Abstract
When DNA replication is stalled, a signal transduction pathway is activated that promotes the stability of stalled forks and resumption of DNA synthesis. In budding yeast, this pathway includes the kinases Mec1 and Rad53. Here we report that the Mediator protein Mrc1, which is required for normal DNA replication and for activation of Rad53, is present at replication forks. Mrc1 initially binds early-replicating sequences and moves along chromatin with the replication fork. Blocking initiation of DNA replication blocks Mrc1 loading onto origins, providing an explanation for why so many mutants in DNA replication show checkpoint defects. In the presence of replication blocks, we find that Mec1 is recruited to regions of stalled replication, where it encounters and presumably phosphorylates Mrc1. Mutation of the canonical Mec1 phosphorylation sites on Mrc1 prevents Mrc1 phosphorylation and blocks Rad53 activation, but does not alter Mrc1's role in DNA replication. Our results suggest a model whereby in response to DNA replication interference, the Mec1 kinase is recruited to sites of replication blocks and phosphorylates a component of the DNA replication complex, Mrc1, thereby setting up a solid-state Rad53 activation platform to initiate the checkpoint response.
Keywords: Checkpoint, DNA damage, Mediator, replication fork, Rad53, Claspin
Cell survival in the face of DNA damage or replication stress depends on the presence of intact checkpoint mechanisms (Zhou and Elledge 2000). In the budding yeast Saccharomyces cerevisiae, such a surveillance mechanism exists, which allows a cell to deal with the problem of stalled replication forks during S phase. When replication forks stall because of DNA damage or insufficient nucleotide levels, cells respond by activating a signal transduction pathway that delays entry into mitosis as long as the task of DNA synthesis remains unfinished (Allen et al. 1994; Weinert et al. 1994). In addition to delaying cell cycle progression, cells also delay late origin firing (Weinreich and Stillman 1999) and activate a transcriptional response (Huang et al. 1998; Gasch et al. 2001) directed toward alleviating replication stress and repairing any resultant DNA damage. A fourth and critical response is to stabilize the replication complex itself to facilitate the re-establishment of fork progression after the stress has been adequately ameliorated (Desany et al. 1998; Lopes et al. 2001; Tercero and Diffley 2001).
To initiate the replication stress response, three groups of proteins are required. The first class consists of the PI3-family kinase ATR-homolog Mec1 (Sanchez et al. 1996) and its associated ATRIP homolog Ddc2 (Paciotti et al. 2000; Rouse and Jackson 2000; Wakayama et al. 2001). The second group is a class consisting of the RFC-like proteins Rad24 and Ctf18 (Naiki et al. 2001); the PCNA-like proteins Rad17, Mec3, Ddc1 (Zhou and Elledge 2000); and perhaps other unidentified redundant molecules. This class is redundant in S. cerevisiae but is absolutely required in other species such as Schizosaccharomyces pombe and mammals. The third group consists of components intimately associated with replication itself. Factors such as Pol2 (Navas et al. 1995), Dpb11 (Araki et al. 1995; Wang and Elledge 1999), Rfc5 (Sugimoto et al. 1997), Dbf4/Cdc7 (Toyn et al. 1995), and Drc1 (Kamimura et al. 1998; Wang and Elledge 1999) all have roles in replication in addition to their requirement for the initiation of the replication stress response. The precise nature of the coordination among these three groups of proteins is not fully understood, and the role that the replication components play in checkpoint initiation remain unknown, although it has been suggested that they control the strength of the checkpoint signal by controlling the number of active replication forks (Shimada et al. 2002; Wang and Elledge 2002).
A key downstream component of the replication stress signal transduction pathway is the Chk kinase Rad53, which becomes hyperphosphorylated and activated in response to DNA damage or DNA replication stress (Allen et al. 1994; Weinert et al. 1994; Sanchez et al. 1996; Sun et al. 1996). Rad53 activation is critical for all four branches of the replication stress response and is dependent on the coordination of the three groups of proteins described above. Although the activation of Rad53 requires Mec1/Ddc2, Mec1/Ddc2 alone is insufficient for activation in response to replication or DNA damage stress. Instead, a new class of checkpoint proteins called Mediators is required to facilitate the activation of the Chk kinases. The prototypical Mediator is Rad9, which is essential for the activation of Rad53 in response to DNA damage (Weinert and Hartwell 1988; de la Torre-Ruiz et al. 1998). Rad9 is subjected to phosphorylation by the two PIK family members in budding yeast, Mec1 and Tel1 (Vialard et al. 1998). This phosphorylated form of Rad9 is found in a complex with Rad53, and it appears to stimulate the autoactivation, in trans, of Rad53 (Vialard et al. 1998; Gilbert et al. 2001). The interaction between phosphorylated Rad9 and Rad53 is direct and is mediated by the C-terminal FHA domain of Rad53 (Sun et al. 1998). Additionally, mutant Rad9, which lacks key phosphorylatable SQ and TQ residues, fails to bind Rad53 in response to MMS, fails to stimulate Rad53 activity, and confers an enhanced sensitivity to MMS (Schwartz et al. 2002).
Although Rad9 is required to activate Rad53 in response to DNA damage, it has no role in responding to DNA replication stress in wild-type cells. This suggests that a different Mediator must exist to activate Rad53 in response to replication stress. Recently such a novel Mediator, Mediator of the Replication Checkpoint (MRC1), was identified that is required for Rad53 activation (Alcasabas et al. 2001; Tanaka and Russell 2001). mrc1 mutants demonstrate a significant delay in the timing of Rad53 activation when replication is blocked. It is thought that in the absence of an immediate response, replication stress causes DNA damage in mrc1 mutants, which then activates Rad9 and the DNA-damage checkpoint to subsequently activate Rad53. In the absence of Rad9, mrc1 mutants show no activation of Rad53 (Alcasabas et al. 2001). Mrc1 is related to the Claspin protein, which mediates Chk1 activation in response to replication blocks in Xenopus extracts (Kumagai and Dunphy 2000).
Several observations suggest that Mrc1 mediates Rad53 activation in a fashion similar to that of Rad9. First, like Rad9, Mrc1 is hyperphosphorylated in response to replication blocks and contains a number of SQ and TQ residues, which are sites of phosphorylation by kinases of the Mec1 family (Kim et al. 1999). Second, in Schizosaccharomyces pombe, Mrc1's phosphorylation is entirely due to the Mec1 homolog Rad3, suggesting that scMrc1 might be phosphorylated on its SQ and TQ residues by Mec1. Finally, Mrc1 shows sequence relationship to Crb2, the S. pombe equivalent of Rad9 and, to a lesser extent, to Rad9 itself (Alcasabas et al. 2001). This relationship suggests that MRC1- and RAD9-related genes may have evolved from a common ancestor during evolution and may work through a common mechanism to activate Rad53.
A second phenotype of mrc1 mutants is a slow S phase during which the DNA-damage response is spontaneously activated (Alcasabas et al. 2001). Although this slow replication could be a manifestation of the inability of mrc1 mutants to activate Rad53, an equally plausible explanation is that Mrc1 plays a role in DNA replication itself. Here we provide evidence that the role of Mrc1 in mediating the replication stress response is separable from this role in DNA replication. We provide evidence that the SQ and TQ residues of Mrc1 are essential for its role in the replication checkpoint but not in DNA replication itself, strengthening the idea that Mrc1 activates Rad53 in a fashion similar to that of other Mediator proteins. We further demonstrate that Mrc1 associates with chromatin during S phase and that Mrc1 localizes to replication forks, demonstrating its role in replication as a direct participant of the fork complex. Last, we show that the checkpoint-defective allele of MRC1 is competent to load onto chromatin and travel with replication forks. Thus, Mrc1 is a Mediator that rides on replication forks and is in a primary position to sense and respond to DNA replication stress and activate the checkpoint response.
Results
SQ and TQ residues of Mrc1 are important for viability in the face of replication stress
In S. pombe and S. cerevisiae, Mrc1's phosphorylation depends on the ATR homologs Rad3 and Mec1, respectively (Alcasabas et al. 2001; Tanaka and Russell 2001). Mrc1 contains a loosely defined SQ/TQ cluster domain that represents target phosphorylation sites for kinases of the ATR class. Mrc1 is also required for normal replication in S. cerevisiae. It is unclear, however, whether it plays a direct role in DNA replication or affects it indirectly through its ability to regulate Rad53. Furthermore, it is unclear whether the phosphorylation of Mrc1 in response to replication stress is important for activation of Rad53, as might be expected if it were playing an analogous role to Rad9 in Rad53 activation, or whether this phosphorylation regulates Mrc1's role in DNA replication.
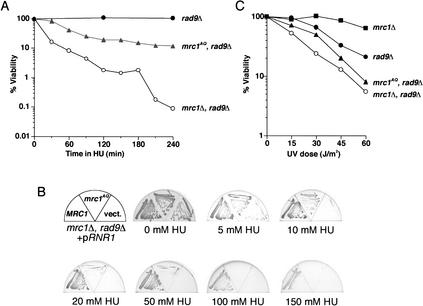
If Mrc1 behaves like Rad9, then eliminating its SQ and TQ motifs should block Rad53 activation. Furthermore, if Mrc1 has a checkpoint-independent role in DNA replication, specifically eliminating its ability to activate Rad53 should leave its replication function intact. Therefore, we eliminated the potential Mec1 phosphorylation sites in Mrc1 by mutating the 17 SQ and TQ motifs to AQ throughout the molecule. This mutant allele, mrc1AQ, along with wild-type MRC1 and vector controls were introduced into mrc1Δ rad9Δ double mutants, and the ability of these cells to respond to replication stress caused by an acute dose of hydroxyurea (HU) was examined. rad9Δ was included in this background to eliminate a back-up pathway that can contribute to survival in the absence of MRC1. Cells containing vector alone were very sensitive to HU killing, displaying 3 logs of killing during the 4-h time course of the experiment, whereas cells carrying MRC1 displayed no loss of viability, as previously determined (Fig. 1A; Weinert and Hartwell 1993; Alcasabas et al. 2001). mrc1AQ mutants were much more resistant to killing than vector alone, but showed a 10-fold greater sensitivity to killing than MRC1 cells.
Figure 1.
Compromised viability of mrc1AQ mutants in the face of replication stress. (A) mrc1Δ rad9Δ cells carrying wild-type MRC1 (Y1133; •), mrc1AQ (Y2296; ▴), or vector alone (Y1131; ○) were arrested in G1 with α-factor, released into YPD containing 200 mM HU at 30°C, and assessed for viability over time. (B) mrc1Δ rad9Δ cells carrying either MRC1 (Y1133), mrc1AQ (Y2296), or vector alone (Y1131) were struck onto YPD plates containing the indicated amounts of HU and grown at 30°C for 3 d. (C) mrc1Δ rad9Δ cells carrying RAD9 (Y1132; ▪), MRC1 (Y1133; •), mrc1AQ (Y2296; ▴), or vector alone (Y1131; ○) were exposed to increasing doses of UV radiation and assessed for viability.
The results of the acute HU stress experiment were mirrored when the cells were examined for their ability to withstand chronic exposure to HU (Fig. 1B). mrc1Δ rad9Δ mutants carrying a wild-type copy of MRC1 are able to grow on plates containing 150 mM HU, but double mutants lacking any allele of MRC1 die with as little as 10 mM HU, similar to rad53Δ cells. Again, mutants carrying the mrc1AQ allele demonstrate an intermediate level of growth in the constant presence of HU.
Thymidine dimers induced by ultraviolet (UV) radiation also induce the replication stress response (Navas et al. 1996). Consequently, we tested the ability of the mrc1AQ allele to mediate survival in response to UV radiation. Similar to what was observed with acute HU exposure, mrc1Δ rad9Δ double-mutant cells carrying the mrc1AQ allele demonstrated reduced viability relative to MRC1 cells (Fig. 1C). Therefore, we conclude that the SQ and TQ residues of Mrc1 are important for maintaining viability when cells are stressed during replication. However, Mrc1 has important functions in response to replication stress that do not depend on SQ/TQ phosphorylation, as the mrc1AQ mutant retains considerable resistance to HU relative to the mrc1-null mutants. It should also be noted that mutants carrying a wild-type RAD9 allele demonstrate the best survival in response to UV radiation. This is likely because Mrc1 may mediate a stress response only to those dimers encountered during replication, whereas Rad9 may be able to respond to UV light throughout the cell cycle (Navas et al. 1996).
mrc1AQ mutants are replication-checkpoint-defective and fail to activate Rad53 in response to replication stress
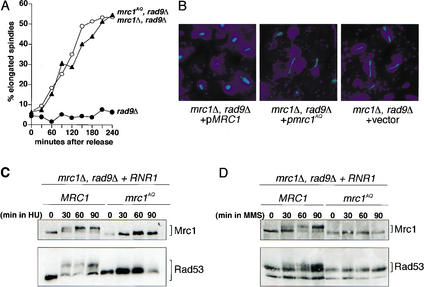
MRC1 is required to prevent mitotic entry in response to replication stress and to arrest cells with short spindles and undivided nuclei (Alcasabas et al. 2001). This is consistent with its ability to regulate Rad53, which is also required for these functions. To determine the extent to which mrc1AQ was capable of mediating the replication checkpoint, we examined the S-phase checkpoint in cells arrested in G1 with α-factor and released into media containing 200 mM HU. MRC1 cells demonstrate an appropriate replication stress response when exposed to HU and maintain short mitotic spindles over a 4-h time course (Fig. 2A,B). In contrast, cells containing either the mrc1Δ or the mrc1AQ allele fail to arrest the cell cycle, and they elongated their spindles with indistinguishable kinetics (Fig. 2A,B). Furthermore, the degree and kinetics of spindle elongation are comparable to those exhibited by rad53Δ cells (Desany et al. 1998).
Figure 2.
mrc1AQ fails to activate Rad53 in response to replication stress. (A) mrc1Δ rad9Δ cells carrying MRC1 (Y1133; •), mrc1AQ (Y2296; ▴), or vector alone (Y1131; ○) were arrested in G1 with α-factor and released into YPD containing 200 mM HU at 30°C. Samples were removed at the indicated times and processed for tubulin staining to assess spindle elongation. (B) Representative fluorescence microscopy images from the experiment described in A. Tubulin staining (green) and DAPI staining (blue) represent the mitotic spindle and chromatin masses, respectively. (C) mrc1Δ rad9Δ cells carrying either MRC1 (Y1133) or mrc1AQ (Y2296) were arrested in G1 with α-factor and released into YPD containing 200 mM HU at 30°C. Samples were removed at the indicated times, and both Rad53 and Mrc1 phosphorylation were assessed by Western blot. (D) mrc1Δ rad9Δ cells carrying either MRC1 (Y1133) or mrc1AQ (Y2296) were arrested in G1 with α-factor and released into YPD containing 0.1% MMS at 30°C. Samples were removed at the indicated times, and both Rad53 and Mrc1 phosphorylation were assessed by Western blot.
We subsequently investigated whether the spindle elongation defect in HU that resulted from the mrc1AQ allele was due to a failure to activate Rad53. As shown in Figure 2C, mrc1Δ rad9Δ double mutants that carry wild-type MRC1 exhibit strong phosphorylation of Mrc1 itself and a robust Rad53 mobility shift when exposed to HU, indicating activation of Rad53. In contrast, mrc1AQ mutants fail to undergo the typical Mrc1 mobility shift and fail to mediate Rad53 activation. Together, the spindle elongation defect and the failure to potentiate Rad53 phosphorylation indicate that the mrc1AQ allele is as defective in mediating the DNA replication stress response as the null allele. Consequently, we have demonstrated a functional link between the SQ and TQ residues of Mrc1 and the activation of the replication stress response.
In addition to responding to replication stress induced by nucleotide depletion, the Mec1–Mrc1–Rad53 pathway also responds to DNA damage from alkylating agents. MRC1 is required for the intra-S-phase checkpoint, which prevents late origin firing in response to MMS treatment and promotes fork stability. We have examined the mrc1AQ mutant and found that it can no longer delay DNA synthesis when treated with MMS (data not shown). Furthermore, in response to MMS, mrc1AQ mutants failed to phosphorylate Mrc1 and failed to activate Rad53 (Fig. 2D). Therefore, we conclude that phosphorylation of Mrc1 is important for both the response to nucleotide depletion and the response to alkylation of DNA. During the course of this work, we constructed several intermediate mrc1 alleles lacking subsets of SQ and TQ sites and observed that no single residue was essential for function. Instead, there appeared to be a gradual decline in Mrc1's checkpoint function the more SQ and TQ residues were absent (data not shown). This suggests that multiple sites are phosphorylated on Mrc1 and contribute to Rad53 activation.
The checkpoint and replication phenotypes of mrc1Δ are separable
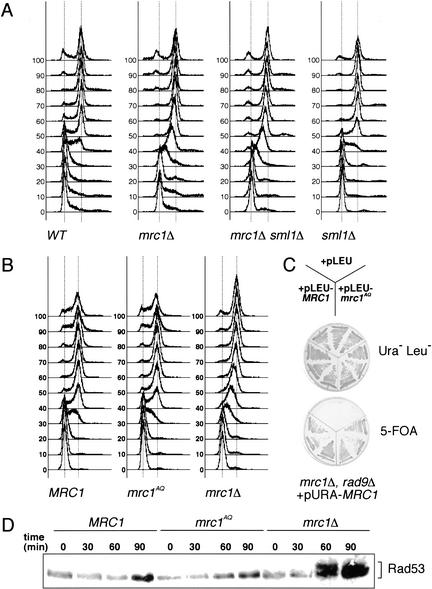
As shown previously, mrc1Δ mutants display two phenotypes (Alcasabas et al. 2001): an inability to activate checkpoint signaling during the S phase, and a slow and damage-inducing S phase. It is possible that Mrc1 promotes replication because it is needed for the Rad53-dependent destruction of Sml1 during S phase. To further define Mrc1's role in replication, we examined S-phase progression in an mrc1Δ sml1Δ mutant. If the slow S phase in mrc1Δ mutants is caused by a failure to degrade Sml1, then a wild-type rate of replication should be restored in mrc1Δ sml1Δ double mutants. The rate of replication in the double mutant, however, is similar to that of mrc1Δ single mutants (Fig. 3A, note the 50-min time point). Thus, elimination of Sml1 does not correct the slow-replication phenotype in the mrc1Δ background, suggesting that this slow S phase is caused by an Rad53-independent role of Mrc1.
Figure 3.
mrc1AQ is competent for DNA replication. (A) The defect of Mrc1 during S phase is not suppressed by deleting SML1. Wild-type (WT; Y2305), mrc1Δ (Y2306), mrc1Δ sml1Δ (Y2307), and sml1Δ (Y2308) cells were arrested in G1 with α-factor and released into YPD at 30°C. Samples were removed at the indicated times and subjected to FACS analysis. The first and second broken vertical lines in each profile represent the 1n and 2n peaks of DNA content, respectively. (B) mrc1AQ mutants undergo a normal S phase. mrc1Δ (Y1127) cells carrying MRC1 (pMRC1), mrc1AQ (pAO138), or vector alone (pRS416) were arrested in G1 with α-factor and released into normal YPD at 30°C. Samples were removed at the indicated times and subjected to FACS analysis. The broken lines represent the peaks of DNA content as in A. (C) mrc1AQ mutants can suppress the lethality of mrc1Δ rad9Δ mutants. mrc1Δ rad9Δ cells carrying MRC1 on a URA3 vector (Y2297) were transformed with MRC1 (pAO122), mrc1AQ (pAO139), or empty LEU2 (pRS416) vectors. The strains containing both URA3 and LEU2 vectors were struck onto 5-FOA plates to select for cells that lost the MRC1-containing URA3 vector. Growth of the strains on plates selecting for both plasmids is shown as a control for the effect of the LEU2 plasmid on viability. (D) mrc1AQ mutants do not induce DNA damage during S phase. The strains used in B were arrested in G1 with α-factor and released into YPD at 30°C. Samples were removed at the indicated times, and Rad53 activation was assessed by mobility shift on a Western blot.
If the role of Mrc1 in replication is not to regulate Rad53, the mrc1AQ allele should maintain the DNA replication function. Therefore, we examined the mrc1AQ allele during a normal unperturbed round of replication. mrc1Δ mutants display a slow progression through the S phase by FACS (Fig. 3B). These cells do not fully replicate their DNA until 80 min after release from G1. In contrast, wild-type and mrc1AQ cells progress normally through S phase, largely completing replication 60 min after release from G1.
mrc1Δ mutant cells incur DNA damage during their normal S phase and require Rad9 for viability (Alcasabas et al. 2001). We hypothesized that this damage creates a requirement for the RAD9 response for survival and that mrc1AQ mutants, which undergo a normal S phase, should not require RAD9 for survival. We examined this using a plasmid shuffle assay and discovered that vector alone could not relieve the requirement of mrc1 rad9 double mutants for the plasmid carrying MRC1 and URA3 (Fig. 3C). However, vectors expressing MRC1 or the mrc1AQ mutant could relieve the requirement, indicating that mrc1AQ mutants do not require RAD9 for survival.
Because of the DNA damage incurred during the S phase, mrc1Δ mutants also exhibit a RAD9-dependent Rad53 phosphorylation during replication. If mrc1AQ mutants fail to become damaged, Rad53 activation should not occur. Unlike mrc1-null alleles, wild-type and mrc1AQ cells exhibit no Rad53 activation (Fig. 3D). Therefore, we conclude that the mrc1AQ mutant represents a separation-of-function allele that is competent for MRC1's replication role but is specifically defective for its replication checkpoint role.
Mrc1 moves with replication forks
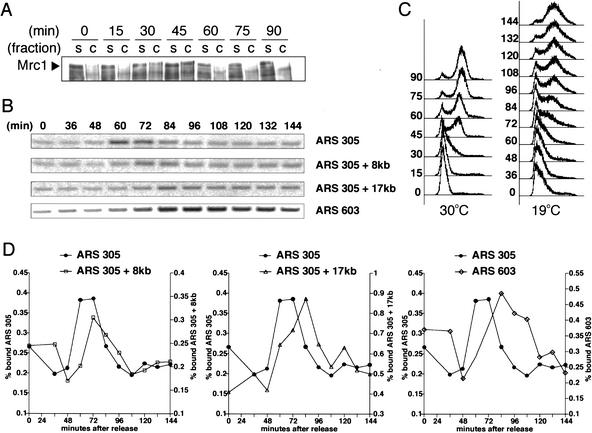
Mrc1's role in DNA replication is not an indirect consequence of its ability to regulate Rad53. Consequently, we hypothesized that Mrc1 plays a direct role in replication, perhaps as a component of the replication complex. To examine Mrc1's S-phase role, we looked at Mrc1's association with chromatin throughout the cell cycle. Mrc1 associates with chromatin as cells begin DNA synthesis at 30 min postrelease and comes off chromatin by 60 min, after DNA synthesis is complete (Fig. 4A). Thus, Mrc1 associates with chromatin during S phase, consistent with a role in DNA replication.
Figure 4.
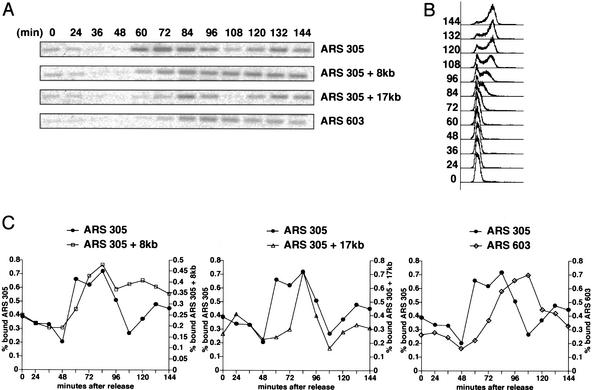
Mrc1 associates with replication forks. (A) Wild-type cells were arrested in G1 with α-factor and released into YPD at 30°C. Samples were removed at the indicated times, and total cellular protein was separated into soluble (s) and insoluble (c) fractions. The presence of Mrc1 in the two fractions was assessed by Western blot. (Right) Cells were also processed for FACS analysis. (B) Cells containing a MYC13-tagged genomic copy of MRC1 (Y1134) were arrested in G1 with α-factor and released into YPD at 19°C. Samples were taken at the indicated times and subjected to formaldehyde cross-linking. The DNA that coimmunoprecipitated with Mrc1–MYC13 was analyzed for the presence of the ARS305 locus, DNA 8 kb and 17 kb centromeric to this origin (ARS 305 + 8 kb and ARS 305 + 17 kb, respectively), and the ARS603 locus by PCR. (C) FACS profiles for cultures at 30°C and 19°C, which were used in A and B, respectively. (D) The PCR products in A were quantified and compared with those obtained from whole-cell DNA to determine the fraction of total DNA brought down in the IP. The percentage bound is represented graphically for ARS305 (•), ARS305 + 8 kb (□), ARS305 + 17 kb (▵), and ARS603 (⋄).
Mrc1 protein could coat all chromatin in an unspecific fashion to facilitate replication. However, if it is associated with the replication fork, it should show specificity in the sequences to which it binds. To examine this issue, we used chromatin immunoprecipitation (ChIP) assays in which proteins are cross-linked to DNA, immunoprecipitated, and analyzed for associated DNA by PCR (Aparicio et al. 1997). Cells expressing Mrc1–Myc were released into the cell cycle at 19°Cto slow fork movement, and samples were prepared at the indicated times and analyzed for Mrc1-associated sequences. Mrc1 showed periodic association with several different sequences. It bound sequences at an early origin, ARS305, with maximal binding at 60 and 72 min after release. At 8 kb away from ARS305, binding peaked at 72 min and declined later than did binding at the origin. Mrc1 associated with regions of the chromosome 17 kb downstream from the origin at 84 min after release. In addition, Mrc1 associates with ARS603, a late firing origin, with significantly delayed kinetics relative to earlier replicating sequences. These experiments demonstrated that, rather than a static association with chromatin, Mrc1 associates with early and late origins of replication in a dynamic fashion similar to that previously observed for Mcm4 and Pol2 (Fig. 4B–D; Aparicio et al. 1997).
Checkpoint activity of Mrc1 is not required for its localization to replication forks
Because the mrc1AQ allele of Mrc1 is able to effect a normal and damage-free S phase, we hypothesized that the mutant protein would travel with replication forks in a fashion similar to that of the wild-type allele and other replication components. We therefore replaced the wild-type allele of Mrc1 in the genome with the mrc1AQ allele and epitope-tagged its Cterminus, with the intention of performing ChIP experiments on this strain. As predicted, the mrc1AQ version of Mrc1 moves with the replication forks in a fashion similar to that of wild-type Mrc1 (Fig. 5).
Figure 5.
mrc1AQ moves with replication forks. (A) Cells containing a MYC13-tagged genomic copy of mrc1AQ (Y2298) were arrested in G1 with α-factor and released into YPD at 19°C. Samples were taken at the indicated times and subjected to formaldehyde cross-linking. DNA that coimmunoprecipitated with mrc1AQ–MYC13 was analyzed for the presence of the ARS305 locus, DNA 8 kb and 17 kb centromeric to this origin (ARS 305 + 8 kb and ARS 305 + 17 kb, respectively), and the ARS603 locus by PCR. (B) FACS profile for the culture. (C) The PCR products in A were quantified and compared with those obtained from whole-cell DNA to determine the fraction of total DNA brought down in the IP. The percentage bound is represented graphically for ARS305 (•), ARS305 + 8 kb (□), ARS305 + 17 kb (▵), and ARS603 (⋄).
Initiation of replication is required for Mrc1's localizing to replication forks
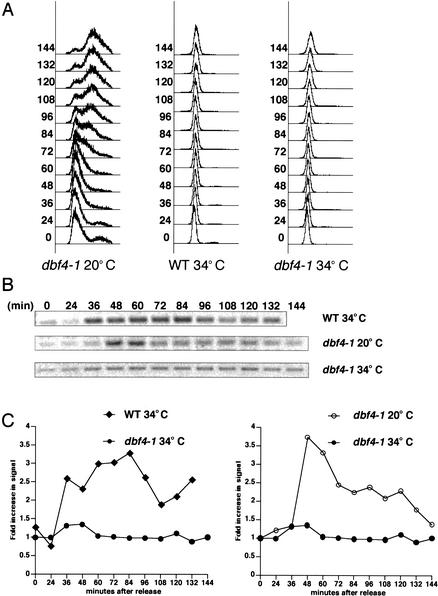
The timing of Mrc1's association with the origins of replication and interorigin regions suggests that Mrc1 is not part of the pre-RC, but rather loads onto the chromatin only once the origins are activated and replication has ensued. If this were true, then blocking initiation of replication should block Mrc1 association. To test this idea, we examined Mrc1's association with an early origin of replication in cells lacking functional Dbf4, a regulatory subunit of the Cdc7 kinase that is critical for origin melting, conversion of the pre-RC into the active replication complex, and replication initiation. G1 wild-type and dbf4 mutant cells were released from a G1 block into media containing 100 mM HU, and ChIP analysis was carried out. Because these experiments cannot be carried out at low temperature owing to the need to inactivate the Ts dbf4 mutant, HU was used to slow down replication so that a kinetic analysis could be carried out at a higher temperature. At the restrictive temperature, wild-type cells showed strong Mrc1 association with the early origin, whereas dbf4-1 cells loaded significantly less Mrc1 onto the early origin of replication (Fig. 6A,B). As a control, Mrc1 is shown to associate with the early origin of replication in dbf4-1 cells at the permissive temperature of 20°C. Further evidence that Mrc1's association with origins is dependent on Dbf4 function is provided by the fact that Mrc1 does not associate with late origins of replication when wild-type cells are treated with HU (data not shown). HU treatment ultimately inactivates Dbf4 kinase before it acts at late origins of replication. Consequently, if Mrc1's origin association depends on Dbf4, we would not expect to see Mrc1's association with late origins in HU when Dbf4 is inactive.
Figure 6.
Replication initiation is required for Mrc1's chromatin association. dbf4-1 cells containing a genomic MRC1–MYC13 construct (Y2299) were arrested in G1 with α-factor and released into YPD at 20°C. Wild-type and dbf4-1 cells with a genomic MRC1–MYC13 construct (Y1134 and Y2299, respectively) were arrested in G1 with α-factor and released into YPD with 100 mM HU at 34°C. Samples were taken at the indicated times and subjected to FACS analysis (A) and to formaldehyde cross-linking (B). DNA coimmunoprecipitating with Mrc1–MYC13 was analyzed for the presence of ARS305 DNA by PCR. (C) The PCR products in B were quantified and compared with those obtained from input DNA to determine the fraction of total DNA brought down in the IP. The fold increase in signal over time from the 0-min and 24-min average baseline is represented for dbf4-1 cells at 34°C(•), wild-type (WT) cells at 34°C(•), and dbf4-1 cells at 20°C(○).
Mec1 localizes to regions of stalled forks
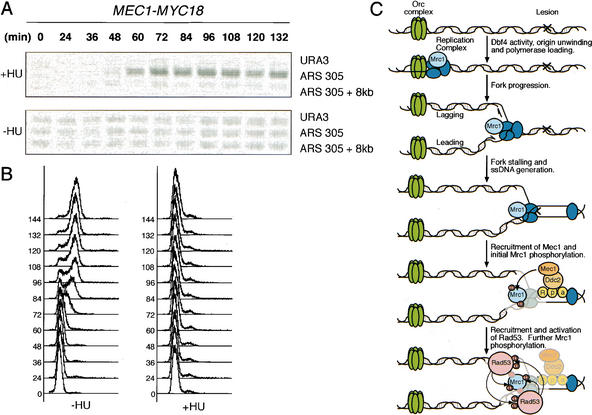
Our results indicate that Mrc1 is a component of the replication fork complex and in response to DNA replication blocks becomes phosphorylated by Mec1 and subsequently by Rad53, which Mrc1 activates. However, it is unknown how Mec1 recognizes Mrc1 in response to stress. It has been demonstrated previously that Mec1 localizes to sites of DNA damage (Kondo et al. 2001; Melo et al. 2001). If our model is correct, Mec1 should be recruited to regions of stalled replication, bringing it into the proximity of Mrc1. To test this, we performed chromatin immunoprecipitations on MEC1–MYC18 cells (a generous gift from M. Longhese, Dipartimento di Biotecnologie e Bioscienze, Universit degli Studi di Milano-Bicocca, Milan, Italy) that had been treated with HU. Mec1 strongly localizes to regions of replication in the presence but not the absence of HU (Fig. 7B). Regions of DNA whose replication is prevented by HU do not bind Mec1 in the presence of HU, showing position specificity of Mec1 recruitment.
Figure 7.
Mec1 is recruited to sites of DNA replication interface. Wild-type cells with a genomic MEC1–MYC18 construct (YLL447.32/1A) were arrested in G1 and released at 19°C into YPD or YPD containing 200 mM HU. Aliquots were removed at the indicated times and subjected to formaldehyde cross-linking (A) and to FACS analysis (B). DNA that associated with Mec1–MYC18 was analyzed for the presence of origin-associated (ARS 305) and nonorigin (ARS 305 + 8 kb, URA3) sequences by PCR. (C) A model for the role of Mec1 and Mrc1 in the activation of Rad53 in response to DNA replication stress. DNA replication begins at origins of replication, which are constitutively bound by the Orc complex. Dbf4 activity promotes the replication complex and Mrc1 to assemble at the origin and begin the task of replication. Once a replication blocking lesion (×) is encountered or deoxyribonucleotides are depleted, the replication complex stalls and abnormal structures are generated. These abnormal structures contain stretches of Rpa-coated single-stranded DNA, which recruits Mec1/Ddc2 kinase to the site of stalled replication (Zou and Elledge 2003). Mec1/Ddc2 then phosphorylates Mrc1, which is uniquely located to transduce the stress signal. This phosphorylation on Mrc1 allows it to mediate the activation of Rad53, which may subsequently further phosphorylate Mrc1.
Discussion
Mrc1 is a mediator of the DNA replication stress response. It is required for the proper activation of Rad53 in response to agents that impede DNA replication such as nucleotide depletion and alkylation damage. However, here we demonstrate that Mrc1 is not merely a transducer of stress information, but is also an active participant in DNA replication itself. Our analysis indicates that Mrc1 loads onto origins and travels along with the replication fork, where it carries out its replication function. The ability of Mrc1 to load onto chromatin is dependent on the activity of proteins required for the activation of DNA replication. Therefore, Mrc1 loading is a late function. It is not clear whether Mrc1 recognizes a particular structure present at forks or whether it associates along with other replication proteins like polymerases. However, as a component of the replication fork complex, Mrc1 is in a perfect position to either sense replication stress, or provide a ready target at the fork for the Mec1/Ddc2 complex. It is likely that in addition to activating Rad53, Mrc1's function at the fork is altered by its phosphorylation, although we have no direct evidence for this. It is important to note that Mrc1 plays an important checkpoint-independent role in maintenance of replication capacity when forks are stalled, because the mrc1AQ mutant is much less sensitive to HU than the mrc1-null mutant. At present, Mrc1 is the only known checkpoint phosphorylation target present at the replication fork. Recently Claspin was shown to associate with chromatin during S phase (Lee et al. 2003). Based on our results, we feel it is likely that Claspin will be localizing to the actual replication fork like Mrc1.
Mrc1 is hyperphosphorylated in response to replication stress. We have shown previously that this phosphorylation is dependent on both the Mec1 and Rad53 kinases (Alcasabas et al. 2001). Thus, two models for Mrc1 phosphorylation exist. The first proposes that Mrc1 is phosphorylated solely by Rad53 once this kinase has been activated in an Mec1-dependent fashion. The second model presents the idea that an initial phosphorylation of Mrc1 by Mec1 allows for the activation of Rad53, which in turn further phosphorylates Mrc1 and causes the resultant mobility shift. Here we show that the kinase responsible for initiating Mrc1 phosphorylation is likely to be Mec1/Ddc2, as mutation of the consensus phosphorylation sites for this kinase, the SQ/TQ motifs, eliminates the mobility shift of Mrc1. Furthermore, elimination of the SQ/TQ motifs renders Mrc1 incapable of activating Rad53 but does not affect Mrc1's ability to localize to replication forks. If the first model above were true, then one would expect to see Rad53 activation in mrc1AQ cells and a subsequent failure of Rad53 to phosphorylate Mrc1, which was not observed. Consequently, we feel that the dependence of Mrc1's mobility shift on Rad53 is likely to be a secondary consequence of Mrc1 activation by Mec1/Ddc2. This is consistent with the Rad3-dependent, Cds1-independent modification of S. pombe Mrc1 in response to replication blocks (Alcasabas et al. 2001).
Our data suggest a model (Fig. 7C) for the function of Mrc1 in activation of the DNA replication checkpoint in S. cerevisiae. As DNA replication is initiated, Mrc1 loads onto active DNA replication forks and travels along with polymerases during DNA replication. At the fork, Mrc1 plays a role that prevents replication problems, possibly replication fork collapse, and the generation of DNA damage. Once the replication fork encounters a blocking lesion on the template DNA, the replication fork stalls. At this point, the abnormal structure of the stalled replication fork recruits Mec1/Ddc2, and this kinase phosphorylates Mrc1 on SQ/TQs, generating a solid-state Rad53 activation machine on the surface of the replication fork. In S. pombe, Mrc1 has been shown to interact with the FHA domain of Cds1, and we believe that Mrc1 phosphorylation likely activates Rad53 either by recruiting Rad53 to Mec1/Ddc2 for direct phosphorylation or by promoting Rad53 autophosphorylation as has been proposed for Rad9 (Sun et al. 1998; Gilbert et al. 2001; Schwartz et al. 2002). Activated Rad53 then further phosphorylates Mrc1, facilitating Mrc1's mobility shift. Rad53 presumably phosphorylates other fork components, contributing to maintenance of fork stability. We and others have observed that Rad53 is capable of phosphorylating SQ/TQ sequences, and we have found that Rad53 is capable of phosphorylating Mrc1 prepared in baculovirus, but this phosphorylation is greatly reduced when the Mrc1AQ protein is used as a substrate (data not shown). Therefore, like Rad9, phosphorylation of Mrc1 is required for its ability to activate Rad53 and mediate the DNA replication stress response. It is possible that once activated, Rad53 phosphorylation of Mrc1 provides additional surfaces for further Rad53 activation, a feed-forward activation loop.
Because Mrc1 is required for the DNA replication checkpoint and DNA replication is required for Mrc1 loading, the regulation of Mrc1 chromatin loading might provide an explanation for why mutants in so many DNA replication proteins are defective for the DNA replication checkpoint in S. cerevisiae. These mutants, such as dbf4, may fail to load Mrc1 onto chromatin so there is less Mrc1 signaling capacity on chromatin when DNA replication is stressed. Thus, the key recipient of DNA replicational stress is localized to replication forks by the replication proteins themselves. It will be interesting to determine what structures or proteins Mrc1 associates with at the fork to determine what exactly it is sensing or regulating during DNA replication.
Materials and methods
Media and growth conditions
Yeast cells were grown at 30°C unless otherwise noted. In the ChIP experiments, dbf4-1 cells and the wild-type controls were grown at 25°C before being shifted to the indicated temperature. Rich and SC media were made as per Kaiser et al. (1994), and the carbon source in all experiments was glucose. In experiments in which cells were arrested in G1, exponentially growing cultures were arrested with α-factor as previously described (Desany et al. 1998).
Strains and plasmids
For a complete list of the strains and plasmids used for this work, see Table 1. pAO138 containing the mrc1AQ allele was created by using Gene Editor (Promega) and Quick Change (Stratagene) site-directed mutagenesis kits to change the SQ and TQ residues in pMRC1 to AQ. The genomic region of pAO138 was sequenced to verify that no extra mutations were created during the mutagenesis process. pAO139 was constructed by subcloning the 5014-bp XhoI fragment from pAO138 into pRS415. pAO122 was constructed by subcloning the MRC1-containing 5014-bp XhoI fragment from pMRC1 into pRS415. Y2298 was constructed in a stepwise fashion. First, a PCR-based method was used to precisely replace the wild-type MRC1 ORF in Y300 with URA3. The resultant strain was transformed with the 4036-bp fragment of pAO138 digested with XhoI and PacI, which contains the genomic sequence of mrc1AQ, and 5-FOA resistant colonies were selected. A PCR-based method was then used to tag the Cterminus of mrc1AQ with the MYC13::his5+ cassette (Longtine et al. 1998). Y2299 was constructed by mating Y2300 with Y2301 and selecting His+, temperature-sensitive spores. Strains Y2305 through Y2308 were derived from spores that were dissected from a cross of Y1127 with Y2309, a version of Y300 that had SML1 replaced by the his5+ cassette (Longtine et al. 1998).
Table 1.
Strains and plasmids used in this study
| Strain | Genotype | Source |
|---|---|---|
| Y300 | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 | Allen et al. 1994 |
| Y1127 | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 mrc1 Δ-3::his5+ | Alcasabas et al. 2001 |
| Y1131 | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 rad9Δ::his5+mrc1Δ-2::HIS3 +pBAD070 | Alcasabas et al. 2001 |
| Y1132 | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 rad9Δ::HIS3 mrc1Δ-2::HIS3 + pBAD070 + YCp50-RAD9 | Alcasabas et al. 2001 |
| Y1133 | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 rad9Δ::HIS3 mrc1Δ-2::HIS3 + pBAD070 + pMRC1 | Alcasabas et al. 2001 |
| Y1134 | MATa trp1-1, ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 HIS::MRC1-MYC13 | Alcasabas et al. 2001 |
| Y2296 | MATa trp1-1, ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 rad9Δ::HIS3 mrc1Δ-2::HIS3 + pBAD070 + pAO138 | This study |
| Y2297 | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 rad9Δ::HIS3 mrc1Δ-2::HIS3 + pMRC1 | This study |
| Y2298 | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 HIS::mrclAQMYC13 | This study |
| Y2299 | MATa trp1-1, ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 HIS::MRC1-MYC13 dbf4-1 | This study |
| Y2300 | MATα trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 HIS::MRC1-MYC13 | This study |
| Y2301 | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 dbf4-1 | This study |
| Y2305 | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 (from Y1127 × Y2309) | This study |
| Y2306 | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 mrc1Δ-3::his5+ (from Y1127 × Y2309) | This study |
| Y2307 | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 mrc1Δ-3::his5+sml1Δ::his5+ (from Y1127 × Y2309) | This study |
| Y2308 | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 sml1Δ::his5+ (from Y1127 × Y2309) | This study |
| Y2309 | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 smllΔ::his5 | This study |
| YLL447.32/1A | MATa trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 MEC1-MYC18::LEU2::mecl | Paciotti et al. 2000 |
| Plasmid | Relevant markers | Source |
|---|---|---|
| pBAD070 | AprTRP1 GAP-RNR1 | Desany et al. 1998 |
| pMRC1 | AprURA3 MRC1 | Alcasabas et al. 2001 |
| YCp50-RAD9 | AprURA3 RAD9 | S. Plon (Baylor College of Medicine, Houston, TX) |
| pAO122 | AprLEU2 MRC1 | This study |
| pAO138 | AprURA3 mrclAQ | This study |
| pAO139 | AprLEU2 mrclAQ | This study |
HU and UV-killing assays
To determine viability in response to acute doses of HU, cells were grown in selective media until they reached log phase. Cells were then arrested in G1 and released into rich media containing 200 mM HU. Aliquots were removed from each culture at the indicated time points, plated on YPD plates, and allowed to grow at 30°C for 3 d. To determine UV sensitivity, the indicated strains were grown to log phase, and an equal number of cells from each culture were spread on YPD plates and irradiated with the indicated dose of UV radiation.
Immunofluorescence for microtubule visualization
To visualize microtubule structures, cells were fixed and stained as described previously (Allen et al. 1994). Briefly, cells were fixed in 5% formaldehyde and washed in EtOH. After being rehydrated in PBS, cells were spheroplasted and stained with the rat monoclonal YOL1/34 anti-tubulin antibody. Cells were then incubated with FITC-conjugated goat anti-rat secondary antibody (Sigma). Cells were briefly stained with 4,6-diamidino-2-phenylindole (DAPI) to visualize DNA.
Western blotting
Samples were prepared for Western blotting using TCA precipitation (Longhese et al. 1997). Precipitated protein was resuspended in 1 M Tris base and SDS loading buffer. Samples were boiled, and solubilized proteins were fractionated by 8% SDS-PAGE, transferred onto Protran membranes (Schleicher and Schuell), and detected using polyclonal α-Mrc1 or α-Rad53 antibodies as described previously (Alcasabas et al. 2001).
Bulk chromatin fractionation
Bulk chromatin fractionation was performed as previously described (Zou and Stillman 1998), with the following exceptions. EDTA-free protease inhibitor tablets (Roche) were used to inhibit protein degradation, and cells were lysed in 0.3% Triton X-100 rather than 1.0%.
ChIP assays
Chromatin immunoprecipitations were performed essentially as described (Strahl-Bolsinger et al. 1997), with the following exceptions. Cells were fixed with 1% formaldehyde at room temperature for 30 min. Following fixing and quenching with 125 mM glycine, cells were washed once with PBS. After lysis, whole lysate was sonicated 4× for 15 sec each, and the sonicated lysate was then clarified by two rounds of centrifugation in an Eppendorf 5417C microcentrifuge at 4°C. Immunoprecipitations were performed on the clarified lysate using Protein A Sepharose beads (Amersham-Pharmacia) and 9E10 monoclonal anti-MYC antibody (Jackson Immunochemicals).
PCR analysis
For PCR analysis, 2/50 of the immunoprecipitated DNA and 1/1000 of the input DNA were used. PCR reactions were carried out in 50-μL reactions using primers at a concentration of 3 μM and Taq polymerase with the appropriate buffer system (Invitrogen). All reactions were for 25 cycles, except for that of ARS603 in MRC1 cells, which was for 27 cycles. PCR products were resolved on 2.5% agarose gels and quantified using NIH Image 1.61 software.
The primers used to determine the presence of certain regions of chromatin were as follows: ARS 305-1 (CTCCGTTTTTAGC CCCCCGTG), ARS 305-2 (GATTGAGGCCACAGCAAGAC CG), ARS 305 + 8 kb-1 (GGTGGTGGAGAAGCGGTTCAA AG), ARS 305 + 8 kb-2 (CCGCTCGTACCCGCTCCTGA), ARS 305 + 17 kb-1 (CAGTTTAGTTGACCCAAGGC), ARS 305 + 17 kb-2 (CAAGTGCCCTCATTGTTCAG), ARS 603-1 (CTCTTT CCCAGATGATATCTAGATGG), ARS 603-2 (CGAGGCTAA ATTAGAATTTTTGAAGTC), URA3-1 (ATCCCTTCCCTTT GCAAATAGTCC), URA3-2 (AAGTAACAAAGGAACCTAG AGGCC).
Acknowledgments
We thank S. Bell, S. Plon, O. Aparicio, O. Cohen-Fix, M. Longhese, M. Tetzlaff, D. Cortez, and A. Alcasabas for helpful discussions and reagents. This work was supported by National Institutes of Health Grant GM44664 to S.J.E. S.J.E. is an Investigator with the Howard Hughes Medical Institute, The Welch Chair of Biochemistry, and Senior Scholar of the Ellison Foundation. A.J.O. was supported by the NIGMS T32 GM08307-11 training grant.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1098303.
References
- Alcasabas A.A., Osborn, A.J., Bachant, J., Hu, F., Werler, P.J., Bousset, K., Furuya, K., Diffley, J.F., Carr, A.M., and Elledge, S.J. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958–965. [DOI] [PubMed] [Google Scholar]
- Allen J.B., Zhou, Z., Siede, W., Friedberg, E.C., and Elledge, S.J. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes & Dev. 8: 2401–2415. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Weinstein, D.M., and Bell, S.P. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell 91: 59–69. [DOI] [PubMed] [Google Scholar]
- Araki H., Leem, S.H., Phongdara, A., and Sugino, A. 1995. Dpb11, which interacts with DNA polymerase II(ε) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl. Acad. Sci. 92: 11791–11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ruiz M.A., Green, C.M., and Lowndes, N.F. 1998. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 17: 2687–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany B.A., Alcasabas, A.A., Bachant, J.B., and Elledge, S.J. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes & Dev. 12: 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A.P., Huang, M., Metzner, S., Botstein, D., Elledge, S.J., and Brown, P.O. 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell 12: 2987–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C.S., Green, C.M., and Lowndes, N.F. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8: 129–136. [DOI] [PubMed] [Google Scholar]
- Huang M., Zhou, Z., and Elledge, S.J. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605. [DOI] [PubMed] [Google Scholar]
- Kaiser C., Michaelis, S., and Mitchell, A. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kamimura Y., Masumoto, H., Sugino, A., and Araki, H. 1998. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell. Biol. 18: 6102–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.T., Lim, D.S., Canman, C.E., and Kastan, M.B. 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274: 37538–37543. [DOI] [PubMed] [Google Scholar]
- Kondo T., Wakayama, T., Naiki, T., Matsumoto, K., and Sugimoto, K. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294: 867–870. [DOI] [PubMed] [Google Scholar]
- Kumagai A. and Dunphy, W.G. 2000. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell 6: 839–849. [DOI] [PubMed] [Google Scholar]
- Lee J., Kumagai, A., and Dunphy, W.G. 2003. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol. Cell 11: 329–340. [DOI] [PubMed] [Google Scholar]
- Longhese M.P., Paciotti, V., Fraschini, R., Zaccarini, R., Plevani, P., and Lucchini, G. 1997. The novel DNA damage checkpoint protein ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 16: 5216–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino, C., Pellicioli, A., Liberi, G., Plevani, P., Muzi-Falconi, M., Newlon, C.S., and Foiani, M. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561. [DOI] [PubMed] [Google Scholar]
- Melo J.A., Cohen, J., and Toczyski, D.P. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes & Dev. 15: 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki T., Kondo, T., Nakada, D., Matsumoto, K., and Sugimoto, K. 2001. Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol. Cell. Biol. 21: 5838–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas T.A., Zhou, Z., and Elledge, S.J. 1995. DNA polymerase ε links the DNA replication machinery to the S phase checkpoint. Cell 80: 29–39. [DOI] [PubMed] [Google Scholar]
- Navas T.A., Sanchez, Y., and Elledge, S.J. 1996. RAD9 and DNA polymerase ε form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes & Dev. 10: 2632–2643. [DOI] [PubMed] [Google Scholar]
- Paciotti V., Clerici, M., Lucchini, G., and Longhese, M.P. 2000. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes & Dev. 14: 2046–2059. [PMC free article] [PubMed] [Google Scholar]
- Rouse J. and Jackson, S.P. 2000. LCD1: An essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 19: 5801–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y., Desany, B.A., Jones, W.J., Liu, Q., Wang, B., and Elledge, S.J. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271: 357–360. [DOI] [PubMed] [Google Scholar]
- Schwartz M.F., Duong, J.F., Sun, Z., Morrow, J.S., Pradhan, D., and Stern, D.F. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9: 1055–1065. [DOI] [PubMed] [Google Scholar]
- Shimada K., Pasero, P., and Gasser, S.M. 2002. ORC and the intra-S-phase checkpoint: A threshold regulates Rad53p activation in S phase. Genes & Dev. 16: 3236–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht, A., Luo, K., and Grunstein, M. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 11: 83–93. [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Ando, S., Shimomura, T., and Matsumoto, K. 1997. Rfc5, a replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol. Cell. Biol. 17: 5905–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Fay, D.S., Marini, F., Foiani, M., and Stern, D.F. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes & Dev. 10: 395–406. [DOI] [PubMed] [Google Scholar]
- Sun Z., Hsiao, J., Fay, D.S., and Stern, D.F. 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281: 272–274. [DOI] [PubMed] [Google Scholar]
- Tanaka K. and Russell, P. 2001. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3: 966–972. [DOI] [PubMed] [Google Scholar]
- Tercero J.A. and Diffley, J.F. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557. [DOI] [PubMed] [Google Scholar]
- Toyn J.H., Johnson, A.L., and Johnston, L.H. 1995. Segregation of unreplicated chromosomes in Saccharomyces cerevisiae reveals a novel G1/M-phase checkpoint. Mol. Cell. Biol. 15: 5312–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialard J.E., Gilbert, C.S., Green, C.M., and Lowndes, N.F. 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17: 5679–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T., Kondo, T., Ando, S., Matsumoto, K., and Sugimoto, K. 2001. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. and Elledge, S.J. 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 96: 3824–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics 160: 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T.A. and Hartwell, L.H. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241: 317–322. [DOI] [PubMed] [Google Scholar]
- ____. 1993. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics 134: 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T.A., Kiser, G.L., and Hartwell, L.H. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes & Dev. 8: 652–665. [DOI] [PubMed] [Google Scholar]
- Weinreich M. and Stillman, B. 1999. Cdc7p–Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 18: 5334–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.B. and Elledge, S.J. 2000. The DNA damage response: Putting checkpoints in perspective. Nature 408: 433–439. [DOI] [PubMed] [Google Scholar]
- Zou L. and Elledge, S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA–ss DNA complexes. Science 300: 1542–1548. [DOI] [PubMed] [Google Scholar]
- Zou L. and Stillman, B. 1998. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science 280: 593–596. [DOI] [PubMed] [Google Scholar]