Abstract
Telomerase contains an essential RNA, which includes the template sequence copied by the reverse transcription action of telomerase into telomeric DNA. Using phylogenetic comparison, we identified seven conserved sequences in telomerase RNAs from Kluyveromyces budding yeasts. We show that two of these sequences, CS3 and CS4, are essential for normal telomerase function and can base-pair to form a putative long-range pseudoknot. Disrupting this base-pairing was deleterious to cell growth, telomere maintenance, and telomerase activity. Restoration of the base-pairing potential alleviated these phenotypes. Mutating this pseudoknot caused a novel mode of shifting of the boundaries of the RNA template sequence copied by telomerase. A phylogenetically derived model of yeast TER structure indicates that these RNAs can form two alternative predicted core conformations of similar stability: one brings the CS3/CS4 pseudoknot spatially close to the template; in the other, CS3 and CS4 move apart and the conformation of the template is altered. We propose that such disruption of the pseudoknot, and potentially the predicted telomerase RNA conformation, affects polymerization to cause the observed shifts in template usage.
Keywords: telomerase, yeast telomerase RNA; telomerase RNA pseudoknot; telomerase RNA secondary structure; telomerase template miscopying
Telomerase adds short DNA repeats onto eukaryotic chromosome ends, compensating for losses caused by incomplete replication or degradation (Blackburn 1994). The essential core components of this specialized reverse transcriptase are the telomerase RNA (TER) and a catalytic protein reverse transcriptase (TERT; Blackburn 2000). A short templating sequence in TER is repeatedly copied by TERT onto the telomere 3′-ends. Telomerase RNAs, which are much longer than their templating domain sequences, appear to share a common core secondary structure, consisting of the template region and a pseudoknot, in ciliates and vertebrates (for review, see Blackburn 2000; Comolli et al. 2002; Chen and Greider 2003; Theimer et al. 2003). Mutagenesis experiments have identified several functionally important bases in the nontemplate regions of several TERs (for review, see Blackburn 2000).
Because addition of a mutated telomeric sequence can have deleterious consequences for the cell, synthesis of the correct DNA sequence onto telomeres by telomerase is critical in vivo. Template copying starts near the 3′-end of the templating domain sequence, and proceeds to its 5′ boundary. The templating domain includes the template bases that are copied, plus a short sequence at its 3′-end (a repeat of the RNA sequence immediately preceding the 5′ boundary of the template), which is used to align the DNA primer to be elongated but is not itself normally copied. In the budding yeast Kluyveromyces lactis, the 5′ boundary of the template is normally demarcated by a TER helix (here called Helix H1; see below) located immediately 5′ of the template (Tzfati et al. 2000). The structure, but not the sequence, of Helix H1 is important for this function (Tzfati et al. 2000). In telomerases of ciliates, the 5′ boundary is set by the position of a conserved nearby sequence that interacts with the TERT protein (Lai et al. 2002). Mutating these 5′-boundary-determining TER elements alters the sequence synthesized by telomerase (Prescott and Blackburn 1997; Tzfati et al. 2000; Lai et al. 2002). Point mutations within the templating sequence of Tetrahymena TER can affect both the 5′ and 3′ boundaries used in vitro (Gilley and Blackburn 1996). However, TER sequences outside the templating domain that affect 3′-boundary definition have not previously been reported.
Here, we present evidence that two phylogenetically conserved sequences, identified by phylogenetic comparison among budding yeast telomerase RNAs and called CS3 and CS4, base-pair to form a long-range pseudoknot. Unexpectedly, CS3 and CS4 mutations also caused shifts in the 3′ boundary of the template used by telomerase. The mutations caused deleterious cellular growth consequences comparable to those previously seen in human, yeast, and Tetrahymena cells when mutant repeats were added to telomeres via direct mutation of the template sequence itself (Yu et al. 1990; Gilley et al. 1995; Krauskopf and Blackburn 1996; Kirk et al. 1997; Smith and Blackburn 1999; Chan et al. 2001; Guiducci et al. 2001; Kim et al. 2001). This is the first demonstration that specific TER sequences not included in the templating domain or its boundary element can cause template miscopying. From phylogenetic modeling of yeast TER structure, we propose that CS3 and CS4 are brought close to the template spatially by a set of RNA–RNA helices.
Results
A phylogenetically conserved set of primary sequences in Kluyveromyces telomerase RNAs
Segments totaling about half of the ∼1.3-kb TERs of the budding yeasts Kluyveromyces lactis and Saccharomyces cerevisiae are partially or completely dispensable for telomerase function (Roy et al. 1998; Livengood et al. 2002). We cloned and sequenced TER genes from five additional Kluyveromyces yeast species that, together with K. lactis, form the “Kluyveromyces marxianus” cluster. Each TER gene encoded a single stable transcript (Supplementary Fig. 1AS). We mapped the 5′- and 3′-ends of the RNAs and determined their lengths to vary among the species from 928 nucleotides in Kluyveromyces nonfermentans to 1315 nucleotides in Kluyveromyces dobzhanskii. Alignment of the sequences allowed a phylogenetic tree to be drawn for the six Kluyveromyces species (Supplementary Fig. 1BS), and, despite the overall low sequence identity, identified seven conserved TER sequences, named CS1–CS7 (Fig. 1).
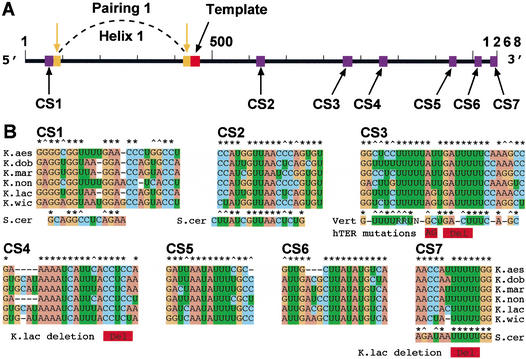
Figure 1.
Telomerase RNAs in six Kluyveromyces budding yeast species. (A) Linear map of the 1268-nucleotide-long telomerase RNA of K. lactis. (Red box) Templating domain sequence. (Purple boxes) Primary sequences CS1–CS7 conserved in the six Kluyveromyces species. (Orange boxes and downward arrows) The evolutionarily conserved long-range base-pairing [previously called Pairing 1 (Tzfati et al. 2000), now called Helix H1, this work]. (B) Comparisons of CS1–CS7 primary sequences among the six Kluyveromyces species analyzed. CS1, CS2, and CS7 are shown aligned with their S. cerevisiae counterparts. CS3 is compared with the vertebrate telomerase RNA CR2 consensus sequence (Chen et al. 2000). Shown in red underneath the sequences are human disease-associated mutations associated with dyskeratosis congenita (AG; Vulliamy et al. 2001) and aplastic anemia (Del; Vulliamy et al. 2002), and two previously analyzed K. lactis mutations (Roy et al. 1998). Conserved bases (*) or conserved purines or pyrimidines (^) are conserved in at least five out of six sequences. “R” denotes a pyrimidine (C or U) and “N” denotes any nucleotide.
Aligning the nucleotide sequence of TLC1, the S. cerevisiae TER, with those of the six Kluyveromyces TERs identified a few short sequences with varying degrees of similarity. CS1 (see Fig. 1A) falls in a TLC1 region required for binding of Est2p, the TERT protein of S. cerevisiae (Livengood et al. 2002). Block substitution of the TLC1 CS1 in vivo resulted in short but stable telomeres (Seto et al. 2002). CS2 is in a bulged-stem secondary structure that is fully conserved among all seven TERs from the Kluyveromyces species and S. cerevisiae and is a binding site of Est1p essential for telomerase function (Livengood et al. 2002; Seto et al. 2002). Finally, CS7 shows sequence identity to the essential Sm binding site in TLC1 (Seto et al. 1999). Like the TLC1 Sm site, CS7 matches the Sm consensus sequence, and is located near the 3′-end of each Kluyveromyces TER (7–17 bases from the 3′-end mapped as described in Materials and Methods). Various mutations in K. lactis CS7, including a five-base deletion (shown in Fig. 1B), reduced TER RNA to undetected levels and abolished telomerase function in vivo (J. Roy and E.H. Blackburn, unpubl.).
CS5 and CS6 are located within a partially dispensable segment of the K. lactis TER (Roy et al. 1998). However, CS4 is located in a region previously found to be essential for K. lactis telomerase function: several small deletions and substitutions in and near CS4 (one such deletion is indicated in Fig. 1B) abolished telomerase activity in vivo and in vitro (Roy et al. 1998).
Alignment of CS1–CS7 with previously identified conserved sequences in 35 species of vertebrate TERs (Chen et al. 2000) revealed sequence similarity between the Kluyveromyces CS3 and the vertebrate highly conserved CR2. The consensus sequence for CR2 is shown under the CS3 sequence in Figure 1B. CR2 can pair to form part of the predicted pseudoknot structure in vertebrate TERs (Chen et al. 2000). A two-base substitution in human CR2, the apparent disease-causing mutation in a human family with hereditary dyskeratosis congenita (Vulliamy et al. 2001), largely abolishes telomerase enzymatic activity in vitro (Comolli et al. 2002), as does a four-base deletion in the same portion of CR2 that has been associated with aplastic anemia (Vulliamy et al. 2002; H. Ly, E.H. Blackburn, and T.G. Parslow, in prep.). These human CR2 mutations are indicated in Figure 1B.
Base substitutions in CS3 and CS4 reduce or abolish telomerase function in vivo
We focused on the two yeast TER conserved sequences CS3 and CS4. To determine the effects of mutating them on telomerase action in vivo, we used a previously developed system that marks the action of a mutant telomerase by including a single-base mutation (called Bcl) in the TER template (Roy et al. 1998). The Bcl mutation is incorporated to the nascent telomeric repeats, generating BclI restriction sites, but otherwise is phenotypically silent and does not affect telomere length or telomerase activity (McEachern et al. 2002). Thus the BclI mutation can be included along with any mutation of interest in a test TER gene, to monitor the action of telomerase (Roy et al. 1998). Diminished telomerase activity causes decreased Bcl repeat incorporation and shorter telomeres. Complete lack of telomerase activity results in no BclI repeats, severe telomere shortening, and senescence. Survivors then emerge that maintain their telomeres by an alternative recombination pathway with a characteristic profile of heterogeneous telomeric restriction fragments (McEachern and Blackburn 1996). The incorporation of BclI sites and the appearance of the telomeric DNA profile are analyzed by Southern blot analysis of genomic DNA (Fig. 2A), or by cloning and sequencing telomeres.
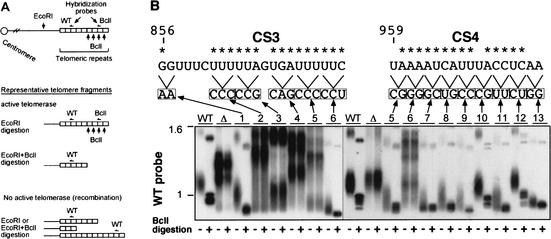
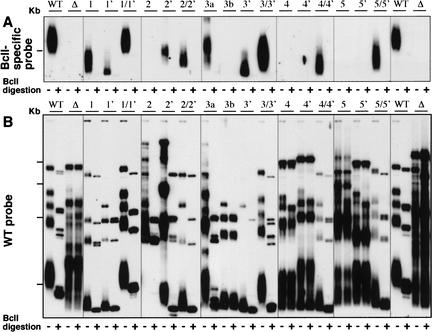
Figure 2.
Sequences CS3 and CS4 in K. lactis telomerase RNA are important for telomerase action and telomere maintenance in vivo. (A) Schematic showing a telomeric restriction fragment, and digests and probes used to analyze telomeres in the Bcl-marked system for telomerase RNA mutants. Also shown on the map are the positions of wild-type (WT) and Bcl-specific probes. (B) Southern blotting analysis of telomeric restriction fragments in K. lactis expressing different CS3- and CS4-region-scanning substitution mutants. Genomic DNA was prepared from K. lactis strains (deleted for the chromosomal TER gene and carrying the different TER alleles on a plasmid; see bottom part of figure) at their sixth passage. The control strains carry the phenotypically wild-type, BclI-marked TER gene (WT), or no insert (Δ) on a plasmid (Tzfati et al. 2000). DNA was digested with EcoRI restriction endonuclease (- lanes) or double-digested with EcoRI and BclI (+ lanes), separated on a 1% agarose gel, and vacuum-blotted. Hybridization was carried out as described previously (Tzfati et al. 2000), first with a BclI-specific oligonucleotide probe (data not shown), and then with a wild-type telomeric sequence probe. For simplicity, only the portion of the gel with a group of seven similarly sized telomeric EcoRI restriction fragments (McEachern et al. 2002) in WT K. lactis is shown. The positions of the scanning substitution mutations tested in the CS3 and CS4 regions are indicated above the autoradiograph. Asterisks indicate conserved residues (see Fig. 1A).
To further dissect the functions of CS3 and CS4, first we analyzed the effects of scanning substitution mutations made along them (Fig. 2B). The scanning mutations revealed that CS3 and CS4 were both essential for normal telomere length maintenance, Bcl incorporation, and cell growth. The effects of the CS3 and CS4 mutations correlated well with the regions of sequence conservation. Strikingly, transition substitutions as small as 2 nucleotides could completely abolish telomerase activity in vivo, as shown in Figure 2B. The levels of all the mutant RNAs were wild type (Supplementary Fig. 2CS). Hence, CS3 and CS4 are essential for telomerase function.
Base-pairing between CS3 and CS4 is required for telomerase function
Vertebrate CR2 is located in an essential pseudoknot element whose specific role is yet unknown (Chen et al. 2000; Martin-Rivera and Blasco 2001). Aided by secondary structure predictions made by the computer programs X2s (Juan and Wilson 1999) and Mfold (see below), we found a potential pseudoknot element formed by long-range base-pairing between CS3 and CS4 (Fig. 3A). The formation of such a pseudoknot structure is consistent with the secondary structure predictions (see below; Fig. 7, below), in which CS3 and CS4 is each located within a conserved secondary structure locale: a loop closed off by phylogenetically supported base-pairing. We tested the pseudoknot prediction directly, by making mutations in CS3 and CS4 designed to disrupt and restore their putative base-pairing interaction. Five sets of such “P” mutant substitutions were made in CS3 and CS4 (Fig. 3B). For each of the five P mutant sets, the substitution in either CS3 or CS4 (P single mutant) was tested separately, and also combined in the same mutant TER gene, to restore the predicted CS3–CS4 pairing (P double mutant). We replaced the wild-type gene in K. lactis cells with each mutant TER gene, and analyzed in vivo telomerase action by effects on cell growth, telomere length, the extent of action of the mutant telomerase on telomeres, and telomeric recombination event frequencies. We also analyzed the steady-state cellular TER RNA levels and in vitro activity of telomerases containing such mutated RNAs.
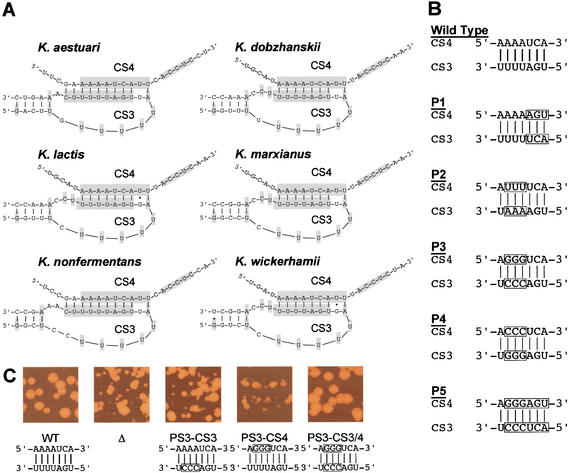
Figure 3.
Disruption of CS3/CS4 base-pairing. (A) Predicted pairing and local secondary structure of CS3 and CS4 in the six Kluyveromyces species analyzed. Conserved CS3 and CS4 bases (Fig. 1B) are shaded. (B) Five sets of P substitution mutations used to test disruption and restoration of pairing between CS3 and CS4 in K. lactis TER. (Boxed nucleotides) Sequences substituted in the P mutations tested. P1, P2, and P4 are transversions, the P3 mutations are transitions, and P5 combines P1 and P3. Each CS3 and CS4 P mutation was tested separately (single mutant), and in combination to restore predicted CS3/CS4 base-pairing (double mutant). (C) Disruption of CS3/CS4 base-pairing impairs cell colony growth. Representative colony phenotypes for P3 CS3 or CS4 single mutants, and for CS3/4 compensatory double mutants, shown compared with colonies of wild-type (WT) or ter1D deletion (Δ) control strains.
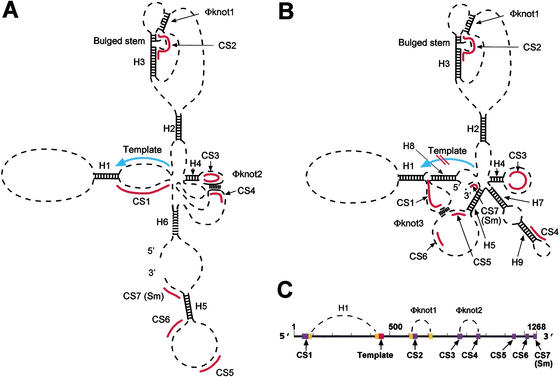
Figure 7.
Two predicted conformations of Kluyveromyces TER RNA. (A) Conformation A. Top-ranked helices predicted by X2s program and Pseudoknots 1 and 2 are indicated. (B) Conformation B, which has comparable overall free energy predicted by X2s program and shares some top-ranked helices with Conformation A, as well as additional helices, but lacks Pseudoknot 2. (C) Linear map indicating positions of Helix H1 (Tzfati et al. 2000) and the pseudoknots.
Mutating TER can impair either the expression or assembly of stable telomerase RNP (Roy et al. 1998; Beattie et al. 2000). However, as with the scanning mutants (Supplementary Fig. 2C), all these P mutants had wild-type TER RNA levels in the usual fractions containing telomerase activity after column chromatography (Fulton and Blackburn 1998; Roy et al. 1998; data not shown). This was consistent with previous findings, in which TER gene mutations in the CS3 and CS4 regions did not decrease the levels of total TER or telomerase RNP in vivo (Roy et al. 1998).
Colony growth of each mutant was assessed by serially restreaking single colonies of four independent transformant clones for 12 restreaks, and recording colony growth at each streaking. If telomeres become too short during cell division in the absence of telomerase, cells undergo senescence. If the structure of the telomere terminus is impaired through mutation in the telomeric DNA sequence or in telomere end-binding proteins (Krauskopf and Blackburn 1996; Karlseder et al. 2002; Lei et al. 2002; Mitton-Fry et al. 2002), telomeres can become uncapped. Such uncapping of telomeres can elicit arrest of cell division, high rates of recombination in and near the telomeric repeats, and/or telomeric fusions, with resulting deleterious effects on chromosome stability and cell growth (Kirk et al. 1997; for review, see McEachern et al. 2000). All 10 single mutations that disrupted the potential base-pairing between CS3 and CS4 caused growth defects. These were alleviated either nearly completely (P1 and P3 double mutants) or significantly (P2, P4, and P5 double mutants) by restoration of the base-pairing in the double mutants (Fig. 3C).
P4 and P5 single mutants underwent senescence indistinguishably from a TER deletion mutant streaked and analyzed in parallel with each CS3/CS4 mutant set. In these strains, large “survivor” colonies emerged in which telomeres have been shown to be maintained, in the absence of telomerase function, by a recombinational mechanism (McEachern and Blackburn 1996). P1, P2, and P3 single mutants from the outset continuously grew slowly, but with no apparent senescent phase, and showed high variability in colony sizes and rough colonies. Examples of the colony phenotypes for the P3 set of single and double mutations, compared with a wild-type and a TER deletion strain, are shown in Figure 3C.
The effects of the different CS3 and CS4 mutations on cell growth correlated well with their effects on telomerase action on telomeres in vivo. We used the Bcl marker mutation in the TER template described above (see Fig. 2A). For each P mutant set, the incorporation of BclI sites into telomeres in vivo in the single mutants was reduced compared with wild-type and with its corresponding compensatory double mutant (Fig. 4A). The P4 and P5 single mutations completely abolished the in vivo action of telomerase, with no incorporation of Bcl repeats being detectable even after 12 restreaks (Fig. 4A, lanes 4,4′,5,5′; data not shown). The P4 and P5 double mutants had partially restored in vivo activity, as shown by incorporation of BclI sites and the lack of telomeric recombination, although telomeres were relatively short (Fig. 4A,B, lanes 4/4′,5/5′).
Figure 4.
Base-pairing disruption impairs telomerase action and telomere maintenance in vivo. Southern blotting analysis of the telomeric restriction fragments of the CS3 and CS4 pairing mutants shown in Figure 3B. Digests, wild-type (WT) and Bcl-specific probes, and methods are as described in Figure 2. The P mutants are indicated above each set of lanes. Lanes 1 and 1′ indicate the P1 single mutants, lane 1/1′ indicates the P1 double mutant, and so on. Lanes 3a and 3b are two independent transformant lines of the P3 CS3 mutant (Fig. 3B). For simplicity, in A only the portion of the gel with a group of seven similarly sized telomeric EcoRI restriction fragments (McEachern et al. 2002) in WT K. lactis is shown. All the telomeric restriction fragments are shown in B.
In the P1 single mutants, Bcl sequence incorporation was much lower than wild type and telomeres were very short. Restoration of the potential CS3/CS4 base-pairing in the P1 double mutant restored telomere length and incorporation of BclI repeats to close to wild type (Fig. 4, lanes 1,1′,1/1′). The P2 mutation set had a somewhat different effect: in both P2 single mutants, telomeres became highly recombinagenic even though BclI sites were clearly incorporated (Fig. 4, lanes 2,2′). As previously described (McEachern and Blackburn 1995), such recombination events included gene conversions between sub-telomeric regions, thereby changing the pattern of restriction fragments, and recombination among the terminal telomeric repeat tracts, which became highly heterogeneous in length. Hence, the P2 single-mutant telomeres exhibited the greatly increased recombination that is one hallmark of compromised capping. In contrast, the compensatory P2 double mutants showed minimal recombination over the course of the 12 serial restreaks (Fig. 4, lane 2/2′; data not shown). The P3 CS3 single mutant showed some clonal variability: in one transformant clone (Fig. 4A, lane 3b), telomerase action in vivo was apparently abolished, although in the other transformant clone studied, very low activity was seen as assayed by BclI site incorporation (Fig. 4A, lane 3a). In the 3a clonal line, however, this mutant telomerase failed to protect against high rates of recombination at the telomeres (Fig. 4B, lane 3a). Again, as with the other four P mutant sets, telomere length and Bcl incorporation were both significantly improved by restoration of the CS3/CS4 pairing potential in the double P3 mutant (Fig. 4, lane 3/3′). Together, these findings provide strong genetic evidence that in vivo base-pairing between CS3 and CS4 is important for TER function.
Effects of CS3 and CS4 mutations on in vitro telomerase activity
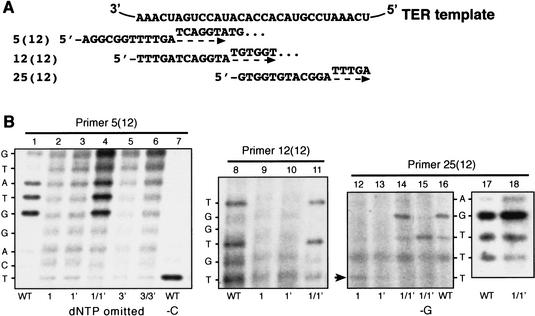
We analyzed the in vitro enzymatic activity of telomerase assembled in vivo in cells expressing only the CS3 and/or CS4 mutant TER. With the three different DNA substrate primers shown in Figure 5A and all four dNTPs, including radiolabeled dTTP, wild-type telomerase synthesized the previously characterized elongation products, forming a characteristic series of labeled bands in denaturing gel electrophoresis, as described previously (Fig. 5B, lanes 1,8,16,17; data not shown; Fulton and Blackburn 1998; Tzfati et al. 2000). In contrast, the P1 and P3 single mutations reduced enzyme activity, with little if any synthesis of these products (Fig. 5B, lanes 2,3,5,9,10,12,13). The background bands in these mutant lanes were RNase-insensitive and hence not attributable to telomerase activity, as previously described (Fulton and Blackburn 1998; Tzfati et al. 2000). A low amount of the +1-nucleotide product from primer 25(12) was seen with the P1 CS3 mutant (Fig. 5B, lane 12, arrow) but not with the P1 CS4 mutant (Fig. 5B, lane 13). This result is consistent with both the low overall in vivo activity of both P1 single mutants and the slightly higher action of the P1 CS3 mutant telomerase than the P1 CS4 mutant in vivo (Fig. 4, cf. lanes 1 and 1′). Restoration of the CS3/CS4 pairing by the P1 double mutation, which restored telomerase function in vivo (Fig. 4, lane 1/1′) also restored the efficiency and expected patterns of telomerase product formation in vitro (Fig. 5B, lanes 4, 11,14,15,18). Therefore, the in vivo functioning of the P1 mutants closely reflected their inherent enzymatic activity.
Figure 5.
Effects of CS3 and CS4 pairing mutations on in vitro telomerase activity. (A) Schematic of in vitro assay for K. lactis telomerase activity using three 12-nucleotide-long DNA primers that can base-pair at different places along the templating domain RNA sequence of K. lactis TER, as described previously (Fulton and Blackburn 1998). The primers used for in vitro activity reactions in B are shown aligned below the BclI TER RNA template sequence. When supplied with such a DNA primer substrate, and appropriate labeled and unlabeled dNTPs, elongation along the template toward its 3′-end extends the primer, producing a series of short elongation products that can be resolved as well-defined bands by electrophoresis in a DNA sequencing gel (Fulton and Blackburn 1998; Tzfati et al. 2000). Dashed arrows indicate the direction of DNA primer extension by polymerization along the template, with the first nucleotides added indicated above the arrow for each primer. (B) Disruption of CS3/CS4 base-pairing impairs polymerization by K. lactis telomerase. Telomerase activity was assayed in vitro (Fulton and Blackburn 1998). Telomerase reactions were incubated in the presence of all four dNTPs, or with dCTP or dGTP omitted as indicated below the lanes. Up the sides of the panels are shown the nucleotides predicted to be incorporated at each position, starting at the first nucleotide added to the primer terminus on polymerization along the template.
Telomerase RNA mutations in CS3 and CS4 cause shifted template usage in vivo
The P3 CS3 single mutant had very low in vitro activity, consistent with its in vivo phenotypes; however, the in vitro activity of the compensatory double P3 mutant 3/3′ was also low (Fig. 5B, lane 6), despite its considerably improved in vivo function (Fig. 4, lane 3/3′). This suggested that the cellular phenotypes may not result solely from reduced telomerase catalytic activity. Another defect in telomeres that can cause cellular growth phenotypes is a change in the telomeric DNA sequence added by telomerase. Therefore, we analyzed the telomeric DNA sequences in the P1–P5 mutants. Telomeric DNA was cloned by a method we reported previously (Tzfati et al. 2000), which uses ligation of an anchor primer to the end of the telomeric DNA to capture the telomerase-synthesized telomeric DNA strand extending to its 3′-most terminal nucleotide. As described previously (Tzfati et al. 2000), the telomeric DNA clones obtained by this method from cells expressing the P-mutant TERs had total lengths and extents of incorporation of BclI sequences that corresponded to those seen by Southern blotting analyses, validating that these clones were representative of the in vivo telomere populations. Thus, for example, the P3 CS3 single-mutant transformant line whose telomeres had shown no BclI incorporation by Southern blotting analyses (see Fig. 4, lane 3b) also had no BclI sites in its cloned telomere sequences. The cloned and sequenced telomeres from a different P3 CS3 mutant clonal transformant line, which showed very low Bcl incorporation and high recombination (see Fig. 4, lane 3a), showed the expected interspersion of the BclI-site-containing repeats within the tracts of otherwise wild-type repeats (data not shown).
The most striking novel feature caused by the P mutations was the appearance of truncated telomeric repeats embedded within tracts of full-length BclI repeats. The sequences of the truncated repeats stopped short of the normal 3′ and 5′ template boundaries, indicative of shifted template usage. Examples are shown in Figure 6. We therefore propose that base changes in CS3 and CS4 distort the spatial relationship between the telomerase active site and the template to cause premature termination of copying before the 5′-end of the template is reached. This, in turn, forces the alignment process to rely on any base-pairing that can form between the truncated 3′-end of the elongation product and the template, causing the shifts to a more central portion of the template. This type of shifted template usage was distinct from that caused by disrupting Helix H1, in which copying extended beyond the normal 5′ boundary of the template, and also the 3′-end of the template shifted to a more central location in the template, as described previously (Tzfati et al. 2000; Supplemental Material; Supplementary Fig. 3S).
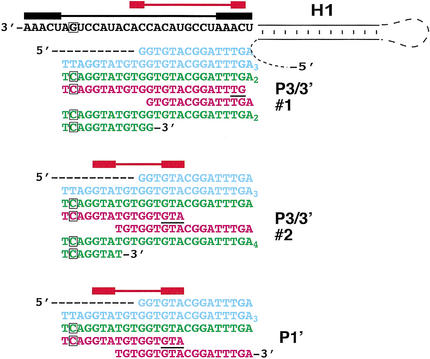
Figure 6.
Mutating CS3 and CS4 decreases the incorporation of BclI-marked repeats onto telomeres and causes shifted template usage in vivo. PCR products were cloned and sequenced. Examples of telomere clone sequences from cells expressing P mutants. The TER RNA sequences are depicted in black, and the telomeric DNA sequences are colored blue for wild-type repeats, green for Bcl repeats added by the mutant telomerase (the phenotypically silent BclI-marker mutation in the RNA and the corresponding incorporated DNA sequences are boxed), and magenta for DNA sequence indicative of shifted template usage. Above the RNA sequence, the horizontal black bar denotes the maximal template sequence, containing the short repeated sequence at both ends (thicker black bars) thought to be required for realignment and synthesis of multiple telomeric repeats by wild-type telomerase; the red bars are the corresponding regions for the mutant Ters.
A phylogenetically conserved set of helices in K. lactis yeast telomerase RNA predicts two alternative conserved core secondary structures
The finding that interaction between CS3 and CS4 (both located ∼400 nucleotides from the template) affects template usage prompted us to examine their possible spatial relationship to the template. Aided by the identification of CS1–CS7 as a guide to sequence alignment, we examined the entire sequences of all six Kluyveromyces spp. TERs for phylogenetic evidence of any predicted core secondary structure. We used the X2s computer program (Juan and Wilson 1999), which predicts RNA helices based on a scoring system that combines phylogenetic covariation, preserving base-pairing, and calculated helical stability to iteratively search for helices that were strongly predicted in all six TERs. We identified nine such top-ranked conserved helices, H1–H9, plus two paired regions that each forms a long-range pseudoknot (Pseudoknots 1 and 2). Helices H1–H9, ranked in order, have scores from 44 to 20 in the X2s program. In other RNAs, comparably scoring helices have correlated strongly with proven helices (Juan and Wilson 1999). Helices H1–H9 all fall within regions of the RNA previously shown experimentally to be important for K. lactis TER function (Roy et al. 1998). The top-ranked helix, H1, is the previously named long-range Pairing 1 located immediately 5′ of the template that demarcates the 5′ boundary of the template (Tzfati et al. 2000). We have not experimentally further tested Helices H2–H9. Pseudoknot 1 has a score of 22. One stem in Pseudoknot 2 is strongly supported by covariation, as shown in Figure 3A. Although the other stem of Pseudoknot 2, the CS3/4 pairing, does not rank high in the X2s program because of its strong primary sequence conservation, as described above, we have shown experimentally that the CS3 and CS4 pairing forms.
Helices H1–H6 and Pseudoknots 1 and 2 together constrain the Kluyveromyces spp. TERs in a predicted conserved partial core structure (Conformation A; Fig. 7A), as indicated by using the X2s program in combination with the Mfold prediction program. This structure was also supported by the Mfold prediction performed directly on these RNAs. A second structure of comparable overall calculated energy, Conformation B (Fig. 7B), was also well supported by both the Mfold and X2s programs, for all the Kluyveromyces spp. TERs. Conformations A and B share Helices H1–H5. In Conformation B, (1) Helix H6 and predicted Pseudoknot 2 are replaced by alternative Helices H7 and H9; (2) a partial pairing of the templating domain sequence forms Helix H8; and (3) CS3 becomes unpaired from CS4, with CS4 becoming engaged in Helix H9 (Fig. 7B). Figure 7C shows a linear map of the paired sequences that form H1 and the two predicted pseudoknots, and their positional relationship to the template and conserved sequences CS1–CS7.
Discussion
Here we have presented evidence that a pair of conserved telomerase RNA sequences, CS3 and CS4, and base-pairing between them, are important for the function of budding yeast telomerase. This pairing may be involved in the overall conformation of the yeast telomerase RNA, as it is present in one predicted secondary structure we have called Conformation A.
Consistent with their strong phylogenetic conservation, the primary sequences of CS3 and CS4, as well as their pairing, are also clearly a determinant of their functionality. Restoring the normal number, but not the base identity, of the seven paired bases in some CS3/CS4 double mutants did not fully restore telomerase activity or normal template usage. In contrast, only the base-pairing between a different pair of sequences, which form Helix H1 immediately next to the template, is needed for its role in demarcating the 5′ boundary of the template (Tzfati et al. 2000). Furthermore, each of the CS3 and CS4 mutations caused highly specific effects, either abolishing or reducing telomerase activity, or causing a distinctive novel type of template shift. Hence, we propose that the CS3/4 sequences are involved in a specific TER structure that influences the spatial relationship of the enzymatic active site to the TER templating domain sequence.
Altering a structure that involves CS3 and CS4 distant in space from the template and active site could cause an allosteric change that is transmitted through the telomerase RNP to the active site. Alternatively, the secondary and tertiary structure of TER could bring CS3 and CS4 spatially close to, or even directly in contact with, the template and active site, possibly as in proposed Conformation A (Fig. 7A). Further structural studies will be necessary to distinguish between these models.
The ciliate and vertebrate TERs contain a shared pseudoknot, which in vertebrates contains sequence CR2, with some homology to CS3. It has been suggested from in vitro structural probing of in vitro transcribed Tetrahymena telomerase RNA that its pseudoknot may be dynamic (Bhattacharyya and Blackburn 1994). In addition, recent structural analysis of the human telomerase RNA pseudoknot also suggests that its CR2-containing stem may exist in alternative paired and unpaired states (Antal et al. 2002; Comolli et al. 2002; Theimer et al. 2003). Analysis of K. lactis telomerase RNA by nondenaturing gel electrophoresis has also provided evidence for conformational flexibility of this RNA (see Supplemental Material; Supplementary Fig. 4S). Our evidence is consistent with the hypothesis that Conformation A is not maintained when the CS3–CS4 pairing is disrupted.
We propose that during rounds of template copying, the conformation of TER may change with each elongation cycle. Specifically, TER may begin a cycle of template copying in Conformation A, and termination of synthesis and unwinding of the newly extended DNA product from the RNA template may be promoted by conversion of TER into Conformation B. This would be predicted to replace the DNA-template RNA base pairs with the intramolecular RNA–RNA base-pairing that forms Helix H8, which encompasses several of the template bases. Such replacement of base-pairing partners could be energy-neutral, and serve to eject the newly synthesized DNA from the template.
We recently showed that intermolecular pairing of the CR2 pseudoknot stem can mediate functional dimerization of human telomerase (Ly et al. 2003). Whether Pseudoknot 2 is the ortholog of the pseudoknot in ciliates and vertebrates is not known. However, the common structural themes in telomerase RNA that continue to emerge across multiple eukaryotic phylogenetic groups hint that underlying functional aspects of this RNA, besides its ability to template telomeric DNA, are conserved.
Materials and methods
Cloning of TER homolog genes from Kluyveromyces
K. lactis was cloned previously (McEachern and Blackburn 1996), and TER1 genes from the other Kluyveromyces spp. were produced as described in the Supplemental Material. All TER sequences were verified by independent PCR amplification and sequencing of the full-length genes from genomic DNAs.
Sequence alignment and secondary structure prediction
TER sequences were aligned, and a phylogenetic tree was generated using the computer program CLUSTALX. Conserved helixes were predicted using the computer program X2s (Juan and Wilson 1999). The five top-scored helices were used to constrain secondary structure prediction by RNA Mfold version 2.3 with the folding temperature at 30°C (http://www.bioinfo.rpi.edu/_zukerm). The GenBank accession numbers are AY151277, AY151278, AY151279, AY151280, and AY151281 for the Kluyveromyces aestuarii, Kluyveromyces dobzhanskii, Kluyveromyces marxianus, Kluyveromyces nonfermentans, and Kluyveromyces wickerhamii telomerase RNAs, respectively.
Southern and Northern blotting analyses
These were performed as described in the Supplemental Material. Southern blots to analyze telomeres were hybridized first with a BclI-specific oligonucleotide probe (KTElBcl, GATCAG GTATGTGG) at 40°C, washed at 40°C with 0.1 M [Na+], and then with a wild-type telomeric sequence probe (Klac1-25, AC GGATTTGATTAGGTATGTGGTGT), hybridized, and washed at 50°C. Blots were exposed to film or to PhosphorImager (Molecular Dynamics).
In vitro telomerase activity assays, PCR amplification, and cloning of telomere fragments and native gel electrophoretic analyses of telomerase RNP analysis
These were done as described previously (Fulton and Blackburn 1998; Roy et al. 1998; Tzfati et al. 2000).
Acknowledgments
This work was supported by National Institutes of Health grant GM26259 to E.H.B., a Human Frontiers Postdoctoral Fellowship to Y.T., and an HHMI predoctoral fellowship to Z.A.K. We thank V. Juan and C. Wilson for help with use of the X2s program and members of the Blackburn Lab for helpful discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1099403.
Footnotes
Supplemental material is available at http://www.genesdev.org.
References
- Antal M., Boros, E., Solymosy, F., and Kiss, T. 2002. Analysis of the structure of human telomerase RNA in vivo. Nucleic Acids Res. 30: 912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie T.L., Zhou, W., Robinson, M.O., and Harrington, L. 2000. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell 11: 3329–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A. and Blackburn, E.H. 1994. Architecture of telomerase RNA. EMBO J. 13: 5721–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E.H. 1994. Telomeres: No end in sight. Cell 77: 621–623. [DOI] [PubMed] [Google Scholar]
- ____. 2000. The end of the (DNA) line. Nat. Struct. Biol. 7: 847–850. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Chang, J., Prescott, J., and Blackburn, E.H. 2001. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 11: 1240–1250. [DOI] [PubMed] [Google Scholar]
- Chen J.L. and Greider, C.W. 2003. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 22: 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.L., Blasco, M.A., and Greider, C.W. 2000. Secondary structure of vertebrate telomerase RNA. Cell 100: 503–514. [DOI] [PubMed] [Google Scholar]
- Comolli L.R., Smirnov, I., Xu, L., Blackburn, E.H., and James, T.L. 2002. A molecular switch underlies a human telomerase disease. Proc. Natl. Acad. Sci. 99: 16998–17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton T.B. and Blackburn, E.H. 1998. Identification of Kluyveromyces lactis telomerase: Discontinuous synthesis along the 30-nucleotide-long templating domain. Mol. Cell. Biol. 18: 4961–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D. and Blackburn, E.H. 1996. Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol. Cell. Biol. 16: 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D., Lee, M.S., and Blackburn, E.H. 1995. Altering specific telomerase RNA template residues affects active site function. Genes & Dev. 9: 2214–2226. [DOI] [PubMed] [Google Scholar]
- Guiducci C., Cerone, M.A., and Bacchetti, S. 2001. Expression of mutant telomerase in immortal telomerase-negative human cells results in cell cycle deregulation, nuclear and chromosomal abnormalities and rapid loss of viability. Oncogene 20: 714–725. [DOI] [PubMed] [Google Scholar]
- Juan V. and Wilson, C. 1999. RNA secondary structure prediction based on free energy and phylogenetic analysis. J. Mol. Biol. 289: 935–947. [DOI] [PubMed] [Google Scholar]
- Karlseder J., Smogorzewska, A., and de Lange, T. 2002. Senescence induced by altered telomere state, not telomere loss. Science 295: 2446–2449. [DOI] [PubMed] [Google Scholar]
- Kim M.M., Rivera, M.A., Botchkina, I.L., Shalaby, R., Thor, A.D., and Blackburn, E.H. 2001. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc. Natl. Acad. Sci. 98: 7982–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk K.E., Harmon, B.P., Reichardt, I.K., Sedat, J.W., and Blackburn, E.H. 1997. Block in anaphase chromosome separation caused by a telomerase template mutation. Science 275: 1478–1481. [DOI] [PubMed] [Google Scholar]
- Krauskopf A. and Blackburn, E.H. 1996. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383: 354–357. [DOI] [PubMed] [Google Scholar]
- Lai C.K., Miller, M.C., and Collins, K. 2002. Template boundary definition in Tetrahymena telomerase. Genes & Dev. 16: 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Baumann, P., and Cech, T.R. 2002. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry 41: 14560–14568. [DOI] [PubMed] [Google Scholar]
- Livengood A.J., Zaug, A.J., and Cech, T.R. 2002. Essential regions of Saccharomyces cerevisiae telomerase RNA: Separate elements for Est1p and Est2p interaction. Mol. Cell. Biol. 22: 2366–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly H., Xu, L., Rivera, M.A., Parslow, T.G., and Blackburn, E.H. 2003. A role for a novel “trans-pseudoknot” RNA–RNA interaction in the functional dimerization of human telomerase. Genes & Dev. 17: 1078–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Rivera L. and Blasco, M.A. 2001. Identification of functional domains and dominant negative mutations in vertebrate telomerase RNA using an in vivo reconstitution system. J. Biol. Chem. 276: 5856–5865. [DOI] [PubMed] [Google Scholar]
- McEachern M.J. and Blackburn, E.H. 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376: 403–409. [DOI] [PubMed] [Google Scholar]
- ____. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes & Dev. 10: 1822–1834. [DOI] [PubMed] [Google Scholar]
- McEachern M.J., Krauskopf, A., and Blackburn, E.H. 2000. Telomeres and their control. Annu. Rev. Genet. 34: 331–358. [DOI] [PubMed] [Google Scholar]
- McEachern M.J., Underwood, D.H., and Blackburn, E.H. 2002. Dynamics of telomeric DNA turnover in yeast. Genetics 160: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry R.M., Anderson, E.M., Hughes, T.R., Lundblad, V., and Wuttke, D.S. 2002. Conserved structure for single-stranded telomeric DNA recognition. Science 296: 145–147. [DOI] [PubMed] [Google Scholar]
- Prescott J. and Blackburn, E.H. 1997. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes & Dev. 11: 528–540. [DOI] [PubMed] [Google Scholar]
- Roy J., Fulton, T.B., and Blackburn, E.H. 1998. Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes & Dev. 12: 3286–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto A.G., Zaug, A.J., Sobel, S.G., Wolin, S.L., and Cech, T.R. 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180. [DOI] [PubMed] [Google Scholar]
- Seto A.G., Livengood, A.J., Tzfati, Y., Blackburn, E.H., and Cech, T.R. 2002. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes & Dev. 16: 2800–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.D. and Blackburn, E.H. 1999. Uncapping and deregulation of telomeres lead to detrimental cellular consequences in yeast. J. Cell Biol. 145: 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer C.A., Finger, L.D., Trantirek, L., and Feigon, J. 2003. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc. Natl. Acad. Sci. 100: 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfati Y., Fulton, T.B., Roy, J., and Blackburn, E.H. 2000. Template boundary in a yeast telomerase specified by RNA structure. Science 288: 863–867. [DOI] [PubMed] [Google Scholar]
- Vulliamy T., Marrone, A., Goldman, F., Dearlove, A., Bessler, M., Mason, P.J., and Dokal, I. 2001. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413: 432–435. [DOI] [PubMed] [Google Scholar]
- Vulliamy T., Marrone, A., Dokal, I., and Mason, P.J. 2002. Association between aplastic anaemia and mutations in telomerase RNA. Lancet 359: 2168–2170. [DOI] [PubMed] [Google Scholar]
- Yu G.L., Bradley, J.D., Attardi, L.D., and Blackburn, E.H. 1990. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 344: 126–132. [DOI] [PubMed] [Google Scholar]