant is supported by the fact that in this mutant the 5oriΔ YAC has a reduced rate of GCRs but still has a high total YAC loss rate.
Discussion
Here we report a simple YAC-based assay to identify mutated genes that elevate the rate of gross chromosomal rearrangements. Other assays for measuring GCRs have already been described, but their use has been limited to the analysis of previously characterized mutations in candidate genes (Kolodner et al. 2002). The sensitivity and ease of the YAC assay make it possible to screen unbiased genome-wide collections of mutants. The success of the screen is evident from three features of the mutants identified to date. First, all mutants are defective for a gene encoding a protein required for essential DNA metabolic functions. Therefore, our mutants are likely to identify proteins directly involved in GCR formation. Second, three of the four genes identified in our screen, YCS4, ORC5, and ORC3, have not been associated previously with GCRs. In addition, whereas previous studies suggested that GCR is caused by perturbing the checkpoint function of Dpb11p, here we present evidence that GCR is also caused by perturbing its elongation function. Therefore, all of the genes from our screen identify new candidates and pathways for GCR formation. The value of these successes is further augmented by the fact that our screen for GCR mutants using the YAC assay is not saturated either for essential (tested here) or nonessential genes (not yet tested). Therefore, it is likely that additional genes and pathways required for chromosome integrity can be discovered using the YAC assay.
GCRs of the YAC in the mre11Δ mutant are 5 orders of magnitude greater than observed at the end of Chromosome V (this study; Chen and Kolodner 1999). Yet, the nonessential target for the YACs (0.3–1.6 Mb) is only ∼7-to 40-fold greater than Chromosome V (∼43 kb). Thus, the increased sensitivity of the YAC for GCRs is unlikely to be explained solely by an increased target size for random DSBs. Rather, we suggest that mre11Δ and many other potential mutants may cause nonrandom DSBs as a result of improper DNA metabolism associated with localized DNA elements such as origins and replication fork barriers. These elements are more likely to be included in a large target like the YAC. In addition, we show that the YACs often undergo significant processing after the initial DSBs. If this is also the case for Chromosome V, then many DSBs that formed initially in the nonessential region might lead to loss of genes in the neighboring essential region and not be recovered.
One alternative explanation for the high rate of GCRs observed for the YAC is that these events are mostly interstitial deletions promoted by repetitive sequences within the human or mouse DNA inserted in the YAC. However, this is extremely unlikely because the YAC terminal markers that are lost in the GCR events are not flanked by insert DNA but are embedded in telomere-proximal vector sequences. Furthermore, at least 50% of GCRs on the YAC are Rad52-independent (D. Huang and D. Koshland, unpubl.). Therefore, the GCRs in the YAC are likely to be terminal deletions or translocations generated by pathways of microhomology recombination or de novo telomere addition (Chen et al. 1998; Chen and Kolodner 1999; Myung et al. 2001a,b; Myung and Kolodner 2002). Although it will be interesting to analyze the sequence of the GCRs on the YAC, these sequences may be of a limited use because the ability of the YAC to tolerate significant processing post-DSB means that the final GCR may be far removed from the initial events that led to its formation.
Origin number, ORC, and chromosome integrity
In this study, we show that the number of origins on a YAC can both positively and negatively influence its integrity. In the dpb11-27 mutant, higher origin number on the YAC suppresses GCR. We also show that this allele is compromised for S-phase progression. Taken together with previous studies implicating Dpb11p in polymerase switching, we suggest that this function of Dpb11p may be needed to allow replication through unusual structures. When compromised forks stall, GCRs may ensue. Higher origin number may ensure that a neighboring fork will be sufficiently close to rescue a stalled fork before it degenerates to a chromosome break. The inhibition of the assembly of prereplicative complexes also causes GCR (Lengronne and Schwob 2002; Tanaka and Diffley 2002). The investigators suggest a similar model in which fewer active origins decrease the probability of the rescue of a stalled fork by a neighboring fork. Thus, increased origin density on chromosomes may have been selected during evolution to rescue stalled forks that occur when functions like Dpb11p become limiting/perturbed as a result of stochastic events or environmental changes.
However, selective pressures against increasing origin numbers must also exist because in budding yeast, as in many organisms, origins are spaced approximately every 40 kb (Newlon and Burke 1980; Tanaka et al. 1996; Poloumienko et al. 2001). Here, we show that YACs with more origins have a higher rate of GCR in orc5-70 and orc3-70 mutants. These mutants provide the first example of conditions in which a chromosome with more origins is less stable and suggest that origins themselves can generate labile structures that are prone to breakage. In wild-type cells, spontaneous or environmental conditions that mimic the defects in orc5-70 and orc3-70 mutants will promote these labile origins and preferentially cause GCRs on chromosomes with more origins. Thus, elucidating the ORC defect in these mutants may provide important insight into the evolutionary forces that select against increased origins on chromosomes.
Chromosome integrity defines a potential new function for ORC
The phenotypes of orc5-70 are unlike any other initiation defect. This allele has no detectable defect in the frequency of initiation by either 2D gel analysis or plasmid loss assay, yet it stimulates GCRs. In contrast, the orc5-1 allele does show a significant reduction in initiation frequency by both the plasmid loss assay and 2D gel analysis but does not induce GCRs. Furthermore, the response of these two alleles to removing origins is opposite. Adding origins to a YAC stabilizes it in the orc5-1 mutant but destabilizes it in the orc5-70 mutant. Reducing the formation of the prereplicative complexes by manipulating cyclin-dependent kinase activity has been shown to cause GCRs (Tanaka and Diffley 2002). However, under these conditions, adding origins to the marker chromosome stabilizes it, like orc5-1 but unlike orc5-70. Based on these phenotypic differences, we suggest that orc5-70 is likely to define a new function for ORC distinct from its role in controlling initiation frequency or assembly of the prereplicative complexes.
An important clue to this putative novel function of ORC is the fact that the orc5-70 allele alters a conserved amino acid within the Walker A ATP-binding motif. Consistent with our analysis, previous characterized mutations in this Walker A site are competent for ORC activities in vitro and initiation in vivo (Klemm et al. 1997; Chesnokov et al. 2001). Thus, ATP binding of Orc5p does not appear to be required for the core initiation function of ORC. One model has suggested that this ATPase might be used to inactivate ORC. In this light, Orc5 ATP binding may be part of a regulatory system that promotes Orc1p ATPase to inactivate a fired ORC after successful initiation, or dissemble an unfired ORC upon its replication from a neighboring fork. When this regulatory system is compromised, the persistent ORC could cause labile DNA structures such as a longer-lived ssDNA at the origin, stalled forks (blockage of a neighboring fork by an unfired ORC), or forks within forks (late activation of an unfired origin after the neighboring fork passes). Any of these labile structures could give rise to GCR. Other models are certainly possible; nonetheless, the CID function of Orc5p ATP binding provides the first in vivo function for this motif and an important new handle to elucidate its molecular function.
Materials and methods
Media and reagents
Cells were grown in YPD medium or supplemented minimal medium as indicated. Dextrose (Sigma) was added to 2% final solution. We purchased α-factor and hydroxyurea from Sigma, antibodies (YOL1/34, a rat anti-tubulin, and goat anti-alpha rat-FITC) from Harlan Bioproduct for Science, 5-fluoroorotic (5-FOA) from Bio Vectra, ethyl-methane-sulfonate (EMS) from Sigma, and G418 from GIBCO-BRL.
YAC strain construction
In strains used for EMS mutagenesis, the ADE2 gene was disrupted with the kanMX cassette by using the PCR-based method. The oligonucleotide sequences were 5′-CTAAGTA CATCCTACTATAACAATCAAGAAAAACAAGAAAATCG GACAAAACAATCAAGTCAGCTGAAGCTTCGTACGCT-3′ and 5′-GATGTACAAGTATATCAATAAACTTATATATTAC TTGTTTTCTAGATAAGCTTCGTAACCGCATAGGCCACT AGTGGATCTG-3′. The ADE2 gene was integrated into the vector sequences of pYAC4 at the end of the right arms of the YACs, PA3 and yWSS1572, to generate PA3-1 and yWSS1572-1. The ADE2 gene replaced pYAC4 sequence from 950 to 1042 bp. The oligonucleotide sequences used for PCR-based integration were 5′-CGGCCTCAACCTACTACTGGGCTGCTTCCTAAT GCAGGAGTCGCATAAGGGAGAGCGTCGACCGGAACT TGACTAGCGCACTACCAGTATATC-3′ and 5′-CTCCAGC GAAAGCGGTCCTCGCCGAAAATGACCCAGAGCGCTGC CGGCACCTGTCCTACGGGACACCTGTAAGCGTTGATT TCTATG-3′. The MRE11 gene was disrupted with the kanMX cassette by using the PCR-based method described by Ritchie and Petes (2000).
YACs were introduced into yeast strains (Table 1) by kar1 mating as described by Spencer and Hieter (Hugerat et al. 1994; Spencer et al. 1994). Transformants that have the correct telomere markers were examined for the presence of intact YAC by pulsed field gel electrophoresis (PFGE) and Southern analysis.
Qualitative assays for telomere marker loss
Papillation assay: Cells with YACs (PA3-1 or yWSS1572-1) were patched onto SC-URA plates and incubated at 23°C overnight. Cells were replica-plated onto YPD plates first then immediately replica-plated from YPD plates onto SC-TRP-URA 5-FOA plates (0.72 g of CSM-TRP-URA powder from Bio 101, 40 mg of uracil, 1 g of 5-FOA per liter). Papillae were counted after incubation at 23°C for 3–4 d.
Sector assay: Cells with YACs (PA3-1 or yWSS1572-1) were streaked for singles on the complete synthetic plates with limiting adenine as described previously (Koshland and Hieter 1987) except 3.76 mg/3 L (final concentration) adenine was added. Plates were incubated at 23°C for 7–10 d to allow for better color development.
Quantitative assays for telomere marker loss and total YAC loss
Single colonies from SC-URA plates that select for the YAC telomere marker (URA3) were picked and resuspended in 1 mL of water. These cell suspensions were diluted and plated onto rich media, YPD plates such that ∼150–200 colonies formed on each plate. Colonies on the YPD plates were replica-plated to SC-TRP and SC-URA, and growth was scored after incubation at 23°C for 3–4 d. The phenotype of these colonies allowed us to distinguish three types of cells present in the colony grown under selection for the telomere marker: those with the full-length YAC (TRP+ URA+), with no YAC (trp- ura-), or with the YAC but lost the telomere marker (TRP+ ura-). The percentage of cells with telomere marker (TRP+ ura-) or entire YAC loss in media selective for the full-length YAC is a direct measure of the rate of these events when there is no phenotypic lag. When there is phenotypic lag, this percentage will be an overestimate of the rate. However, because all measures of telomere loss and YAC loss were done using the same markers, the contribution of the phenotypic lag will be the same and the relative rates (fold difference) the same as when there was no lag.
cid- mutant isolation and cloning CID genes by complementation
PA3-1 or yWSS1572-1 cells were mutagenized with EMS and screened for temperature-sensitive lethal (ts) mutants at 37°C. ts lethal mutants were then tested for increased telomere marker loss by the papillation assay and sector assay. After one backcross, a fresh (unmutagenized) YAC was introduced into candidate cid- mutants by kar1 mating. Mutants in which the cid- phenotype was unlinked to the YAC were then backcrossed five times following the ts phenotype. A fresh YAC was again introduced into ts mutants, and linkage of the ts phenotype and the cid- phenotype was determined.
A subset of cid- mutants was transformed with a library DNA C10(F. Spencer and P. Hieter, unpubl.). Transformed cells were grown at 37°C to isolate genomic DNA inserts that complement the ts phenotype. The sequence of the ends of the complementing insert was determined by sequencing across the junction of the C10 vector and insert. The intervening sequence and corresponding open reading frames were determined using the Saccharomyces Genome Database.
γ-irradiation
Wild-type cells with PA3-1 or yWSS1572-1 were grown in synthetic medium lacking uracil to select for the YAC to mid-log phase at 23°C. Cells were arrested with 5 μg/mL α-factor for 3 h. Then the cell culture was divided into four equal portions and irradiated with 0, 2, 8, and 20 krad. Cell viability was measured after cells were incubated at 23°C for 1 h with YPD medium with 0.1 mg/mL pronase and plated on YPD plates. The rate of YAC telomere marker loss was determined as described above.
Cell cycle analyses and S-phase checkpoint assay
Cultures were grown in YPD medium to mid-log phase at 23°C. Cells were arrested with 5 μg/mL α-factor for 3 h. For cell cycle analysis, cells were either released from α-factor arrest with YPD medium with 0.1 mg/mL pronase at 23°C or shifted to 37°C for 30 min and then released from α-factor arrest by washing cells with prewarmed YPD medium (37°C) with 0.1 mg/mL pronase at 37°C. An aliquot of cells was withdrawn every 20 or 30 min and prepared for flow cytometry (Yamamoto et al. 1996). For the S-phase checkpoint, cells were treated as described above except medium upon release from α-factor arrest contained 0.1 mg/mL pronase and hydroxyurea (0.2 M final concentration). An aliquot of cells was withdrawn after 90 min or 120 min and fixed with 3.6% formaldehyde (final concentration) at room temperature for 60 min. Fixed samples were processed for immunofluorescence (Kilmartin and Adams 1984).
Determination of plasmid loss rates
Plasmid loss rates were determined in cells carrying pDK plasmids with one or eight ARSs (Hogan and Koshland 1992) using the formula: loss rate = 1 - 10m, where m = [log(Ff) - log(Fi)]/n, F(i) being the initial and F(f) the final fraction of cells containing the plasmid after n doublings. The fraction of plasmid-containing cells was determined by plating the cells on YPD plates and replica plates onto SC-LEU plates after incubation at 23°C for 3–4 d.
2D gelanalysis
Total yeast DNA was extracted from asynchronized cells or S-phase cells. Conventional 2D gel analysis was performed as described by Brewer and Fangman (Brewer et al. 1988; Brewer and Fangman 1991). The first dimension gels were 0.4% agarose and the second dimension gels were 1.1% agarose. We analyzed the 4207-bp EcoRV restriction fragment for ARS305 and the 5366-bp PstI restriction fragment for ARS306.
Structural analysis of GCRs on the YAC: pulsed field gel electrophoresis method and Southern blot analysis
Total chromosomal DNA agarose plugs were prepared as described by Schwartz and Cantor (1984) and resolved on a 1% agarose gel using Bio-Rad CHEF-DR III system at 6 V/cm for 17 h with a switch time ramped from 20 to 50 sec at 14°C for yWSS1572-1 or 6 V/cm for 22 h with a switch time ramped from 50 to 90 sec at 14°C for PA3-1. Gels were blotted and probed with a pYAC4 sequence that is a 2.3-kb EcoRI and PstI fragment from PBR322.
Acknowledgments
We thank Eileen Hogan, Brigitte Lavoie, Bonnie Brewer, Margaret Hoang, M.K. Raghuraman, and Stephen Bell for technical assistance and reagents. We thank Orna Cohen-Fix, Bonnie Brewer, Margaret Hoang, Stephen Bell, Andy Hoyt, Yixian Zheng, Catherine Huang, Elcin Unal, and Yukan Duan for helpful discussions and critical reading of the manuscript. This work is supported by a grant from Howard Hughes Medical Institution.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1089203.
References
- Araki H., Leem, S.H., Phongdara, A., and Sugino, A. 1995. Dpb11, which interacts with DNA polymerase IIε in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl. Acad. Sci. 92: 11791–11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P. 2002. The origin recognition complex: From simple origins to complex functions. Genes & Dev. 16: 659–672. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman, W.L. 1991. Mapping replication origins in yeast chromosomes. BioEssays 13: 317–322. [DOI] [PubMed] [Google Scholar]
- Brewer B.J., Sena, E.P., and Fangman, W.L. 1988. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 6: 229–234. [DOI] [PubMed] [Google Scholar]
- Burke D.T., Carle, G.F., and Olson, M.V. 1987. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science 236: 806–812. [DOI] [PubMed] [Google Scholar]
- Chen C. and Kolodner, R.D. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23: 81–85. [DOI] [PubMed] [Google Scholar]
- Chen C., Umezu, K., and Kolodner, R.D. 1998. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell 2: 9–22. [DOI] [PubMed] [Google Scholar]
- Chesnokov I., Remus, D., and Botchan, M. 2001. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc. Natl. Acad. Sci. 98: 11997–12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar S.K., Delmolino, L., and Dutta, A. 2001. Architecture of the human origin recognition complex. J. Biol. Chem. 276: 29067–29071. [DOI] [PubMed] [Google Scholar]
- Freeman L., Aragon-Alcaide, L., and Strunnikov, A. 2000. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell. Biol. 149: 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber L.T. and Walker, G.C. 1991. Altering the conserved nucleotide binding motif in the Salmonella typhimurium MutS mismatch repair protein affects both its ATPase and mismatch binding activities. EMBO J. 10: 2707–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E., Avner, P., and Rothstein, R. 1994. Creation of a deletion series of mouse YACs covering a 500 kb region around Xist. NAR 22: 1830–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan E. and Koshland, D. 1992. Addition of extra origins of replication to a minichromosome suppresses its mitotic loss in cdc6 and cdc14 mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 89: 3098–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugerat Y., Spencer, F., Zenvirth, D., and Simchen, G. 1994. A versatile method for efficient YAC transfer between any two strains. Genomics 22: 108–117. [DOI] [PubMed] [Google Scholar]
- Kamimura Y., Masumoto, H., Sugino, A., and Araki, H. 1998. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell. Biol. 18: 6102–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J. and Adams, A. 1984. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 98: 922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm R.D., Austin, R.J., and Bell, S.P. 1997. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell 88: 493–502. [DOI] [PubMed] [Google Scholar]
- Kolodner R.D., Putnam, C.D., and Myung, K. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552–557. [DOI] [PubMed] [Google Scholar]
- Koshland D. and Hieter, P. 1987. Visual assay for chromosome ploidy. In Methods in enzymology Vol. 155 (ed. R. Wu), pp. 351–372. Academic Press, New York. [DOI] [PubMed] [Google Scholar]
- Kuhn R.M. and Ludwig, R.A. 1994. Complete sequence of the yeast artificial chromosome cloning vector pYAC4. Gene 141: 125–127. [DOI] [PubMed] [Google Scholar]
- Lee D.G. and Bell, S.P. 1997. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol. 17: 7159–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A. and Schwob, E. 2002. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol. Cell 9: 1067–1078. [DOI] [PubMed] [Google Scholar]
- Liang C., Weinreich, M., and Stillman, B. 1995. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 81: 667–676. [DOI] [PubMed] [Google Scholar]
- Loo S., Fox, C., Rine, J., Kobayashi, R., Stillman, B., and Bell, S. 1995. The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol. Biol. Cell 6: 741–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H., Sugino, A., and Araki, H. 2000. Dpb11 controls the association between DNA polymerases α and ε and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 20: 2809–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1939. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl. Acad. Sci. 25: 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B. 2000. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 25: 173–178. [DOI] [PubMed] [Google Scholar]
- Myung K. and Kolodner, R.D. 2002. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 99: 4500–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Chen, C., and Kolodner, R.D. 2001a. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076. [DOI] [PubMed] [Google Scholar]
- Myung K., Datta, A., and Kolodner, R.D. 2001b. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104: 397–408. [DOI] [PubMed] [Google Scholar]
- Newlon C. and Burke, W. 1980. Replication of small chromosomal DNAs in yeast. In Mechanistic studies of DNA replication and recombination (eds. B. Alberts and C. Fox), pp. 399–409. Academic Press, New York.
- Pflumm M.F. and Botchan, M.R. 2001. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development 128: 1697–1707. [DOI] [PubMed] [Google Scholar]
- Poloumienko A., Dershowitz, A., De, J., and Newlon, C.S. 2001. Completion of replication map of Saccharomyces cerevisiae chromosome III. Mol. Biol. Cell 12: 3317–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid R.J., Fiorani, P., Sugawara, M., and Bjornsti, M.A. 1999. CDC45 and DPB11 are required for processive DNA replication and resistance to DNA topoisomerase I-mediated DNA damage. Proc. Natl. Acad. Sci. 96: 11440–11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K.B. and Petes, T.D. 2000. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R., Michel, B., and Gangloff, S. 2000. Replication fork pausing and recombination or “gimme a break.” Genes & Dev. 14: 1–10. [PubMed] [Google Scholar]
- Schwartz D.C. and Cantor, C.R. 1984. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37: 67–75. [DOI] [PubMed] [Google Scholar]
- Slijepcevic P. and Bryant, P.E. 1998. Chromosome healing, telomere capture and mechanisms of radiation-induced chromosome breakage. Int. J. Radiat. Biol. 73: 1–13. [DOI] [PubMed] [Google Scholar]
- Spencer F., Hugerat, Y., Simchen, G., Hurko, O., Connelly, C., and Hieter, P. 1994. Yeast kar1 mutants provide an effective method for YAC transfer to new hosts. Genomics 22: 118–126. [DOI] [PubMed] [Google Scholar]
- Tanaka S. and Diffley, J.F. 2002. Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation. Genes & Dev. 16: 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Tanaka, Y., and Isono, K. 1996. Systematic mapping of autonomously replicating sequences on chromosome V of Saccharomyces cerevisiae using a novel strategy. Yeast 12: 101–113. [DOI] [PubMed] [Google Scholar]
- Tennyson R.B., Ebran, N., Herrera, A.E., and Lindsley, J.E. 2002. A novel selection system for chromosome translocations in Saccharomyces cerevisiae. Genetics 160: 1363–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brabant A.J., Fangman, W.L., and Brewer, B.J. 1999. Active role of a human genomic insert in replication of a yeast artificial chromosome. Mol. Cell. Biol. 19: 4231–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brabant A.J., Buchanan, C.D., Charboneau, E., Fangman, W.L., and Brewer, B.J. 2001. An origin-deficient yeast artificial chromosome triggers a cell cycle checkpoint. Mol. Cell 7: 705–713. [DOI] [PubMed] [Google Scholar]
- Wang H. and Elledge, S.J. 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 96: 3824–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Guacci, V., and Koshland, D. 1996. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J. Cell Biol. 133: 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]