Abstract
The RecQ DNA helicases human BLM and yeast Sgs1 interact with DNA topoisomerase III and are thought to act on stalled replication forks to maintain genome stability. To gain insight into this mechanism, we previously identified SLX1 and SLX4 as genes that are required for viability and for completion of rDNA replication in the absence of SGS1–TOP3. Here we show that SLX1 and SLX4 encode a heteromeric structure-specific endonuclease. The Slx1–Slx4 nuclease is active on branched DNA substrates, particularly simple-Y, 5′-flap, or replication forkstructures. It cleaves the strand bearing the 5′ nonhomologous arm at the branch junction and generates ligatable nicked products from 5′-flap or replication fork substrates. Slx1 is the founding member of a family of proteins with a predicted URI nuclease domain and PHD-type zinc finger. This subunit displays weakstructure-specific endonuclease activity on its own, is stimulated 500-fold by Slx4, and requires the PHD finger for activity in vitro and in vivo. Both subunits are required in vivo for resistance to DNA damage by methylmethane sulfonate (MMS). We propose that Sgs1–Top3 acts at the termination of rDNA replication to decatenate stalled forks, and, in its absence, Slx1–Slx4 cleaves these stalled forks.
Keywords: Replication restart, RecQ helicase, DNA topoisomerase, endonuclease, Mus81—Mms4
Genome stability in eukaryotes is maintained in part by the RecQ family of DNA helicases (Chakraverty and Hickson 1999). Founded by the RecQ helicase of Escherichia coli, this family includes the human homologs BLM and WRN, Schizosaccharomyces pombe Rqh1, and Saccharomyces cerevisiae Sgs1. Loss of BLM or WRN leads to increased rates of sister-chromatid exchange or variegated translocation mosaicism, respectively, as well as increased sensitivity to certain DNA-damaging agents (Chaganti et al. 1974; Fujiwara et al. 1977; Vijayalaxmi et al. 1983; Salk et al. 1985). In the yeasts, sgs1 or rqh1 mutants display increased rates of recombination, chromosome loss and missegregation, and a decrease in sporulation efficiency. These strains also display hypersensitivity to a variety of DNA-damaging agents including methyl methanesulfonate (MMS), ultraviolet light (UV), and hydroxyurea (HU; Gangloff et al. 1994; Watt et al. 1996; Stewart et al. 1997; Mullen et al. 2000).
In addition to the conserved helicase domain, these RecQ homologs have large N-terminal extensions that are not well conserved. The N-terminal domain of Rqh1, Blm, and Sgs1 physically interacts with its cognate DNA topoisomerase III (Top3; Maftahi et al. 1999; Johnson et al. 2000; Wu et al. 2000; Fricke et al. 2001; Hu et al. 2001). Top3 is a type I 5′-DNA topoisomerase that displays a weak relaxing activity on negatively supercoiled DNA (Kim and Wang 1992). In addition, eukaryotic Top3 can functionally replace bacterial DNA topoisomerase III in the RecQ-dependent catenation of double-stranded DNA molecules (Harmon et al. 1999). This may be significant as E. coli DNA topoisomerase III is noted for its ability to decatenate pBR322 replication intermediates in vitro (DiGate and Marians 1988). Mutations in TOP3 generate a slow-growth or lethal phenotype in the yeast that is suppressed by loss of the RecQ helicase (Gangloff et al. 1994; Maftahi et al. 1999). A variety of biological roles have been proposed for the eukaryotic RecQ helicases, either alone or in conjunction with Top3. These include roles in the termination of DNA replication (Wang 1991; Rothstein and Gangloff 1995), chromosome segregation (Watt et al. 1995), and the restart of stalled replication forks (Stewart et al. 1997; Doe et al. 2000; Rothstein et al. 2000; Kaliraman et al. 2001; Fabre et al. 2002; Bastin-Shanower et al. 2003).
Synthetic-lethal screens have been used to isolate genes that are functionally redundant with SGS1 in S. cerevisiae (Mullen et al. 2001; Tong et al. 2001). We previously identified six nonessential genes required for viability in the absence of SGS1 or TOP3 and placed them into three phenotypic groups: MMS4 and MUS81, SLX1 and SLX4, and SLX5 and SLX8. Biochemical and genetic studies suggested that these groups encode protein complexes required in parallel pathways for maintaining genome stability (Mullen et al. 2001). One of these complexes, Mus81–Mms4, is a structure-specific endonuclease that cleaves 3′-ssDNA or dsDNA branches from duplex DNA (Kaliraman et al. 2001; Bastin-Shanower et al. 2003). The S. pombe homolog of this complex (spMus81–spEme1) behaves similarly in that it is required for viability in the absence of rqh1+. Although it has been suggested that Mus81 is a component of Holliday junction (HJ) resolvase (Boddy et al. 2001; Chen et al. 2001), other preparations of this enzyme preferentially cleave 3′-flap or replication fork (RF) substrates (Kaliraman et al. 2001; Constantinou et al. 2002; Doe et al. 2002). The SGS1–TOP3 and MUS81–MMS4 pathways appear to intersect downstream of homologous recombination because the synthetic-lethal phenotype of sgs1 mus81 mutants is suppressed in the absence of the RAD52 epistasis-group genes (Fabre et al. 2002; Bastin-Shanower et al. 2003). Consistent with this interpretation, it has been proposed that a 3′-flap intermediate, generated during synthesis-dependent strand annealing (SDSA), is a substrate for either Mus81–Mms4 or Sgs1–Top3 (de los Santos et al. 2001; Fabre et al. 2002; Bastin-Shanower et al. 2003).
SLX1–SLX4 is likely to define a pathway distinct from MUS81–MMS4 because the synthetic lethality of sgs1 slx1 or sgs1 slx4 double mutants is not suppressed in a rad52 background (Fabre et al. 2002; Bastin-Shanower et al. 2003). Loss of SLX1 or SLX4 results in no obvious growth or sporulation defects; however, slx1 or slx4 cells containing a temperature-sensitive allele of SGS1 lose viability at the restrictive temperature and exhibit defects in the completion of rDNA replication (Kaliraman and Brill 2002). These results suggested that Slx1–Slx4 and Sgs1–Top3 play overlapping roles at the termination of rDNA replication. The replication of rDNA may be particularly susceptible in these mutants because of the stalling of forks at the rDNA replication fork barrier (RFB; Brewer and Fangman 1988; Kaliraman and Brill 2002; Versini et al. 2003).
Slx1 is the founding member of a family of proteins that includes E. coli YhbQ and Bacillus subtilis YazA (Aravind and Koonin 2001). This family is defined by a UvrC-intron-endonuclease domain (URI), and the eukaryotic members of the family typically contain a C-terminal PHD-type zinc-finger domain (Aasland et al. 1995; Aravind and Koonin 2001). Slx1 coimmunoprecipitates from yeast extracts with Slx4, suggesting that these proteins function in a complex (Mullen et al. 2001). In this study, we show that recombinant Slx1 and Slx4 proteins form a complex that is active in hydrolyzing 5′ branches from duplex DNA. Cleavage occurs specifically at the branchpoint such that RF or 5′-flap substrates are converted into ligatable nicked DNA. The data are consistent with a model in which the Slx1–Slx4 endonuclease cleaves replication forks that cannot be resolved by Sgs1–Top3.
Results
Recombinant Slx1 and Slx4 form a heteromeric complex
Based on their association in yeast extracts, we tested whether recombinant Slx1 and Slx4 formed a stable complex in vitro. SLX1 and SLX4 were coexpressed in E. coli using a strategy previously used to purify recombinant Mus81–Mms4 (Kaliraman et al. 2001). A plasmid, pNJ6750, was constructed in which the SLX1 and SLX4 open reading frames were placed downstream of separate T7 RNA polymerase promoters. In addition, a hexahistidine tag was placed on the N terminus of Slx4 to aid in protein purification. Complementation assays indicated that the epitope-tagged allele of SLX4 was fully functional in yeast (data not shown). Following transformation of pNJ6750 into the appropriate bacterial strain, extracts from induced cells were fractionated by phosphocellulose chromatography, in which Slx1 and Slx4 were observed to coelute at ∼725 mM NaCl. Peak fractions were further purified by Ni-agarose chromatography, in which the Slx1–Slx4 complex eluted at 500 mM imidazole. The purified fraction consisted of the two subunits plus some Slx4 breakdown products, as judged by SDS–polyacrylamide gel electrophoresis (SDS-PAGE; Fig. 1, lane 1). Analysis of this preparation by scanning densitrometry of Coomassie-blue-stained gels indicated that there was a molar excess of Slx1 over Slx4. We conclude that recombinant Slx1 and Slx4 form a soluble complex that is stable even in the presence of high salt concentrations. Hereafter we refer to this preparation as the Slx1–4 complex.
Figure 1.
Purification of the Slx1–Slx4 complex. Approximately 1.9 μg of Slx1–Slx4, 1.4 μg of Slx4, and 0.5 μg of Slx1 were resolved by 10% SDS-PAGE as indicated, and detected by Coomassie blue staining. The Slx1 (36 kD) and Slx4 (130 kD) subunits are indicated. The ladder of low-intensity bands in lanes 1 and 2 is breakdown products of Slx4. Molecular mass standards (in kilodaltons) are indicated on the left.
Endonuclease activity of the Slx1–4 complex
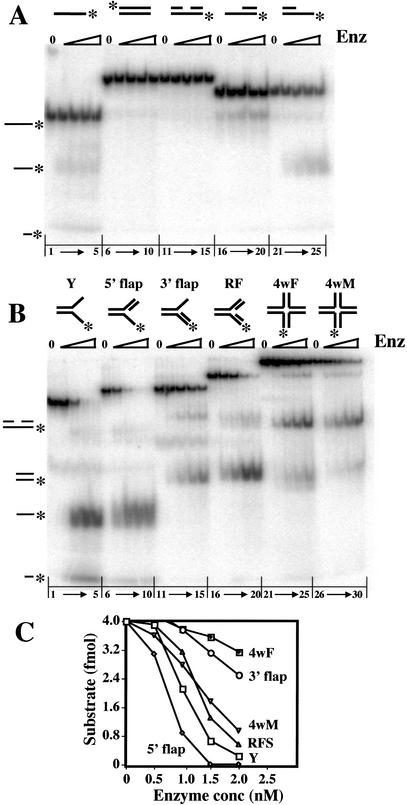
The Slx1–4 complex was incubated with a variety of 5′-[32P]-end-labeled DNA substrates in the presence of Mn2+, and the products were analyzed by native PAGE. As shown in Figure 2A, titrations of Slx1–4 revealed no detectable activity on duplex (Fig. 2A, lanes 6–10), nicked duplex (Fig. 2A, lanes 11–15), or duplex DNA containing a 3′-single-stranded extension (Fig. 2A, lanes 16–20). Low levels of two products migrating at <50 nt were observed when single-stranded DNA (Fig. 2A, lanes 1–5) or duplex DNA containing a 5′-single-stranded extension was used as probe (Fig. 2A, lanes 21–25). This activity was minor, however, when compared with Slx1–4 cleavage of branched DNA substrates. As shown in Figure 2B, Slx1–4 displayed significant, although differing, levels of activity on the following substrates: simple-Y (Y; Fig. 2B, lanes 1–5), 5′ flap (Fig. 2B, lanes 6–10), 3′ flap (Fig. 2B, lanes 11–15), RF (Fig. 2B, lanes 16–20), or a HJ containing a fixed [4wF (4-way fixed); Fig. 2B, lanes 21–25] or mobile [4wM (4-way mobile); Fig. 2B, lanes 26–30] central core. Based on the amount of undigested substrate remaining in each titration, we conclude that a 5′ flap is the preferred substrate for Slx1–4 (Fig. 2C). The Y and RF were also good substrates, although they were not digested to completion as was the 5′ flap. As judged by the migration of their major products, cleavage of the Y, 5′ flap, and RF occurred at or near the branchpoint of the 5′-end-labeled arm. The Y and 5′-flap products also included a fast-migrating band that may be related to Slx1–4 activity on ssDNA as its abundance is substantially reduced using 3′-flap and RF substrates.
Figure 2.
Substrate preferences of the Slx1–4 nuclease. Under standard assay conditions, 4 fmole of the indicated 5′-[32P]-labeled substrate was incubated with 0, 10, 20, 30, or 40 fmole of Slx1–4. The reactions were terminated, and the products were resolved by native 10% PAGE in 1× TBE. (A) Unbranched substrates were assembled from the following oligonucleotides: ssDNA (lanes 1–5), *891; dsDNA (lanes 6–10), *888/896; nicked DNA (lanes 11–15), *891/994/1084; 3′ extension (lanes 16–20), *891/994; and 5′ extension (lanes 16–20), *891/1084. (B) Branched substrates were assembled from the following oligonucleotides: Y (lanes 1–5), 888/*891; 5′ flap (lanes 6–10), 888/*891/992; 3′ flap (lanes 11–15), 888/*891/994; replication fork (lanes 16–20), 888/*891/992/994; fixed HJ (4wF, lanes 21–25), 888/889/890/*891; and mobile HJ (4wM, lanes 26–30), *892/893/894/895. (C) The quantity of undigested substrate remaining in the reactions shown in B was determined and is presented as a function of Slx1–4 concentration. The positions of reaction products are indicated on the left of A and B. An asterisk represents the 5′-[32P] label.
At first glance, the data of Figure 2B could be interpreted to suggest that Slx1–4 digests HJs in a meaningful way. Not only is there significant cleavage activity, especially on the branch-migratable substrate (4wM), but the products appear to migrate at the size expected for nicked duplex DNA. Such a product would be expected from a HJ resolvase like RuvC. Importantly, some models of replication fork restart that invoke a fork regression mechanism make the prediction that cells lacking a RecQ helicase require HJ resolvase for viability (Doe et al. 2000; Boddy et al. 2001). To explore this possibility, we prepared 4wM substrates in which each of the strands was individually labeled, and analyzed their cleavage products by sequencing gel electrophoresis. As shown in Figure 3A, each of the strands was cleaved at multiple sites. By comparison to a chemical sequencing ladder and an oligonucleotide marker of known size, we determined that cleavage occurred almost exclusively within the branch-migratable core of the HJ. These cleavage sites are presented schematically in Figure 3B. It should be noted that in our hands the products of chemical sequencing migrate 1.5–2 nt faster than the corresponding unmodified DNA. For each arm, the sites cluster in the 3′ half of the core; however, there is little evidence of symmetrical cleavage. In fact, several cleavage sites occur outside of the core (e.g., in strands 893 and 895). Another test of authentic resolvase activity is ligation of the cleavage products. Although a control substrate was fully ligated (Fig. 3A, lanes 7,8), no obvious religation of the 4wM cleavage products was observed (Fig. 3A, lanes 9–12). We conclude that the cleavage of HJs by Slx1–4 nuclease is inconsistent with authentic HJ resolvase activity.
Figure 3.
Holliday junction cleavage by Slx1–4. Four substrates were prepared by individually labeling each strand (892, 893, 894, or 895) of a HJ containing a 12-bp migratable core, and incubated with Slx1–4 under standard conditions (lanes 1–4). The digestion products, or a control substrate consisting of nicked duplex DNA (lane 7), were then treated with DNA ligase (lanes 8–12). All samples were subsequently resolved by sequencing gel electrophoresis. A chemical sequencing ladder of oligonucleotide 895 was used as a reference to identify sites of cleavage (lanes 5,6). (B) A schematic diagram of the central region of the HJ is presented summarizing the cleavage results obtained in A. The major (larger arrows) and minor (smaller arrows) sites of cleavage are indicated. The branch-migratable core is shown in gray. R, G + A; Y, T + C.
Mapping the cleavage site of the Slx1–4 endonuclease
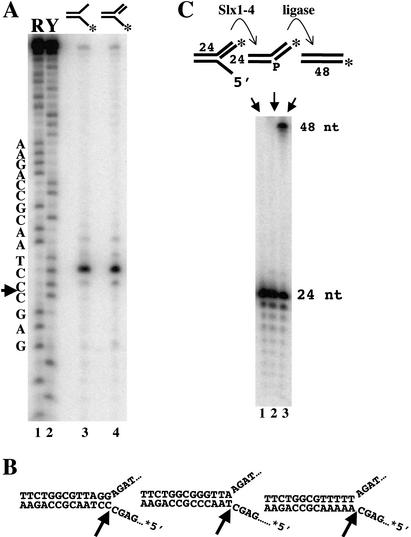
To determine the site of cleavage on Y and 5′-flap substrates, we analyzed their digestion products by sequencing gel electrophoresis. As shown in Figure 4A, these substrates yielded identical products consisting of a single intense band with much weaker bands at ±1 nt. Comparison of this product to a chemical sequencing ladder indicated that cleavage occurred at the sequence 5′-AGC↓CCT-3′. This interpretation places the cleavage site precisely at the junction between the 5′-ssDNA arm of the Y and the duplex region (Fig. 4B, left). Cleavage at this position should yield a 25-nt 5′-end-labeled product, which was confirmed by comparison to an oligonucleotide marker of known size (data not shown; see below). To determine whether the site of cleavage was affected by its DNA sequence, we assayed Y substrates containing alternate sequences in the duplex region adjacent to the junction. As illustrated in Figure 4B, cleavage occurred exclusively at the ssDNA–dsDNA junction regardless of sequence. We conclude that the cleavage site is determined by structure, not DNA sequence.
Figure 4.
5′-flap cleavage by Slx1–4. (A) The indicated Y and 5′-flap substrates were incubated with Slx1–4, and the reaction products were resolved by sequencing gel electrophoresis (lanes 3,4). Comparison to a chemical sequencing ladder of 5′-[32P]-labeled oligonucleotide 891 (lanes 1,2) revealed that hydrolysis occurred mainly at a single site (5′-GAGC↓CCTA-3′) on both substrates. Substrates were assembled as follows: Y, 888/*891; 5′ flap, 888/*891/992. R, G + A; Y, T + C. (B) Schematic diagrams indicate the sites of cleavage on three 5′-flap substrates of varying sequence adjacent to the branch site. (C) The indicated 5′-flap substrate (888/891/*992) was prepared in which the 24-nt downstream oligonucleotide was 32P-end-labeled (lane 1). The substrate was either incubated with Slx1–4 (lane 2) or incubated with Slx1–4 followed by DNA ligase (lane 3). Products were analyzed by sequencing gel electrophoresis.
The above mapping results predict that hydrolysis of a 5′-flap substrate by Slx1–4 should yield a ligatable nicked duplex provided that the cleaved strand retains a phosphate group on its 5′ terminus. To test this prediction, we prepared a 5′-flap substrate in which the 24-nt oligonucleotide adjacent to the junction was labeled on its 5′-end with 32P (Fig. 4C, top). Following treatment with Slx1–4, the product was incubated with DNA ligase and then analyzed by sequencing gel electrophoresis. As shown in Figure 4C, the initial substrate, or substrate plus Slx1–4, yielded the expected 24-nt band. However, treatment of the substrate with Slx1–4 and DNA ligase yielded a 48-nt product. Treatment of the RF substrate with Slx1–4 and DNA ligase similarly yielded a 48-nt product, whereas incubation of the initial substrates with DNA ligase had no effect (data not shown). We conclude that Slx1–4 cleaves 5′-flap and RF structures to produce ligatable nicked DNA.
Nuclease activity of the Slx1 and Slx4 single subunits
To test whether the individual subunits, Slx1 and Slx4, display nuclease activity on their own, we purified them using essentially the same protocol used to purify the heteromer (Fig. 1, lanes 2,3). Increasing amounts of the purified proteins were incubated with a Y substrate, and the products were analyzed by native gel electrophoresis. As shown in Figure 5A, treatment with maximal amounts of Slx1 yielded a single weak band (product a; Fig. 5A, lane 4) that comigrated with the band produced by the Slx1–4 complex (Fig. 5A, lanes 14–16). In contrast, maximal levels of Slx4 yielded several weak and diffuse bands that were distinct from product a (products b–d; Fig. 5A, lane 8). Specifically, Slx4 cleaved the 5′-arm at least twice to create products b and c and cleaved the unlabeled 3′-arm creating product d. Further analysis indicated that product d arose from two cleavage sites, 6 and 8 nt 3′ of the branchpoint (data not shown). Interestingly, the purified Slx1–4 complex appeared to greatly enhance Slx1 activity; densitometric quantitation of this gel revealed that Slx1-type cleavage activity was stimulated 535-fold in the Slx–4 complex compared with that of Slx1 alone. We attempted to reconstitute Slx1–4 activity by preincubating the Slx1 and Slx4 subunits under various conditions, but these treatments revealed no stimulation of Slx1 (Fig. 5A, lanes 9–12). We conclude that Slx1 is responsible for the major cleavage activity of the Slx1–4 complex. It is unclear whether the low levels of smaller Slx4-like products that were observed following treatment of the Y substrate with Slx1–4 (Figs. 2B, lanes 1–5, 5A, lanes 13–16) arose from cleavage of the initial substrate or product a. Although it is a formal possibility that these weak Slx4-specific activities arise from bacterial nucleases contaminating our preparations, we believe this is unlikely. All three proteins (Slx1, Slx4, and Slx1–4) were purified using similar methods, yet they show differing levels of the Slx4-specific activities. Furthermore, the weak Slx4 activities are enhanced on branched DNA substrates (data not shown) and are therefore unlikely to be caused by a random bacterial endonuclease.
Figure 5.
Slx1 and Slx4 subunits are required in vitro and in vivo. (A, left) The Y substrate was incubated with 0, 10, 100, or 1000 fmole of Slx1 (Slx1 lanes), Slx4 (Slx4 lanes), Slx1 and Slx4 that had been preincubated on ice for 30 min (Slx1 + Slx4), or the copurified Slx1–4 complex (Complex). The reaction products were then analyzed by native gel electrophoresis. Cleavage products are indicated by a lowercase letter at the right. (Right) Schematic diagram illustrating the sites of cleavage giving rise to the indicated products. (B) Strains of the indicated genotype were spotted in 10-fold serial dilutions on YPD plates or YPD plates containing the indicated concentrations of MMS. The plates were photographed following 3 d of growth at 30°C.
Mutations in SLX4 have recently been reported to result in sensitivity to MMS (Chang et al. 2002). To investigate their role in DNA damage tolerance, we observed the growth of slx1 and slx4 cells spotted onto solid media containing MMS. Compared to wild-type cells, the growth of slx1 cells was slightly retarded, whereas slx4 mutants showed little or no growth in the presence of 0.012% MMS (Fig. 5B). These phenotypes were specific to the mutations as a centromeric plasmid containing the corresponding gene complemented this growth defect. These strains had previously been found to be MMS-resistant (Mullen et al. 2001). This discrepancy is likely to be caused by the fact that sensitivity is observed only at high concentrations of MMS; little or no effect was seen when cells were plated on media containing one-half this level of MMS (Fig. 5B) or when 0.012% MMS plates older than 1 d were used (data not shown). The sensitivity of an slx1 slx4 double mutant was equal to that of the slx4 strain, indicating that slx4 is epistatic to slx1 (Fig. 5B). Although the mechanism for this epistasis is not obvious from their biochemical activities, the fact that both subunits are required for DNA-damage tolerance is consistent with the idea that Slx1 and Slx4 function as a complex.
Role of the PHD finger in Slx1 function
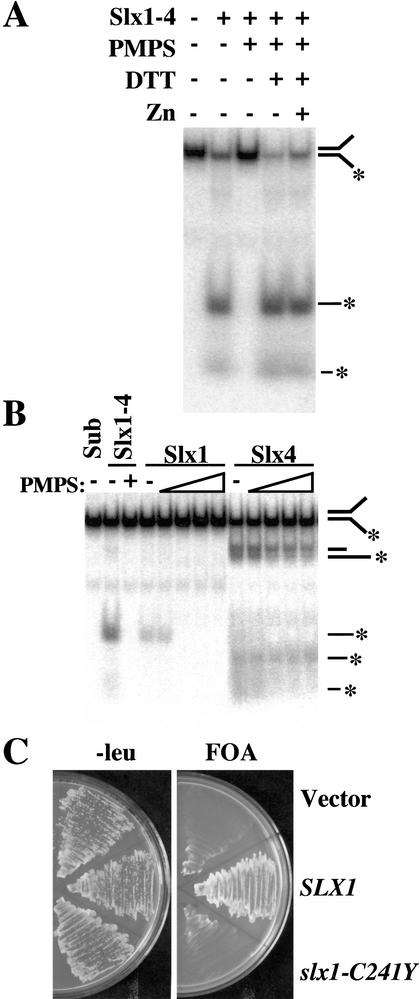
Homologs of Slx1 are found in numerous eukaryotic species, and each contains a putative PHD-type Zn-binding domain (Aravind and Koonin 2001; Mullen et al. 2001). To determine whether this cysteine-rich domain plays a role in Slx1–4 nuclease activity, we tested its sensitivity to p-hydroxymercuriphenylsulfonate (PMPS). PMPS reversibly modifies cysteine residues and has been previously used to inactivate Zn-binding domains found in a number of DNA-binding proteins (Diffley and Stillman 1989; Brill and Bastin-Shanower 1998). Under the conditions of this experiment, Slx1–4 hydrolyzed most of the Y substrate, whereas an equivalent amount of PMPS-treated Slx1–4 was devoid of activity (Fig. 6A). Reactivation of PMPS-modified protein can often be achieved by treatment with a reducing agent, and, indeed, Slx1–4 nuclease activity was completely restored following this treatment (Fig. 6A). The addition of Zn2+ was not required to restore activity, nor did it improve activity. Although these data suggest that Slx1–4 does not require Zn2+ for activity, we cannot rule out the possibility that small amounts of contaminating Zn2+ were present and sufficient to restore activity in this experiment.
Figure 6.
Cysteine residues of Slx1 are required in vitro and in vivo. (A) The Y substrate was incubated with 100 fmole of Slx1–4 that had been subjected to the following treatments as indicated: no treatment, 5 mM PMPS, addition of 10 mM DTT, and addition of 10 μM ZnCl2. The reaction products were then analyzed by native gel electrophoresis. (B) The Y substrate was incubated with 4 pmole of Slx1 or Slx4 that had been treated with 0, 0.2, 0.6, 1.0, 1.4, or 1.8 mM PMPS. Sub, substrate alone; Slx1–4, Slx1–4 complex with or without 0.2 mM PMPS treatment. (C) Strain NJY420 [sgs1-3:TRP1 slx1-11::HIS3 pJM500 (SGS1/URA3)] was transformed with either vector alone (pRS415) or vector containing SLX1 (pKR6129) or slx1-C241Y (pKR6128). Transformants were then streaked on media containing 5-FOA to select against pJM500. Plates were photographed following growth at 30°C for 3 d.
To confirm that the loss of Slx1–4 nuclease activity observed in Figure 6A was due to an effect on Slx1, we tested the effect of PMPS on the individual Slx1 and Slx4 subunits. As expected, Slx1 activity was abolished at 0.6 mM PMPS (Fig. 6B). In contrast, however, Slx4 activity was resistant to PMPS and showed only slight inhibition at the highest level of 1.4 mM PMPS. These results suggest that Slx1–4 nuclease requires unmodified cysteine residues in Slx1 for activity.
To determine whether the PHD domain of Slx1 is functionally important in vivo, we assayed a mutant allele mapping to this domain. Complementation of the synthetic lethality of an sgs1 slx1 strain was tested by transforming plasmid-borne SLX1 alleles into strain JMY420, which carries deletions of SGS1 and SLX1, and a complementing plasmid, pJM500 (SGS1/URA3). Leu+ transformants carrying either the vector alone, or the vector bearing SLX1 or slx1-C241Y, were streaked onto plates containing 5-FOA to select against pJM500. As shown in Figure 6C, SLX1 conferred growth on 5-FOA, whereas the empty vector and slx1-C241Y did not. Taken together, these results indicate that the conserved PHD domain of Slx1 is required for activity in vitro and in vivo.
Discussion
This study demonstrates for the first time that yeast Slx1 exhibits structure-specific endonuclease activity. SLX1 and SLX4 were originally isolated based on their requirement for viability in the absence of SGS1 or TOP3, but their biological function was unknown (Mullen et al. 2001). A clue to their activity was the conservation of an N-terminal URI nuclease domain and a C-terminal PHD-type zinc-finger motif within Slx1 (Aravind and Koonin 2001; Mullen et al. 2001). Based on their coprecipitation from yeast extracts (Mullen et al. 2001), we tested and confirmed that recombinant Slx1 and Slx4 form a complex with a robust structure-specific endonuclease activity. This enhanced activity could not be reconstituted from the individual subunits presumably because of their inability to form a complex in vitro. This behavior is similar to other multisubunit proteins, such as Replication Protein A or Mus81–Mms4, whose subunits must be coexpressed in order for them to form protein complexes (Bochkareva et al. 1998; Kaliraman et al. 2001). Slx1 is likely to be responsible for the endonuclease activity of Slx1–4 complex because both the monomeric and heteromeric forms of Slx1 cleave 5′ flaps from duplex DNA. Furthermore, Slx1 nuclease activity appears to require an intact PHD finger as it was sensitive to cysteine modification in vitro and mutation in vivo. PHD fingers have been identified in proteins of diverse function but are often found in proteins involved in chromatin-mediated transcriptional regulation (Loewith et al. 2000). Thus, it is possible that the PHD domain of Slx1 plays a role in substrate recognition or in mediating the Slx1–Slx4 interaction. Although further experiments will be required to test these possibilities, our results suggest that the Slx1 family defines a set of structure-specific endonucleases.
The function of Slx4 is more puzzling. First, amino acid sequence analysis has identified only one functional domain in Slx4. A putative DNA-binding motif, a SAP domain, is located near the C terminus, at residues 625–659 (Aravind and Koonin 2000). Second, BLAST analysis with S. cerevisiae Slx4 identifies clear homologs only in Saccharomyces species (with scores of E-40 to E-250), suggesting that Slx4 is either unique to this genus or that its sequence evolves rapidly. Interestingly, Saccharomyces castellii Slx4 was found to be only 39% identical (48% conserved) to S. cerevisiae Slx4, whereas S. castellii Slx1 is 54% identical (63% conserved) to S. cerevisiae Slx1. Thus, Slx4 is one of the more rapidly evolving genes in this genus (Cliften et al. 2001), suggesting that more divergent homologs of Slx4 exist in other species. Although Slx4 is required for full Slx1 endonuclease activity in vitro, it must have other biologically important activities because slx4 is epistatic to slx1 with respect to MMS sensitivity. If the only function of Slx4 were to stimulate Slx1 or direct Slx1 to its substrate, perhaps via its SAP domain, one would expect slx1 to be epistatic to slx4. The observed epistasis could be explained by several models. The most obvious possibility is that Slx4 has a function apart from Slx1. This might be a completely novel activity, or Slx4 might serve to stimulate nucleases other than Slx1. Alternatively, Slx1 and Slx4 may be part of a larger complex that is destabilized more by the loss of Slx4 than by the loss of Slx1. Another possibility is that the weak endonuclease activities of Slx4 are more important to the function of the Slx1–4 complex than is the 5′-flap cleavage by Slx1. If Slx4 has roles that are distinct from Slx1 and involve other endonucleases or recombination proteins, then these could be identified biochemically by purifying Slx4 directly from yeast.
Slx1–4 is capable of digesting HJs in vitro; however, we doubt that this activity is biologically relevant. Conventional HJ resolvases, like RuvC or the yeast mitochondrial resolvase Cce1, cleave HJs symmetrically to produce ligatable nicked DNA. Slx1–4 does not cleave HJs symmetrically and does not produce ligatable nicks. Instead, Slx1–4 digests the branch-migratable region of the HJ at multiple sites with a distinct preference for the 3′ side of the branch-migratable core. It has not escaped our notice that this pattern of HJ cleavage is reminiscent of that obtained with the 3′-flap endonuclease, Mus81–Mms4 (Kaliraman et al. 2001). It is particularly revealing that the Mus81-associated endonucleases cleave the branch-migratable region of a HJ at multiple sites with a distinct preference for the 5′ side of the core (Boddy et al. 2001; Chen et al. 2001; Constantinou et al. 2002). This contrast in HJ cleavage preference may correspond to the enzymes' opposite polarities on flaps. For example, flap-specific endonucleases may bind the arms of a HJ at the junction between the fixed sequence and the branch-migratable core because breathing of the core transiently resembles a Y structure. If so, then cleavage consistent with the enzymes' known polarity would result in the observed preference for the 5′ or 3′ side of the core. Such cleavage activity would not be expected to be symmetric because the HJ arms are recognized independently by the flap endonucleases. Finally, if Slx1–4 functioned as a HJ endonuclease in vivo, then one would expect a reduction or elimination of meiotic crossing-over in mutant diploids. Genetic mapping studies indicate that the map distance in each of three intervals is unchanged in slx1/slx1 diploids (data not shown).
What, therefore, is the role of Slx1–4 in the cell, and why does it functionally overlap with Sgs1–Top3? We speculate that eukaryotic cells require these enzymes in response to stalled or converging replication forks. It is generally accepted that converging forks are fully replicated to produce multiply intertwined molecules that are decatenated by DNA topoisomerase II (Sundin and Varshavsky 1981; DiNardo et al. 1984). However, converging forks may arrest prematurely, for example, because of DNA damage, preventing decatenation by Top2. As previously proposed (Wang 1991; Rothstein and Gangloff 1995), the concerted action of a RecQ helicase, like Sgs1, and the nicking-closing activity of Top3 may untwine the parental DNA between the arrested forks to allow subsequent “filling-in” by a DNA polymerase (Fig. 7, left). Precedent for this kind of mechanism comes from studies of DNA topoisomerase III of E. coli, which decatenates replicating pBR322 on its own or catenates dsDNA circles in combination with RecQ (DiGate and Marians 1988; Harmon et al. 1999). As illustrated in Figure 7, the Slx1–4 endonuclease is required in the absence of Sgs1–Top3 or in response to a large number of stalled forks. For simplicity, it is proposed that these enzymes act on the same substrate, with Slx1–4 cleaving the 5′-arms of the converging forks (Fig. 7, right) to produce one doubly nicked chromatid and a broken sister chromatid with 3′ overhangs. This double strand break (DSB) would then be repaired by homologous recombination.
Figure 7.
Model of Sgs1–Top3 and Slx1–4 at the termination of rDNA replication. (Left) When replication forks converge and stall, Sgs1–Top3 helicase–topoisomerase separates the parental strands of the DNA template, allowing replication to finish by a DNA polymerase filling-in reaction (broken lines). (Right) In the absence of Sgs1–Top3, Slx1–4 cleaves the 5′ arm of each replication fork, creating a doubly nicked chromatid and a broken sister chromatid with 3′ overhangs. At the rDNA, this DSB can be repaired by RAD52-independent single-strand annealing (SSA). Arrowheads, 3′-ends.
This model is in agreement with the following experimental results. First, it is consistent with the idea that Slx1–4 and Sgs1–Top3 interact upstream of homologous recombination. Previous studies indicated that Sgs1–Top3 and Mus81–Mms4 interact downstream of the initiation of recombination because sgs1 mus81 synthetic lethality is suppressed in strains lacking homologous recombination (i.e., sgs1 mus81 rad52 strains are viable; Fabre et al. 2002; Bastin-Shanower et al. 2003). In contrast, mutations in RAD52 failed to suppress the synthetic lethality of slx1 sgs1 or slx4 sgs1 strains, implying (1) that Sgs1–Top3 has more than one role in the cell, and (2) that homologous recombination, if required, is needed downstream of the Slx1–4/Sgs1–Top3 interaction. Second, the model of Figure 7 proposes a specific role for Top3 consistent with the synthetic lethality of slx1 top3 strains and the known physical interaction between Sgs1 and Top3. Finally, the redundancy of Slx1–4 and Sgs1–Top3 at the termination of DNA replication explains their synthetic effect on rDNA replication. It has been shown previously that slx4 strains carrying temperature-sensitive alleles of SGS1 replicate the bulk of their DNA normally at the nonpermissive temperature. However, the completion of rDNA replication, as judged by its ability to migrate in pulsed-field gel electrophoresis, is inhibited under these conditions (Kaliraman and Brill 2002). Indeed, recent studies using DNA combing confirm that the completion of rDNA replication is retarded in SGS1 mutants (Versini et al. 2003). The specificity of this replication defect was attributed to the rDNA replication fork barrier (RFB), which prohibits forks from moving in the direction opposite to RNA polymerase I (Brewer and Fangman 1988; Kaliraman and Brill 2002; Versini et al. 2003). Presumably, the large number of forks that stall at the RFB each S-phase makes this locus highly dependent on these redundant mechanisms.
A challenge to this model is that cells lacking SGS1–TOP3 would be expected to require not only SLX1–SLX4 for viability, but the RAD52 pathway of homologous recombination as well. Given that sgs1 rad52 cells are viable with no obvious synthetic effects (Fabre et al. 2002; Bastin-Shanower et al. 2003), this model cannot rely on RAD52-dependent DSB repair. It is known, however, that the single-strand annealing (SSA) pathway of homologous recombination is independent of RAD51, RAD54, RAD55, and RAD57 when breaks occur between large regions of homology (Ivanov et al. 1996) and is independent of RAD52 when breaks occur in the rDNA (Ozenberger and Roeder 1991). Therefore, Slx1–4 may be restricted to the nucleolus, where it would be dedicated to the repair of forks stalled at the rDNA. Such a mechanism could explain the high rates of recombination at the rDNA in sgs1 and top3 strains (Gangloff et al. 1994; Mullen et al. 2000), while predicting a loss of rDNA repeats in these strains.
S. cerevisiae contains two other 5′-branch endonucleases, Rad27/FEN1 and Rad2/XPG, that are required for Okasaki fragment processing and nucleotide excision repair, respectively. Although, preliminary in vitro experiments indicate that Slx1–4 shows no preference for the double-flap structures preferred by Rad27 (Kao et al. 2002; data not shown), it will be interesting to test for potential overlap between these nucleases and Slx1–4.
Materials and methods
Strains and plasmids
The S. cerevisiae strains used in this study are isogenic derivatives of W303–1a (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100; Thomas and Rothstein 1989). Strain construction, growth, and transformation were performed using standard procedures (Rose et al. 1990). Plasmid pKR6128 contains the slx1-2 allele (C241Y) on a BamHI/SalI fragment ligated into pRS415(Sikorski and Hieter 1989). This allele was created by site-directed PCR mutagenesis and changing residue 722 from A to T. Plasmid pKR6129 contains the wild-type SLX1 gene in plasmid pRS415. Strain JMY420 [MATa ade2-1 ura3-1 his3-11,15, trp1-1 leu2-3,112 can1-100 sgs1-3::TRP1 slx1-11::HIS3 pJM500 (SGS1:URA3:ADE3)] was transformed with plasmids pRS415, pKR6128, and pKR6129 to test for complementation of sgs1 slx1 synthetic lethality. We note that both wild-type SLX1 alleles used in this study (from either W303 or a genomic library of unknown origin) encode proteins with a single amino acid difference (isoleucine in place of methionine at position 266) from the Saccharomyces Genome Database (Mullen et al. 2001).
For expression of Slx1 and Slx4 individually, SLX1 and SLX4 were subcloned into the pET28a expression vector, creating plasmids pNJ6124 and pNJ6408, respectively. To express the dimeric Slx1–4 endonuclease, a bicistonic plasmid expressing both His6-Slx4 and Slx1 was created essentially as described for Mms4–Mus81 (Kaliraman et al. 2001). Specifically, SLX1 was subcloned into the pET11a expression vector, creating pJM6110. The SLX4 expression cassette from pNJ6408 was then subcloned into pJM6110 to create pNJ6125.
Expression and purification of recombinant proteins
The expression and purification of His6-Slx1, His6-Slx4, and Slx1–(His6)Slx4 was carried out essentially as described for Mus81–(His6)Mms4 (Kaliraman et al. 2001). Plasmids pNJ6124 (His6-Slx1), pNJ6408 (His6-Slx4), and pNJ6125[Slx1–(His6)Slx4] were transformed into E. coli BL21-RIL cells and grown in 2 L of LB medium containing the appropriate antibiotic at 37°C until OD595 = 0.4. Cells were chilled to 16°C and induced with 0.1 mM IPTG for 16 h. The following steps took place at 0°C or 4°C: Cells were pelleted and resuspended in Buffer B (25 mM HEPES, 1 mM EDTA, 0.01% NP-40, 10% glycerol, 0.1 mM PMSF, 1 mM DTT) plus 300 mM NaCl, sonicated three times for 1 min, frozen, and thawed, and then centrifuged at 26,900g for 30 min. The supernatant was taken as extract.
For all proteins, the extract was diluted to Buffer B containing 150 mM NaCl and loaded onto a 20-mL phosphocellulose column. The column was washed with 3 column volumes of Buffer B containing 150 mM NaCl, and a 6-column-volume gradient from 150 to 1000 mM NaCl was applied in Buffer B lacking DTT and EDTA. Peak protein fractions were identified by SDS-PAGE and Coomassie blue staining. His6-Slx1 eluted from the phosphocellulose column at ∼900 mM NaCl, His6-Slx4 eluted at 600 mM NaCl, and Slx1–(His6)Slx4 eluted at 725 mM NaCl. The peak fractions were pooled, bound to Ni-Probond resin (Invitrogen) in the presence of 10 mM imidazole, and then washed with 10 column volumes of Buffer N (25 mM Tris-HCl at pH 7.5, 10% glycerol, 500 mM NaCl, 0.01% NP-40, and 0.1 mM PMSF) plus 10 mM imidazole. The column was developed with 7 column volumes of Buffer N containing 50 mM imidazole, 7 column volumes of Buffer N containing 100 mM imidazole, and 4 column volumes of Buffer N containing 500 mM imidazole. His6-Slx1 and Slx1–(His6)Slx4 eluted at 500 mM imidazole. His6-Slx4 eluted at 50 and 100 mM imidazole, although the 100 mM imidazole fractions were purer. Purified proteins were dialyzed and stored at -80°C in Buffer A (25 mM Tris-HCl at pH 7.5, 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 0.01% NP-40) plus 50 mM NaCl.
Nuclease assays
Assays were performed essentially as described (Kaliraman et al. 2001). All reactions (20 μL) contained 4 fmole of probe in 25 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM DTT, 1 mM MnCl2, 0.01% NP-40, and 10% glycerol, and were incubated at 30°C for 30 min. Reaction products were resolved by native (1× TBE, 10%) or denaturing (7 M urea, 10%) PAGE and analyzed by a phosphorimager. IPLab gel densitometry software was used to quantify band intensities. Chemical sequencing ladders were prepared as described (Maxam and Gilbert 1980). PMPS treatment was carried out as described (Brill and Bastin-Shanower 1998) except that inhibition was reversed by dialysis in Buffer A plus 50 mM NaCl and 10 mM DTT.
DNA substrates
DNA substrates were constructed from 5′-[32P]-end-labeled oligonucleotides that were annealed and purified as described (White and Lilley 1996). Oligonucleotide sequences were as previously described (Kaliraman et al. 2001) except for 889 (CT TAAGCCGAAGCTTATCGGTATCTTGCTTACGACGCTA GCAAGTGATC) and 890 (TGATCACTTGCTAGCGTCGTA AGCAGCTCGTGCTGTCTAGAGACATCGA). Sequences for the HJ with a 12-bp branch-migratable core (892/893/894/895) are taken from Connolly et al. (1991).
Acknowledgments
We thank Val Carabetta for technical assistance; Robert Bambara for proteins; Grant Brown for sharing results prior to publication; and Paul Cliften, Mark Johnston, and the Washington University Genome Sequencing Center for providing Saccharomyces genome sequence information. This work was supported by grant AG16637 from the NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1105203.
References
- Aasland R., Gibson, T.J., and Stewart, A.F. 1995. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20: 56–59. [DOI] [PubMed] [Google Scholar]
- Aravind L. and Koonin, E.V. 2000. SAP—A putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25: 112–114. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 11: 1365–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin-Shanower S.A., Fricke, W.M., Mullen, J.R., and Brill, S.J. 2003. Mechanism of Mus81–Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1–Rad10. Mol. Cell. Biol. 23: 3487–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E., Frappier, L., Edwards, A.M., and Bochkarev, A. 1998. The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J. Biol. Chem. 273: 3932–3936. [DOI] [PubMed] [Google Scholar]
- Boddy M.N., Gaillard, P.H., McDonald, W.H., Shanahan, P., Yates III, J.R., and Russell, P. 2001. Mus81–Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman, W.L. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643. [DOI] [PubMed] [Google Scholar]
- Brill S.J. and Bastin-Shanower, S. 1998. Identification and characterization of the fourth single-stranded-DNA binding domain of Replication Protein A. Mol. Cell. Biol. 18: 7225–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti R.S., Schonberg, S., and German, J. 1974. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl. Acad. Sci. 71: 4508–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraverty R.K. and Hickson, I.D. 1999. Defending genome integrity during DNA replication: A proposed role for RecQ family helicases. Bioessays 21: 286–294. [DOI] [PubMed] [Google Scholar]
- Chang M., Bellaoui, M., Boone, C., and Brown, G.W. 2002. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl. Acad. Sci. 99: 16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.B., Melchionna, R., Denis, C.M., Gaillard, P.H., Blasina, A., Van de Weyer, I., Boddy, M.N., Russell, P., Vialard, J., and McGowan, C.H. 2001. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8: 1117–1127. [DOI] [PubMed] [Google Scholar]
- Cliften P.F., Hillier, L.W., Fulton, L., Graves, T., Miner, T., Gish, W.R., Waterston, R.H., and Johnston, M. 2001. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 11: 1175–1186. [DOI] [PubMed] [Google Scholar]
- Connolly B., Parsons, C.A., Benson, F.E., Dunderdale, H.J., Sharples, G.J., Lloyd, R.G., and West, S.C. 1991. Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc. Natl. Acad. Sci. 88: 6063–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou A., Chen, X.B., McGowan, C.H., and West, S.C. 2002. Holliday junction resolution in human cells: Two junction endonucleases with distinct substrate specificities. EMBO J. 21: 5577–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T., Loidl, J., Larkin, B., and Hollingsworth, N.M. 2001. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159: 1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F. and Stillman, B. 1989. Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science 246: 1034–1038. [DOI] [PubMed] [Google Scholar]
- DiGate R.J. and Marians, K.J. 1988. Identification of a potent decatenating enzyme from Escherichia coli. J. Biol. Chem. 263: 13366–13373. [PubMed] [Google Scholar]
- DiNardo S., Voelkel, K., and Sternglanz, R. 1984. DNA topoisomerase II mutant of Saccharomyces cerevisiae: Topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc. Natl. Acad. Sci. 81: 2616–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C.L., Dixon, J., Osman, F., and Whitby, M.C. 2000. Partial suppression of the fission yeast rqh1- phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 19: 2751–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C.L., Ahn, J.S., Dixon, J., and Whitby, M.C. 2002. Mus81–Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 25: 25. [DOI] [PubMed] [Google Scholar]
- Fabre F., Chan, A., Heyer, W.D., and Gangloff, S. 2002. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. 99: 16887–16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke W.M., Kaliraman, V., and Brill, S.J. 2001. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem. 276: 8848–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y., Higashikawa, T., and Tatsumi, M. 1977. A retarded rate of DNA replication and normal level of DNA repair in Werner's syndrome fibroblasts in culture. J. Cell Physiol. 92: 365–374. [DOI] [PubMed] [Google Scholar]
- Gangloff S., McDonald, J.P., Bendixen, C., Arthur, L., and Rothstein, R. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14: 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon F.G., DiGate, R.J., and Kowalczykowski, S.C. 1999. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: A conserved mechanism for control of DNA recombination. Mol. Cell 3: 611–620. [DOI] [PubMed] [Google Scholar]
- Hu P., Beresten, S.F., van Brabant, A.J., Ye, T.Z., Pandolfi, P.P., Johnson, F.B., Guarente, L., and Ellis, N.A. 2001. Evidence for BLM and topoisomerase III α interaction in genomic stability. Hum. Mol. Genet. 10: 1287–1298. [DOI] [PubMed] [Google Scholar]
- Ivanov E.L., Sugawara, N., Fishman-Lobell, J., and Haber, J.E. 1996. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F.B., Lombard, D.B., Neff, N.F., Mastrangelo, M.A., Dewolf, W., Ellis, N.A., Marciniak, R.A., Yin, Y., Jaenisch, R., and Guarente, L. 2000. Association of the Bloom syndrome protein with topoisomerase IIIα in somatic and meiotic cells. Cancer Res. 60: 1162–1167. [PubMed] [Google Scholar]
- Kaliraman V. and Brill, S.J. 2002. Role of SGS1 and SLX4 in maintaining rDNA structure in Saccharomyces cerevisiae. Curr. Genet. 41: 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman V., Mullen, J.R., Fricke, W.M., Bastin-Shanower, S.A., and Brill, S.J. 2001. Functional overlap between Sgs1–Top3 and the Mms4–Mus81 endonuclease. Genes & Dev. 15: 2730–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao H.I., Henricksen, L.A., Liu, Y., and Bambara, R.A. 2002. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 277: 14379–14389. [DOI] [PubMed] [Google Scholar]
- Kim R.A. and Wang, J.C. 1992. Identification of the yeast TOP3 gene product as a single strand-specific DNA topoisomerase. J. Biol. Chem. 267: 17178–17185. [PubMed] [Google Scholar]
- Loewith R., Meijer, M., Lees-Miller, S.P., Riabowol, K., and Young, D. 2000. Three yeast proteins related to the human candidate tumor suppressor p33ING1 are associated with histone acetyltransferase activities. Mol. Cell. Biol. 20: 3807–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maftahi M., Han, C.S., Langston, L.D., Hope, J.C., Zigouras, N., and Freyer, G.A. 1999. The top3+ gene is essential in Schizosaccharomyces pombe and the lethality associated with its loss is caused by Rad12 helicase activity. Nucleic Acids Res. 27: 4715–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A.M. and Gilbert, W. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65: 499–560. [DOI] [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman, V., and Brill, S.J. 2000. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154: 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman, V., Ibrahim, S.S., and Brill, S.J. 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozenberger B.A. and Roeder, G.S. 1991. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol. Cell. Biol. 11: 1222–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D., Winston, F., and Hieter, P. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Rothstein R. and Gangloff, S. 1995. Hyper-recombination and Bloom's syndrome: Microbes again provide clues about cancer. Genome Res. 5: 421–426. [DOI] [PubMed] [Google Scholar]
- Rothstein R., Michel, B., and Gangloff, S. 2000. Replication fork pausing and recombination or “gimme a break.” Genes & Dev. 14: 1–10. [PubMed] [Google Scholar]
- Salk D., Au, K., Hoehn, H., and Martin, G.M. 1985. Cytogenetic aspects of Werner syndrome. Adv. Exp. Med. Biol. 190: 541–546. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter, P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E., Chapman, C.R., Al-Khodairy, F., Carr, A.M., and Enoch, T. 1997. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16: 2682–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin O. and Varshavsky, A. 1981. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: Dissection of the final stages of SV40 DNA replication. Cell 25: 659–669. [DOI] [PubMed] [Google Scholar]
- Thomas B.J. and Rothstein, R. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630. [DOI] [PubMed] [Google Scholar]
- Tong A.H., Evangelista, M., Parsons, A.B., Xu, H., Bader, G.D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C.W., Bussey, H., et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Versini G., Comet, I., Wu, M., Hoopes, L., Schwob, E., and Pasero, P. 2003. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 22: 1939–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayalaxmi, Evans, H.J., Ray, J.H., and German, J. 1983. Bloom's syndrome: Evidence for an increased mutation frequency in vivo. Science 221: 851–853. [DOI] [PubMed] [Google Scholar]
- Wang J.C. 1991. DNA topoisomerases: Why so many? J. Biol. Chem. 266: 6659–6662. [PubMed] [Google Scholar]
- Watt P.M., Louis, E.J., Borts, R.H., and Hickson, I.D. 1995. Sgs1: A eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81: 253–260. [DOI] [PubMed] [Google Scholar]
- Watt P.M., Hickson, I.D., Borts, R.H., and Louis, E.J. 1996. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.F. and Lilley, D.M. 1996. The structure-selectivity and sequence-preference of the junction-resolving enzyme CCE1 of Saccharomyces cerevisiae. J. Mol. Biol. 257: 330–341. [DOI] [PubMed] [Google Scholar]
- Wu L., Davies, S.L., North, P.S., Goulaouic, H., Riou, J.F., Turley, H., Gatter, K.C., and Hickson, I.D. 2000. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 275: 9636–9644. [DOI] [PubMed] [Google Scholar]