Abstract
Activation of the CLB gene cluster through the assembly of Mcm1p–Fkh2p complexes at target promoters is essential for mitotic entry and transition through M phase. We show that the activation of this mitotic transcriptional program is dependent on the recruitment of Ndd1p, a coactivator that performs its essential function by acting through Fkh2p. Although an essential gene, NDD1 is dispensable in cells expressing a truncated form of Fkh2p lacking its C terminus. When phosphorylated on T319, Ndd1p is recruited to CLB cluster promoters by association with the forkhead-associated (FHA) domain of Fkh2p. Substitution of T319 for alanine significantly reduces recruitment of Ndd1p, resulting in loss of normal transcriptional regulation, severe impairment of cell growth, and a budding defect reminiscent of cells with a Cdk-Clbkinase deficiency. Finally, we show that phosphorylation of T319 and recruitment of Ndd1p to CLB2 and SWI5 promoters is dependent on Cdc28-Clbkinase activity. These data provide a model describing the activation of G2/M transcription through the phosphorylation of Ndd1p by Cdc28-Clb kinase activity.
Keywords: CLB cluster, Cdk activity, mitotic transcription, forkhead transcription factor, FHA domain, cell cycle
In the budding yeast Saccharomyces cerevisiae, over 800 genes are cell-cycle regulated at the transcriptional level (Spellman et al. 1998), accounting for >10% of the entire number of genes in the yeast genome. Each of these genes belongs to a transcriptional wave, or cluster, that is coregulated such that the genes are activated and repressed together during specific cell-cycle intervals. At least 10 gene clusters have been identified that generate identifiable and partially overlapping waves of transcriptional activity associated with the different cell-cycle phases/transitions (for review, see Futcher 2002).
Closely associated with the G2-to-M-phase transition is the activation of at least 30 genes, collectively known as the “CLB cluster” (Spellman et al. 1998). The function of genes in this cluster is associated with late cell-cycle events such as cytokinesis, mitosis, cell-wall remodeling, sister chromatid separation, and establishment of events for the ensuing G1 phase (for review, see Breeden 2000; Jorgensen and Tyers 2000). Closely associated with the CLB cluster is the presence of a common regulatory motif in the 5′ regulatory region of these genes (Lydall et al. 1991; Althoefer et al. 1995; Maher et al. 1995; Spellman et al. 1998). This element is both necessary and sufficient for cell-cycle regulation of the CLB cluster and consists of an Mcm1p-binding site, immediately flanked by a site for forkhead transcription factors (Althoeffer et al. 1995; Maher et al. 1995; Koranda et al. 2000; Kumar et al. 2000; Pic et al. 2000). The establishment of G2–M transcriptional periodicity involves the Mcm1p-dependent recruitment of the forkhead transcription factor Fkh2p (Hollenhorst et al. 2000; Koranda et al. 2000; Kumar et al. 2000; Pic et al. 2000; Zhu et al. 2000) that binds CLB cluster regulatory elements throughout the cell cycle (Althoefer et al. 1995; Koranda et al. 2000). Another closely related forkhead factor, Fkh1p, can maintain periodic transcription of these genes in the absence of Fkh2p, but unlike its relative, does not appear to bind cooperatively with Mcm1p (Kumar et al. 2000; Hollenhorst et al. 2002). While deletion of either FKH1 or FKH2 has no major impact on cell-cycle progression, simultaneous deletion causes severe morphological defects and delayed mitotic entry (Hollenhorst et al. 2000; Kumar et al. 2000; Pic et al. 2000; Zhu et al. 2000). This phenotype closely resembles that seen for various mutants defective in Cdc28p-Clbp kinase activity (Surana et al. 1991; Amon et al. 1993), consistent with a role for Fkh2p in the regulation of mitotic cyclin levels. Although functionally redundant, Fkh1p and Fkh2p normally operate on different subsets of target genes in vivo (Hollenhorst et al. 2002). Fkh2p appears to be the primary regulator of the cell-cycle-regulated CLB cluster, while Fkh1p has a distinct role in silencing genes involved in cell-type determination (Hollenhorst et al. 2000; Sun et al. 2002).
Fkh1p and Fkh2p are quite distinct from other members of the forkhead family in budding yeast as they belong to a subfamily that is classified according to a common domain, known as the forkhead-associated (FHA) domain. This domain facilitates the multimerization of FHA-containing polypeptides with target molecules phosphorylated on threonine residues (for review, see Durocher and Jackson 2002). FHA-domain-containing proteins have a variety of roles involved in processes including cell-cycle checkpoint control, DNA repair, signal transduction, transcriptional regulation, and premRNA splicing (for review, see Li et al. 2000; Durocher and Jackson 2002). Sequence alignments define the core FHA domain as a motif spanning ∼75 amino acids (Hofmann and Bucher 1995), although fully functional FHA domains are likely to span up to 180 residues (Liao et al. 1999; Hammet et al. 2000). Structural studies modeled around the Rad53p–FHA1,2 reveal that the core FHA domain consists of a β sandwich consisting of two twisted antiparallel β sheets (Liao et al. 1999; Durocher et al. 2000). The binding site for pThr epitopes is composed of two adjacent loops connecting the β sheets. The five most highly conserved FHA domain residues are located in these loops and all appear to be important for binding pThr ligands (Li et al. 2000; Pike et al. 2001). Based on studies with short peptides, ligand specificity of FHA domain binding by pThr peptides is mediated through distinct short-sequence motifs flanking both sides of a central pThr residue (Durocher et al. 2000).
Activation of the CLB cluster also requires the activity of NDD1, an essential gene that was originally identified as a high copy-number suppressor of strains carrying the temperature-sensitive (ts) cdc28-1n allele (Loy et al. 1999). Hence, Ndd1p was originally proposed to act in a mitotic pathway involving CDC28. Several lines of evidence support the idea that Ndd1p works in collaboration with Mcm1p and forkhead transcription factors to control the periodic transcription of CLB cluster genes. For example, the requirement for NDD1 can be bypassed by simultaneous deletion of FKH2, implying that Ndd1p works through Fkh2p (Koranda et al. 2000). Moreover, ectopic expression of NDD1 enhances levels of CLB cluster transcription (Loy et al. 1999), and its recruitment to CLB2 and SWI5 promoters is dependent on the presence of either Fkh1p or Fkh2p (Koranda et al. 2000). Unlike Mcm1p and Fkh2p that bind throughout the cell cycle, Ndd1p is recruited to CLB2 and SWI5 promoters only during G2–M (Koranda et al. 2000), suggesting that it may have a role in the periodic activation of these genes.
The mechanism by which CLB cluster genes are controlled is not understood but is likely to involve the activity of mitotic cyclin-dependent kinase (Cdk) activities (Amon et al. 1993; for review, see Breeden 2000; Jorgensen and Tyers 2000). The activation of mitotic Cdk activities has two main effects on transcriptional control in budding yeast. First, it triggers the inactivation of transcription associated with earlier cell-cycle events, such as G1–S progression, by a negative feedback loop. Failure to activate mitotic Clb kinase activities in G2 allows genes such as CLN1,2 to remain transcriptionally active during G2 (Amon et al. 1993). The second role for mitotic cyclins is to promote transcription of genes encoding the mitotic cyclins themselves and other genes that make up the CLB cluster (Amon et al. 1993). Hence, Clbp-dependent kinase activity is thought to be required for CLB1,2 transcription during G2–M and defines the existence of a positive feedback loop where Clb-associated Cdk activity is required for CLB transcription. It is not certain, however, if Clb kinases act directly on the CLB promoter or by some other mechanism. To resolve this issue, it will be important to determine if Clb kinases target Mcm1p, Fkh2p, or Ndd1p as part of this regulatory pathway. This is a particularly relevant issue because Fkh2p recently has been shown to be phosphorylated during G2/M, consistent with it being targeted by mitotic Cdk activities (Pic et al. 2000). This overall scheme of CLB cluster regulation is attractive because it not only explains how transcription of the CLB cluster is maintained during G2–M, but also can explain how transcription is shut down at the end of mitosis when Clbkinase activities collapse (Surana et al. 1993).
In this report, we characterize the mechanism by which Ndd1p participates in the activation of CLB cluster genes during G2–M phases of the cell cycle. For mitotic transcription to occur, Ndd1p must be recruited to promoters through the FHA domain of Fkh2p. This requires phosphorylation of Ndd1p on T319 by mitotic Cdk activity, consistent with earlier models proposing a role for Clb kinases in the activation of G2–M transcription. This work provides a mechanism for how CLB cluster genes, such as CLB2 and SWI5, are activated in an Ndd1p-dependent manner and why this requires Clb kinase activity.
Results
The essential role of Ndd1p acts through the C terminus of Fkh2p
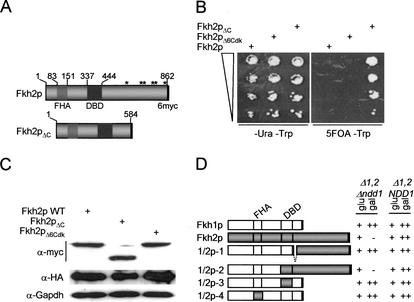
Genetic interactions between NDD1 and FKH2, but not FKH1, indicate that the essential role of Ndd1p is specifically linked to Fkh2p (Koranda et al. 2000). This suggested to us that the specificity of this functional relationship is mediated by structural differences between Fkh1p and Fkh2p. The most obvious possibility was a role for the C-terminal extension of Fkh2p, which extends for ∼278 amino acids and is conspicuous by the presence of six potential Cdk phosphorylation sites (Fig. 1A). To test this possibility, a Δfkh1 Δndd1 strain carrying a plasmid expressing Ndd1p was modified at the FKH2 locus so that it expressed a C-terminal truncation of Fkh2p or derivatives of Fkh2p carrying alanine substitutions at S/T residues within each canonical Cdk phosphorylation site (Fig. 1A). These experiments were performed in a Δfkh1 background so that the activity of Fkh2p could be directly evaluated and not masked by Fkh1p's redundant function. As expected, curing of the pADH1.NDD1 by growth on 5-FOA was lethal in a strain expressing wild-type Fkh2p (Koranda et al. 2000) and in all strains carrying single or multiple alanine substitutions (Fig. 1B). However, truncation of Fkh2p, resulting in loss of its C-terminal extension, completely abolished the requirement for Ndd1p (Fig. 1B). The morphology and growth rate of the Δndd1 fkh2ΔC strains were comparable to that of wild-type strains and independent of the FKH1 status (Fig. 1; data not shown). Moreover, the DNA binding activity of Fkh2pΔC was indistinguishable from that of wild-type Fkh2p as judged by band shift analysis and by chromatin immunoprecipitation (ChIP) assays (data not shown). Each mutant was expressed at comparable levels (Fig. 1C).
Figure 1.
The essential function of Ndd1p depends on the C terminus of Fkh2p. (A) Features of Fkh2p are indicated including the DNA-binding domain (DBD), the FHA (forkhead-associated) domain, six canonical Cdk phosphorylation sites (*) in the C-terminal extension, and the C-terminal 6myc tag. The truncated form of Fkh2p (Fkh2pΔC), lacking the C-terminal extension and potential Cdk phosphorylation sites, is also depicted. (B) S600 (FKH2), S601 (FKH2ΔC), and S602 (FKH2ΔCdk) were grown on selective medium (-ura -trp) or on -trp 5-FOA plates to cure for the pADH.NDD1 plasmid. Note that only the strain expressing Fkh2pΔC can grow in the absence of Ndd1p. (C) Immunoblot analysis showing equivalent expression levels of 6myc-tagged Fkh2p, Fkh2pΔC and Fkh2pΔ6Cdk in strains S600, S601 and S602. Whole-cell lysates (30 μg protein) were resolved on 8% SDS–polyacrylamide gels, blotted, and probed with α-myc 9e10 and α-HA (12ca5) monoclonal antibody. Filters were stripped and reprobed with α-Gapdh monoclonal antibody as a loading control. (D) The C terminus of Fkh2p is sufficient to confer Ndd1p dependency on the function of Fkh1p. The table shows a summary of results that defined the region of Fkh2p that is sufficient to confer Ndd1 dependency on Fkh1p. FKH1, FKH2, or FKH1–FKH2 chimeras, driven by a GAL1 promoter on a YCplac33-based (URA3) plasmid, were tested for Ndd1p-dependent cell growth in MK146 (Δfkh1 Δfkh2 Δndd1) or S610 (Δfkh1 Δfkh2). Fkh1p is shown in white and Fkh2p is shown in gray with respective FHA and DBD indicated. Assays were performed by transforming plasmids into both strains on -ura glucose selective plates, which have a similar morphology and growth rate. Transformants were then struck on selective plates, with either glucose or galactose as the carbon source. Colony formation was evaluated after 4 d growth at 30°C and compared to the growth rate and morphology in the strain transformed with the G.FKH1 plasmid. On galactose, this plasmid produced growth rates and a morphology similar to the congenic FKH1 FKH2 NDD1 strain (++). A - represents lethality in the Δfkh1 Δfkh2 Δndd1 strain as a result of induced Fkh2p or 1/2p-2 expression on galactose and hence, Ndd1p dependency. In cases where NDD1 dependency was apparent (i.e., 1/2p-2), a plasmid expressing Ndd1p was introduced and shown to rescue the lethality.
To complement this analysis, we asked if we could make Fkh1p Ndd1p-dependent by fusing it to the C-terminal region of Fkh2p. To do this, several FKH1–FKH2 hybrid constructs under control of the GAL1 promoter were expressed in congenic Δfkh1 Δfkh2 strains where NDD1 was either intact or deleted. Both strains grew at reduced rates (∼360 min) relative to wild type (∼90 min) or single Δfkh1/Δfkh2 strains and displayed a pseudohyphal morphology (Kumar et al. 2000; data not shown). When forkhead function was restored by expression of FKH1 on galactose, growth rates were comparable to wild-type strains (++) and associated with normal cell morphology. Ndd1p dependency was evaluated in this assay by scoring for lethality on galactose in the Δfkh1 Δfkh2 Δndd1 strain. Ndd1p-independent function would result in normal cell growth (++) and a reversal of the pseudohyphal phenotype on galactose in this strain. The assay was validated by showing that FKH2 expression was lethal in a Δndd1 strain but not in the NDD1 strain (Fig. 1D).
While the C-terminal 288 amino acids of Fkh2p could not confer Ndd1p dependency on Fkh1p (1/2p-1; Fig. 1D), extension of this hybrid so that it now included a contiguous C-terminal region of Fkh2p containing the DNA-binding domain (1/2p-2), rendered Fkh1p function fully dependent on Ndd1p to support cell growth (Fig. 1D). Swapping DNA binding (1/2p-3) or FHA domains (1/2p-4) alone did not reproduce this effect, indicating that the C-terminal region of Fkh2p is the key element targeted by Ndd1p in vivo. These constructs were also tested in the Δfkh1 Δfkh2 deletion strain that has a characteristic morphology and severely retarded growth rates indicative of defective CLB cluster transcription and Clb kinase deficiency. Like the wild-type FKH1 and FKH2 constructs, all of the hybrid constructs tested in this study were capable of rescuing the Δfkh1 Δfkh2 defect, as judged by assessing morphological criteria and growth rates (Fig. 1D), indicating that all hybrids were functional. These results are consistent with a scenario where Ndd1p acts as a transcriptional coactivator with the ability to negate inhibitory effects imposed by the Fkh2p C terminus.
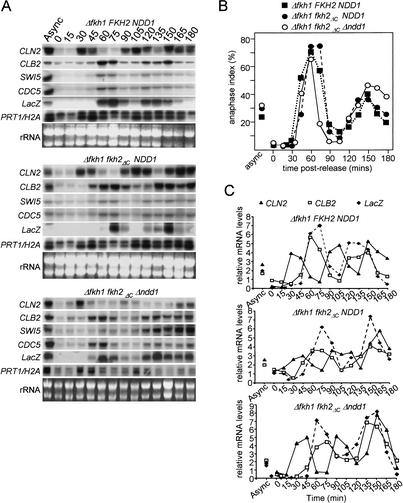
To determine if Ndd1p-independent transcription of the CLB cluster retained cell-cycle regulation, cells were released from an α-factor block and transcript levels monitored by Northern blot analysis over multiple synchronous cell cycles. This analysis showed that in cells expressing Fkh2pΔC, the absence of Ndd1p has no marked impact on either periodicity or the magnitude of CLB cluster transcription (Fig. 2A). Cell synchrony was monitored by scoring cells for the presence of anaphase spindles (Fig. 2B) and transcript levels determined by phosphor-imaging analysis (Fig. 2C). These results indicate that the essential function of Ndd1p is to act through the Fkh2p C terminus and implicate the existence of a second pathway that is able to activate forkhead-dependent (Fkh1p, Fkh2pΔC) transcription at CLB cluster promoters, independently of Ndd1p.
Figure 2.
Ndd1p is not required for normal cell-cycle regulation of CLB cluster genes in strains carrying a C-terminal truncation of Fkh2p. S466 (Δfkh1), S618 (Δfkh1 fkh2ΔC), and S619 (Δfkh1 fkh2ΔC Δndd1) were synchronized with α-factor for 2 h. (A) Following release from the block, mRNA was prepared from cells at 15 min intervals and resolved on 1% formaldehyde-agarose gels, blotted onto nitrocellulose filters, and probed with random-primed 32P-labeled probes. (B) Cell-cycle synchrony was assessed by fixing cells with formaldehyde and staining with anti-tubulin antibodies to visualize mitotic spindles. The percentage of cells with anaphase spindles were scored (200 cells for each time point). (C) mRNA levels were quantitated by PhosphorImaging analysis. Relative levels of each transcript are shown after standardization to the Prt1 loading control.
The FHA domain of FKH2 recruits Ndd1p into complexes
In a previous study, we used DNA affinity chromatography to identify Fkh2p as an SFF-like DNA binding activity (Kumar et al. 2000). Several copurifying polypeptides were identified by this analysis, one of them Mcm1p and another of ∼62 kD that we identified later as being Ndd1p (Kumar et al. 2000; R. Kumar, D. Reynolds, and S. Dalton, unpubl.). This is consistent with other published work showing that Ndd1p is recruited to CLB cluster promoters in vivo (Koranda et al. 2000).
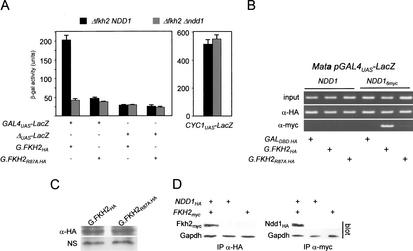
As the FHA domain of Fkh2p was a likely interaction interface for the recruitment of coactivators, we set out to determine if Ndd1p or other proteins could be recruited via the FHA domain. During the course of this work, we noticed that when fused to a GAL4 DNA-binding domain (GALDBD), a truncated form of Fkh2p containing the FHA domain (Fkh2p1–291), but lacking the DNA-binding domain and the C terminus, could activate transcription of a linked GAL4UAS–LacZ reporter gene (Fig. 3A). Because Ndd1p has been shown to possess a potent transactivation domain in a one-hybrid assay (Loy et al. 1999), we reasoned that Ndd1p could be recruited to the GAL promoter through its association with the G.Fkh2p fusion. A second Gal–Fkh2p fusion carrying a single amino-acid substitution at position 87 of Fkh2p (R87A) was also tested as this residue is conserved amongst FHA domains (Hofmann and Bucher 1995) and is known to be critical in the binding of phospho-threonine-containing epitopes (Pike et al. 2001). These experiments were performed in a FKH1 Δfkh2 background because deletion of NDD1 in a FKH2 strain is lethal. Our results indicate that reporter gene transcription is dependent on Ndd1p as LacZ activity was reduced more than fivefold in a congenic Δndd1 strain. Activation of the reporter through G.Fkh2p activity was also dependent on a functional FHA domain as the R87A mutation reduced LacZ reporter activity by approximately fourfold (Fig. 3A). The activity of the CYC1 promoter was not affected by NDD1 deletion, ruling out the possibility that the previous results were part of a general transcriptional response to loss of NDD1 function. Activation of the reporter gene also shows cell-cycle dependency, being high in nocodazole-blocked cells but low in α-factor blocked cells (data not shown), consistent with the activation being Ndd1p-dependent.
Figure 3.
Ndd1p is recruited into complexes in vivo with Fkh2p through its FHA domain. (A) An HA-tagged GAL4DBD fused to the N-terminal 291 amino acids of FKH2 containing the FHA domain (G.FKH2WT), or a mutant (G.FKH2R87A.HA) carrying a single amino acid substitution (R87A) in the FHA domain, was tested for its ability to activate a GAL4UAS–LacZ reporter gene or a reporter lacking a UAS (ΔUAS–LacZ) in S508 (Δfkh2 NDD1) or MK465 (Δfkh2 Δndd1). The activity of an NDD1-independant reporter (CYC1UAS–LacZ) was also assessed in these strains. Each experiment was performed independently three times and the error bars represent the standard error of the mean. (B) The ability of Ndd1p to be recruited to the Fkh2p FHA domain was evaluated by ChIP analysis in NDD1 or NDD16myc strains carrying a reporter plasmid containing a LacZ reporter gene driven by a GAL4UAS. The NDD16myc strain was created by modifying the NDD1 locus so that a 6myc-tagged form of Ndd1p is expressed (Ndd1p6myc). Primers flanking the GAL4-binding sites in the reporter plasmid were used for PCR analysis. (C) Immunoblot analysis of G.Fkh2pHA and G.Fkh2pR87A.HA levels. Whole-cell lysate (30 μg protein) was subject to immunoblot analysis, probing with α-HA (12ca5) monoclonal antibody. NS represents a cross-reacting polypeptide consistently recognized by the HA antibody and which served as a loading control. (D) Detection of Fkh2p–Ndd1p complexes in cell extracts. Cell lysates were prepared from strains expressing 6myc-tagged Fkh2p or 3HA-tagged Ndd1p, or both together, from ADH1-driven CEN-based plasmids. Fkh2p or Ndd1p was immunoprecipitated with myc-9e10 or HA-12ca5 antibody. After immunoblotting, immunoprecipitates were probed with either myc or HA antibody. Amounts of input protein were compared by immunoblot analysis of whole-cell extracts, probing with α-Gapdh antibody.
To directly demonstrate that Ndd1p is recruited to Fkh2p through its FHA domain, ChIP experiments were performed under the same conditions as described for the one-hybrid analysis shown in Figure 3A, except that Ndd1p was 6myc-tagged. These experiments show that Fkh2p1–291 is sufficient for the recruitment of Ndd1p6myc to the GAL4UAS (Fig. 3B). GAL4DBD fused to Fkh2p151–862 was unable to recruit Ndd1p as determined by the ChIP assay, nor was it able to activate the LacZ reporter gene above background levels (data not shown). Hence, neither the DNA-binding domain or the C terminus of Fkh2p is required for Ndd1p recruitment. A specific role for the FHA domain in Ndd1p recruitment was demonstrated by using a G.Fkh2p derivative where the R87A mutation was introduced (Fig. 3B). While this fusion still bound the GAL4UAS, its ability to recruit Ndd1p was severely limited, indicating that a functional FHA domain is required for Ndd1p recruitment. The inability of FHAR87A to recruit Ndd1p cannot be accounted for by altered protein stability, as levels were comparable to the wild-type form (Fig. 3C). We then went on to determine if Ndd1p and Fkh2p could be detected in complexes from whole-cell lysates by immunoprecipitation-immunoblot analysis. This analysis showed that both proteins can be coimmunoprecipitated from cell extracts, indicating that they are assembled into complexes, consistent with the previous 1-hybrid analysis (Fig. 3D). It is not known if the interaction between Fkh2p and Ndd1p is direct or requires an intermediate bridging protein (see Discussion).
T319 phosphorylation is essential for normal Ndd1p function in vivo
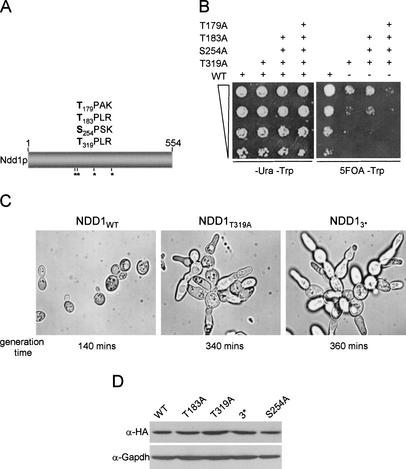
Cell-cycle regulation of G2–M transcription in budding yeast requires the activity of Clb kinases (Amon et al. 1993), raising the possibility that regulators of these genes such as Ndd1p, may be Cdk targets (Breeden 2000; Jorgensen and Tyers 2000). This was of special interest to us because the recruitment of Ndd1p may interact with the FHA domain of Fkh2p through a phosphorylated threonine residue. Ndd1p has four consensus Cdk phosphorylation sites [(S/T)PX(K/R)], three of which contain threonine residues (Fig. 4A). To determine if these sites are important for Ndd1p function, serine or threonine residues in each canonical Cdk site were substituted for alanine. Mutant forms of Ndd1p were expressed from a TRP1, CEN-based plasmid under the control of the ADH1 promoter (pT.NDD13HA) in a Δndd1 strain carrying a wild-type, untagged copy of the NDD1 gene on a CEN-based, URA3 plasmid (pADH.NDD1). Curing of pADH.NDD1 on 5-fluororotic acid (5-FOA) plates then allowed for the functional evaluation of each ndd1 mutant. Of all the single mutants tested, only Ndd1pT319A caused an obvious effect (Fig. 4; data not shown), resulting in a severe growth delay (Fig. 4B) and a severe budding defect similar to that seen in Δfkh1 Δfkh2 double-deletion mutants (Fig. 4C; Hollenhorst et al. 2000; Koranda et al. 2000; Kumar et al. 2000; Zhu et al. 2000). The T319A mutant alone had a similar effect on morphology and growth rate to the triple (Ndd13*; T179A, S254A, T319A) and quadruple mutants (Ndd14*; T179A, T183A, S254A, T319A; Fig. 4B,C). Protein levels of each mutant form of Ndd1p were comparable to wild type (Fig. 4D).
Figure 4.
Mutagenesis of potential Cdk phosphorylation sites in Ndd1p cause severe morphological growth and transcriptional defects. (A) Four consensus Cdk phosphorylation sites in Ndd1p were identified and S/T residues substituted for alanine either one at a time or in combination with other sites. (B) S599 (Δndd1 pADH.NDD1) was transformed with pT.NDD13HA (TRP1) and grown on either -ura -trp selective plates or -trp 5-FOA plates at different cell densities (indicated by graded box). Sites of amino-acid substitutions expressed from pT.NDD13HA (single or multiple) are indicated (+). (C) Bright field photographs (100× magnification) of cells grown in -trp liquid media curing on -trp 5-FOA plates. Shown are strains carrying plasmids for the wild type (NDD1WT), T319A (NDD1T319A), and T183A, S254A, T319A (NDD13*) with associated generation times. (D) Immunoblot analysis showing levels of Ndd1p3HA and mutant derivatives in S599. Thirty micrograms of whole-cell lysate was resolved on a 10% polyacrylamide gel and probed with α-Gapdh and α-HA (12ca5) monoclonal antibody.
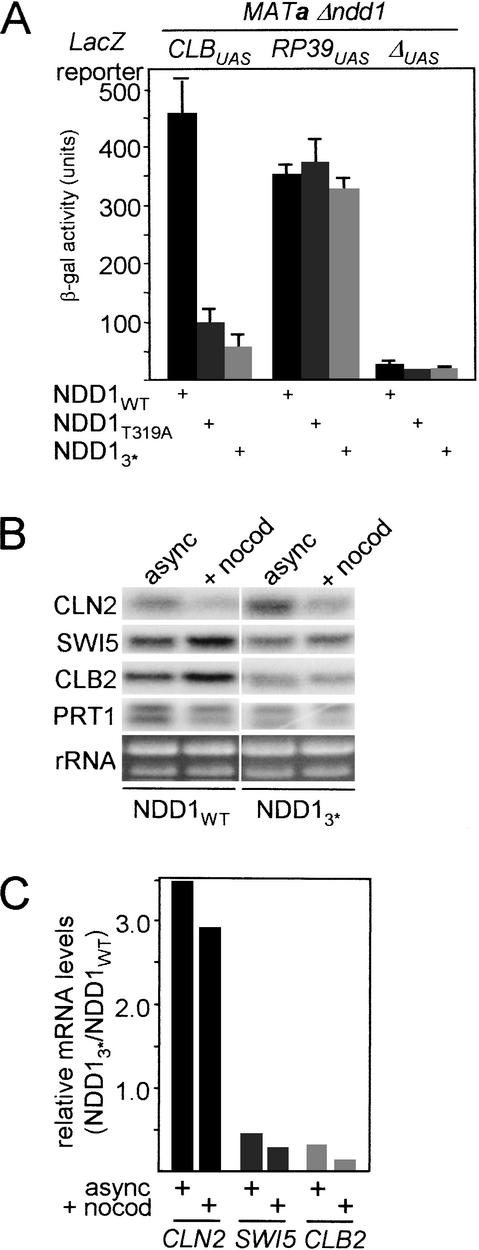
To determine the transcriptional consequences of these mutants, the activity of a CLB2UAS–LacZ reporter gene (Maher et al. 1995; Kumar et al. 2000) was first evaluated in a Δndd1 strain that expressed wild-type Ndd1p, or one of several S/T to alanine mutants from a plasmid. Quantitative β-galactosidase assays showed that activity of the CLB2UAS-reporter was approximately fivefold less active in asynchronous cells carrying the Ndd1pT319A mutant compared to wild type (Fig. 5A). The activity of the RP39UAS was independent of the NDD1 status. These results were corroborated by similar decreases in levels of endogenous SWI5 and CLB2 transcripts levels in asynchronous and nocodazole blocked cells (Fig. 5B). Coinciding with reduced CLB2 and SWI5 transcription, CLN2 transcript levels were elevated 3.5-fold and 2.9-fold in asynchronous and nocodazole blocked cells, respectively, compared to wild-type cells (Fig. 5C). This is likely to be due to low Clb kinase activity, as elevated Clb-kinase activity is required for CLN transcriptional shut-down after completion of G1 (Amon et al. 1993). Under these conditions, axial bud growth is favored over radial bud growth (Lew and Reed 1993), accounting for the abnormal phenotype described. The phenotypic and transcriptional defects described were all associated with mutant forms of Ndd1p expressed from the ADH1 promoter on a CEN-based plasmid. These effects could be suppressed to varying degrees by overexpression of these mutants from a high-copy number plasmid (data not shown). When integrated at the NDD1 locus and under control of its authentic promoter, the T319 mutant was unable to support cell growth (data not shown), presumably because of an acute defect in CLB cluster transcription.
Figure 5.
Transcriptional defects associated with NDD1 phosphorylation site mutants. (A) Liquid culture β-galactosidase assays were performed in S599 carrying 2 μ LacZ reporter plasmids and ADH1-driven forms of NDD1 in YCplac22, as indicated. Assays were performed in triplicate and the results shown as the standard error of the mean. (B) Northern blot analysis of mRNA levels in asynchronous (async) and nocodazole-blocked (+nocod) cells. US262 carrying a YCplac22-based plasmid expressing wild-type NDD1 (NDD1WT) or NDD1 with mutations in three Cdk sites (NDD13*; T183A, S254A, T319A; see A). Flow profiles were difficult to generate in pseudohyphal ndd1 mutant strains so cell-cycle blockade was monitored by morphological criteria. In both the wild-type and T319A mutant strains, nocodazole treatment resulted in >90% of cells bearing a large budded phenotype with unduplicated DNA, duplicated spindle pole bodies, and absence of spindle. In asynchronous cells, the percentage of cells with these characteristics was 15% for wild-type cells and 23% for T319A cells. (C) Quantitation of relative mRNA levels was determined by PhosphorImaging. The data are presented as the ratio between the mRNA level in NDD1 and NDD13* strains after being first normalized against the PRT1 signal.
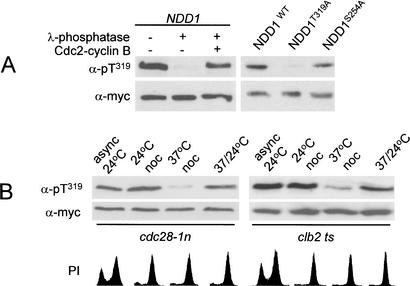
Our data indicate T319 to be a critically important residue involved in Ndd1p function. Because this threonine residue was part of a canonical Cdk phosphorylation sequence, we set out to establish if in fact T169 was phosphorylated in vivo. This was done by first immunoprecipitating 6myc epitope-tagged Ndd1p (Ndd1p6myc) from whole-cell lysates. Imunoprecipitates were then subject to immunoblot analysis by probing with an antiphospho Ndd1pT319 antibody (see Materials and Methods). This antibody recognized immunoprecipitated Ndd1p6myc and, when treated with λ phosphatase, this reactivity was abolished (Fig. 6A). Treatment of the phosphatase-treated immunoprecipitate with purified cdc2–cyclin B restored reactivity with the anti-phospho Ndd1pT319 antibody (Fig. 6A). The specificity of the antibody for pT319 was confirmed as it recognized Ndd1pWT and Ndd1pS254A but not Ndd1pT319A in immunoprecipitates (Fig. 6A). The functionally important T319 residue is therefore phosphorylated in vivo.
Figure 6.
(A) Ndd1pT319 is phosphorylated in vivo. S287 was transformed with YCplac22-based plasmids expressing ADH1-driven forms of NDD16myc that was immunoprecipitated from whole-cell lysate (500 μg protein), resolved on a 10% polyacrylamide gel and probed with α-myc (9e10) or α-pNdd1pT319 (α-pT319) polyclonal antibody. Immunoprecipitates were untreated (-) or treated (+) with λ-phosphatase, cdc2–cyclin B complexes as described in Materials and Methods. (B) Cdc28-Clb kinase activity is required for phosphorylation of Ndd1pT319. Ndd1p6myc immunoprecipitates were subject to immunoblot analysis by probing with α-myc (9e10) or α-pNdd1pT319 (α-pT319). Mata cdc28-1n pNDD1 or Mata clb2 ts pNDD1 were grown at 24°C (async 24°C), at 24°C or 37°C + nocodazole for 2.5 h (24°C noc and 37°C noc, respectively), or at 37°C + nocodazole followed by a shift down to 24°C for 2 h. Cell-cycle profiles were determined by flow cytometry of propidium iodide (PI)-labeled cells.
Cdk-Clb kinase activity is required for Ndd1T319 phosphorylation
To address the question of whether T319 phosphorylation was dependent on Cdk activity, we evaluated Ndd1pT319 phosphorylation status in the cdc28-1nts (Surana et al. 1991) and clb2ts strains grown at either permissive or restrictive temperatures. Asynchronous cells were grown at 24°C, blocked in G1 with α factor, then released at either 24°C or 37°C in the presence of nocodazole. α-pNdd1pT319 antibody was then used to probe Ndd1p6myc immunoprecipitates to evaluate the phosphorylation status of T319. This analysis showed T319 to be phosphorylated under conditions of high Cdc28p-Clb kinase activity but not at the restrictive temperature, when kinase activity was low (Fig. 6B; see Discussion). The temperature effects of the cdc28-1n and clb2 ts alleles are mutation dependent and are not seen in the isogenic wild-type strain (Amon et al. 1993; D. Reynolds and S. Dalton, unpubl.).
T319 is crucial for the recruitment of Ndd1p to CLB cluster promoters
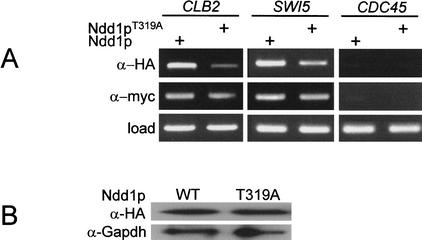
The previous experiments indicate a crucial role for pT319 as a major determinant of Ndd1p activity. To determine if T319 was required for the recruitment of Ndd1p to the SWI5 and CLB2 promoters, chromatin immunoprecipitation (ChIP) analysis was performed on chromatin prepared from nocodazole-blocked cells expressing Fkh2p6myc and either 3HA-tagged Ndd1pT319A or wild-type Ndd1p3HA. Fkh2p6myc served as an internal control as its binding to promoter elements is independent of Ndd1p activity. This analysis showed that recruitment of Ndd1T319A was severely impaired in comparison to wild-type Ndd1p (Fig. 7A). This could not be accounted for by differences in Ndd1p levels (Fig. 7B). Binding of Fkh2p6myc to the SWI5 and CLB2 promoters was comparable in Ndd1p and Ndd1pT319A expressing strains, as expected (Fig. 7A). We performed dilution analysis of PCR products generated from ChIP analysis in Figure 7 and estimate that in the T319A mutant, Ndd1p recruitment is decreased by eightfold and sixfold to the CLB2 and SWI5 promoters, respectively (data not shown), relative to wild-type Ndd1p. In contrast, other mutants such as Ndd1pS254A showed no obvious reduction in recruitment relative to wild-type Ndd1p (data not shown). Reduced recruitment of Ndd1pT319A to promoters therefore accounts for reduced activation of target genes, aberrant morphology and slow growth rates as described in Figures 4 and 5. Similar trends were also seen in similar experiments performed on asynchronous cells (data not shown).
Figure 7.
T319 is required for recruitment of Ndd1p to the SWI5 and CLB2 promoters in vivo. (A) S600 pADH.NDD13HA was grown to exponential phase then arrested with 10 ng/mL nocodazole for 120 min. Cells were fixed with formaldehyde and ChIP analysis performed immunoprecipitating with either α-myc (9e10) or α-HA (12ca5) monoclonal antibodies. PCR analysis was performed to evaluate the recruitment of wild-type or T319A forms of Ndd1p to SWI5 or CLB2 promoters in vivo. Ndd1p does not bind the CDC45 promoter and was used as a negative control. “Load” represents PCR analysis performed on whole chromatin prior to immunoprecipitation and was used to confirm equal input of chromatin material into each assay. (B) Wild-type (WT) Ndd1p and Ndd1pT319A are expressed at comparable levels. Extracts (30 μg protein) from cells expressing 3HA-tagged Ndd1p were subject to immunoblot analysis and probed with α-HA (12ca5) or α-Gapdh monoclonal antibody.
Cdc28-Clb kinase activity is required for Ndd1p recruitment to CLB cluster promoters
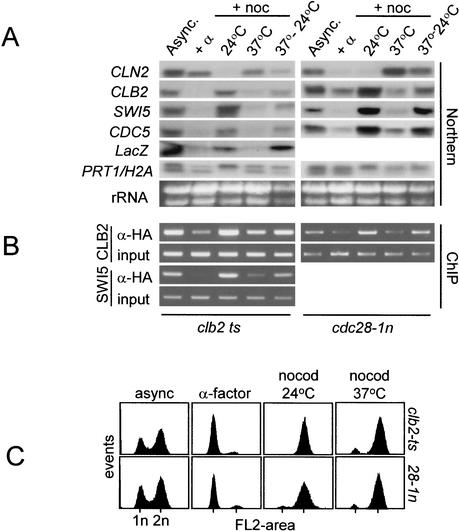
To show that Cdk activity plays a crucial role in the recruitment of Ndd1p to the CLB2 and SWI5 promoters, we performed ChIP analysis in cdc28-1nts and clb2ts strains that expressed triple HA-tagged Ndd1p. The recruitment of Ndd1p to target promoters was compared to the corresponding transcriptional activity of CLB2, SWI5, CDC5, and the CLB2UAS–LacZ reporter genes under conditions of high and low Clb kinase activity. Transcriptional analysis confirms earlier work (Amon et al. 1993) that transcription of the CLB cluster is dependent on Clb kinase activity. Moreover, inclusion of the CLB2UAS–LacZ reporter in this analysis shows this dependency to act through transcriptional control mechanisms since the CLB2UAS is sensitive to changes in Clb kinase activity (Fig. 8A) in a similar manner to endogenous genes.
Figure 8.
Cdc28-Clb kinase activity is required for recruitment of Ndd1p to the SWI5 and CLB2 promoters. A clb2ts strain (S315; Amon et al., 1993) was modified so that it had a CLB2UAS–ubiYLacZ reporter at the URA3 locus. The other ts strain, cdc28-1n, has been described previously (S272; Surana et al. 1992). Each ts strain was grown at permissive temperature (24°C; async.), arrested in G1 with α-factor (+α) at 24°C, blocked at either 24°C or 37°C + nocodazole (+noc/24°C, +noc/37°C) or at 37°C + nocodazole then shifted back to 24°C (as described in Fig. 5A). (A) Northern blots were used to evaluate transcript levels. (B) ChIP analysis as a measure of Ndd1p recruitment to SWI5 and CLB2 promoters in vivo. (C) Cell-cycle profiles of propidium iodide stained cells corresponding to A and B was determined by flow cytometry.
When the recruitment of Ndd1p to CLB2 and SWI5 promoters was evaluated, we found a clear relationship between high Cdc28-Clb kinase activity and high Ndd1p recruitment in both ts strains (Fig. 8B,C). Moreover, the transcriptional activity of these genes correlated with the level of Ndd1p recruitment to target genes in vivo (Fig. 8A,C). This is consistent with the transcriptional activity of CLB cluster genes being dependent on the ability of Fkh2p to recruit Ndd1p to CLB cluster promoters by a Cdc28-Clb–hdependent mechanism. This is likely to involve direct phosphorylation of Ndd1pT319 by Cdc28-Clb kinase.
Discussion
Ndd1p is a Fkh2p-specific coactivator
Although FKH1 and FKH2 are a pair of redundant forkhead transcription factors involved in the regulation of G2–M transcription, Fkh2p is the major regulator of the CLB gene cluster (Hollenhorst 2002; this report). Understanding the normal mechanism of CLB cluster gene activation therefore required an understanding of how Fkh2p is itself regulated. Clues as to the individual roles of each forkhead have been gleaned from subtle differences in the magnitude and periodicity of CLB2 and SWI5 mRNAs in synchronized single Δfkh1 or Δfkh2 deletion strains. Two reports (Hollenhorst et al. 2000; Pic et al. 2000) show that Δfkh1 strains have slightly elevated CLB2 and SWI5 transcripts with slightly higher basal levels in G1, compared to Δfkh2 strains that show a slight decrease in overall mRNA levels, while retaining periodicity. Moreover, Fkh2p has been proposed to function through alternate phases of repression and activation by itself acting as a repressor during G1 and S phase (Koranda et al. 2000).
Because Mcm1p–Fkh2p and not Fkh1p complexes are the main contributors to cell-cycle regulation of the CLB cluster, this study focusesed on how Fkh2p-dependent transcription is activated during G2–M. There are multiple regulatory mechanisms that could potentially account for Fkh2p-dependent periodicity of transcription. For example, because Mcm1p and Fkh2p are constitutively bound to CLB cluster promoters in vivo, it is possible that either of these factors could be subject to cell-cycle regulated posttranslational modification. Alternatively, or in addition to this, some additional factor may be recruited to Mcm1p–Fkh2p complexes to derepress and/or activate these complexes during G2–M. Our work has taken a path to investigate the second alternative, although it is possible that the former scenario is also important. NDD1 seemed an obvious candidate for a role in the Fkh2p-dependent activation of CLB cluster genes during G2–M, as it performs an essential role in the G2–M transition (Althoeffer et al. 1995); it is a rate-limiting regulator of the mitotic transcriptional program (Loy et al. 1999); it genetically and physically interacts with FKH2/Fkh2p on CLB cluster promoters (Koranda et al., 2000); and it has a potent transactivation domain (Loy et al. 1999).
NDD1 is only essential in cells expressing FKH2 (Koranda et al. 2000). Although Fkh2p has an absolute dependency for Ndd1p, the reason for this was not understood at the beginning of this study and was surprising given that Fkh1p did not have a similar requirement, especially as it can also support cell-cycle regulated transcription. A clue to explain differences in the Ndd1p dependency between Fkh1p and Fkh2p lied in the difference in these proteins at their C termini, where Fkh2p has an extension of 278 amino acids. The C terminus of Fkh2p was of particular interest with regard to its regulation because it contains six canonical Cdk phosphorylation sites (Fig. 1A) and because it is phosphorylated late in the cell cycle (Pic et al. 2000). This pointed toward a role for these Cdk sites in Fkh2p function that could distinguish its regulation from that of Fkh1p. However, when each or all of these sites were mutated to alanine, it had no detectable impact on the regulation of CLB cluster genes or the dependency of Fkh2p on Ndd1p. Hence, we were unable to demonstrate any role for the six consensus Cdk sites in the C terminus of Fkh2p. There are several potential MAPK sites in the C terminus of Fkh2p, and it is possible that these are functionally important for G2–M progression. No other consensus Cdk phosphorylation sites were identified in Fkh2p besides those characterized in this study. It would be interesting to determine if Fkh2pΔ6Cdk is still phosphorylated during G2–M.
Deletion of the C-terminal extension of Fkh2p completely abolished the requirement for Ndd1p as fkh2ΔC Δndd1 Δfkh1 cells had a similar generation time and morphology to wild-type cells. This establishes that Fkh2p is the only essential target for Ndd1p. This is consistent with a model where Ndd1p acts through the C-terminal region of Fkh2p and would explain why Fkh1p functions independently of Ndd1p as it lacks an extended C terminus, thus allowing it to interact with other coactivators. To demonstrate that the C terminus was sufficient to confer Ndd1p dependency, we fused Fkh2p574–862 to the C terminus of Fkh1p, with the prediction that the fusion would be Ndd1p dependent. Although the extreme 288 amino acids was not sufficient, further extension to include the Fkh2pDBD (residues 336–862) did render Fkh1p fully dependent on Ndd1p. Further mapping within the C terminus is required to define the exact region required for Ndd1p dependency. As part of this study, we show that FHA domains are interchangeable between Fkh1,2p, with no apparent change in function of the respective forkheads. We also show that the DBD's of Fkh1,2p are interchangeable, consistent with previous findings (Hollenhorst et al. 2000). The ability to swap FHA domains is intriguing because Ndd1p can be recruited to promoters occupied by either Fkh1p or Fkh2p (Koranda et al. 2000), consistent with our findings that the FHA domain is crucial for Ndd1p recruitment in vivo. This raises the possibility that Ndd1p may also act as a coactivator of Fkh1p-dependent transcription in some scenarios.
Taking into account all of the data available, we propose that during G1 and S phase, Mcm1p–Fkh2p complexes are silent because of the absence of a transcriptional activator. It is not possible to conclude that they are subject to active repression in G1/S, based on the evidence available (Hollenhorst et al. 2000; Koranda et al. 2000; Kumar et al. 2000; Pic et al. 2000). During early G2 phase through to M phase, Ndd1p is recruited to Mcm1p–Fkh2p complexes and allows for the transcriptional activation of CLB cluster genes (Koranda et al. 2000; this report). Our data would indicate that Ndd1p performs roles in addition to it neutralizing the Fkh2p C terminus. Recruitment of Ndd1p to the Fkh2pFHA is clearly associated with transcriptional activation (Figs. 3, 5), implying that Ndd1p is likely to function as a transcriptional coactivator. The equivalent mutation in FKH1, which blocks recruitment of Ndd1 to Fkh2p, renders it unable to support activated transcription of CLB cluster genes in Δfkh2 Δndd1 strains (D. Reynolds and S. Dalton, unpubl.). These data suggest that recruitment of a coactivator(s) through an FHA domain-dependent mechanism is also required for activation of Fkh1p.
Our data suggest that Ndd1p is the only coactivator that can bind the Fkh2p FHA domain, while Fkh2p has an intact C terminus. However, we speculate that other coactivators can interact with forkhead FHA domains (Fig. 9), because transcriptional periodicity is maintained in Δndd1 Δfkh2 (Fkh1p-dependent) and Δndd1 fkh2ΔC (Fkh2pΔC-dependent) strains. Under these circumstances, an intact FHA domain is still required (D. Reynolds and S. Dalton, unpubl.) leading us to invoke a role for an as yet uncharacterized Ndd1p-like coactivator. Because FHA domains are interchangeable, we propose that this coactivator could activate through Fkh1p and Fkh2pΔC. This may explain why subtle differences in the periodicity and magnitude of activation are seen in Δfkh1 and Δfkh2 strains. Our model would indicate that although Fkh2p can be activated exclusively by Ndd1p, Fkh1p-dependent transcription utilizes a separate coactivator (possibly in conjunction with Ndd1p). It is unclear why a backup pathway of CLB cluster regulation is in place in cases where Fkh2p and Ndd1p are absent.
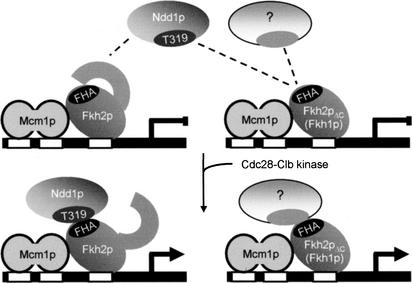
Figure 9.
A model to explain the mechanism for how CLB cluster genes are activated by Ndd1p during G2–M, based on the finding described in this report. The model reconciles how Ndd1p specifically works through Fkh2p and how Cdc28-Clb kinase activity works through a positive feedback loop (Amon et al. 1993) to activate CLB2 transcription, by targeting Ndd1p on T319. The existence of an additional coactivator is also proposed. (Left) The control of CLB cluster genes in wild-type cells. (Right) Redundant control pathways that can operate under conditions where the primary pathway is disrupted.
The mechanism by which Ndd1p can activate Fkh2p through its C terminus is still unclear but may involve the enforcement of a conformational change that inactivates the C terminus. Alternatively, Ndd1p may be insensitive to steric effects (created by the C terminus of Fkh2p) that block the recruitment of other coactivators. The role of Mcm1p in these mechanisms has not been directly addressed in this study. It is interesting however, that Mcm1p favors Fkh2p binding to CLB cluster promoters at the expense of Fkh1p in vivo (Hollenhorst et al. 2002). Together with our data, this invokes a role for Mcm1p in providing Ndd1p dependence on G2–M transcription.
One of the questions that has not been extensively addressed in this report is how transcription of the CLB cluster is initiated. Because Ndd1p is itself cell-cycle regulated, the answer probably lies in its regulation at the transcriptional level. This may involve Clb3,4,5 kinases, which have also been shown to be important for the accumulation of Clb1,2 (Yeong et al. 2002). We have also observed an inverse relationship between Clb2 kinase activity in conditional mutants and Ndd1p transcription (D. Reynolds and S. Dalton, unpubl.). This may involve some mechanism where Clb2 acts through Clb3,4 to suppress NDD1 transcription. This issue awaits further characterization.
FHA-dependent recruitment of Ndd1p to CLB cluster promoters
Our work has established that Ndd1p is recruited to promoters through the FHA domain of Fkh2p and that Ndd1p assembles into complexes with Fkh2p in cell lysates. Whether this is a direct interaction or requires an intermediate bridging protein has yet to be established. Attempts to answer this question are ongoing. Recruitment of Ndd1p by Fkh1p (in Δfkh2 strains; Koranda et al. 2000) together with the ability of the Fkh1p FHA domain to function in a Fkh2p chimera (this report) argues that Ndd1p can be recruited to promoters by Fkh1p in vivo. As described above, we believe that a redundant coactivator can act with Fkh1p making Ndd1p dispensable for its function. This may explain why some CLB cluster genes, such as CDC20, are regulated by an Ndd1p-independent mechanism (Loy et al. 1999). Perhaps CDC20 is controlled through a mechanism involving Fkh1p and a second coactivator, independent of Ndd1p.
The mechanism of Ndd1p recruitment clearly involves T319, probably through a phosphorylation-mediated mechanism. Our work showed, however, that Ndd1p recruitment to CLB2 and SWI5 promoters could still be detected, albeit at a significantly reduced level, in the T319A mutation. It is possible that T319 phosphorylation is not absolutely essential for recruitment but significantly enhances its efficiency. It will be interesting to determine the effects of making a mutant where T319 is substituted for an acidic residue to mimic constitutive phosphorylation. This experiment was not included as part of this study because Ndd1p is itself cell-cycle regulated and is degraded at the completion of mitosis (Loy et al. 1999). Hence, in the event that Ndd1pT319E/G was produced, it is unlikely to have a pronounced effect. Instead, the expression of a stable version of activated Ndd1p would be more desirable, as this would be predicted to deregulate Fkh2p-dependent transcription, perhaps making it constitutive. Hence, Ndd1p activity in the cell cycle is regulated through multiple mechanisms—transcriptional, protein stability, and recruitment into Fkh2p-containing complexes.
Although the C terminus of Fkh2p appears to be an essential target of Ndd1p activity, this region is not required for Ndd1p recruitment. Instead, an intact FHA domain capable of binding pThr epitopes is required. Because Ndd1p recruitment is probably pThr 319-dependent, we were tempted to speculate that Ndd1p may be the pThr ligand for the Fkh2p FHA domain. Although we have no direct physical evidence to support this, several lines of evidence are consistent with this possibility. First, recruitment of Ndd1p to Fkh2p is pThr-dependent. Second, mutations that block FHA–pThr interactions also severely reduce recruitment of phosphorylated Ndd1p. Third, failure to recruit Ndd1p to promoters by Fkh2p results in a transcriptional defect reminiscent of an inactivated forkhead pathway, and finally, deletion of NDD1 has a similar lethal phenotype to cells carrying an R87A mutation at the FKH2 locus (D. Reynolds and S. Dalton, unpubl.). The question of direct interactions between Fkh2p and Ndd1p requires further investigation.
The T319A mutant used in these studies does not completely block the ability of Ndd1p to be recruited to promoters. This is reflected by impaired, but not extinguished transcriptional activity, and a pronounced morphology very similar to Δfkh1 Δfkh2 deletion strains (Hollenhorst et al. 2000; Kumar et al. 2000; Pic et al. 2000; Zhu et al. 2000). When expressed from its own promoter, at its chromosomal locus, Ndd1pT319A was lethal in a Δndd1 background. In contrast, when expressed from a high-copy number plasmid under control of the ADH1 promoter, the aberrant morphology could be partially suppressed. Hence, we believe that the T319A mutant has minimal activity when expressed at elevated levels, and thus acts as a partial loss of function mutant, but not at physiological levels.
A role for Cdc28-Clb kinase activity in CLB cluster gene activation
Amon et al. (1993) proposed that Clb-dependent kinase activity acted in a positive feedback loop to activate and maintain CLB2 (CLB cluster) transcription. We sought to identify the mechanism by which Cdc28-Clb kinases activated G2–M transcription by identifying targets of this kinase that were part of the transcriptional complexes that formed at CLB cluster promoters. As part of this study, we showed that the CLB2UAS is subject to Clb kinase regulation, implicating Mcm1p, forkhead transcription factors and Ndd1p as potential targets. Although we found no role for Cdk regulation through the C terminus of Fkh2p, a critical residue in Ndd1p (T319) required for its function and recruitment to Fkh2p was shown to be phosphorylated by a Cdc28-Clb-dependent mechanism. The ability to phosphorylate this residue in vitro by purified Cdk complexes supports the model that Cdk-Clb kinase acts directly on Ndd1p. It is possible that other components of the transcriptional machinery, including Mcm1p and Fkh2p, are targeted by Cdks through nonconsensus sites or by other cell-cycle regulated mitotic kinases acting in collaboration with Cdk's.
Although Fkh2p-dependent mechanisms control G2–M transcription, we also determined that FKH1-dependent CLB cluster control (i.e., FKH1 Δfkh2) is cell-cycle regulated and dependent on Clb kinase activity (D. Reynolds and S. Dalton, unpubl.). Hence, mitotic Cdk activity has a role in both Fkh1p and Fkh2p-dependent transcriptional control.
An intriguing finding of this work was that Ndd1p T319 phosphorylation was influenced by the cdc28-1n mutant, which in vitro retains kinase activity (Surana et al. 1991). Our results, however, clearly show that its activity in vivo is compromised at the restrictive temperature. The possibility that this is an indirect effect on Ndd1p is unlikely because similar results were obtained in the clb2 ts mutant. The issue of correlating in vitro and in vivo kinase activity is a difficult one, particularly when dealing with conditional mutants in Cdks. It has been proposed that many preformed Cdc28pts-cyclin complexes are not readily inactivated upon shift to the restrictive temperature, leaving the possibility that alleles such as cdc28-1n may be defective in some aspect of kinase activity in vivo, even though they display robust activity in vitro at restrictive temperature (Amon et al. 1997). For example, kinase localization, substrate access, and assembly of Cdks into complexes required for in vivo activity, could account for differences between in vivo and in vitro activity. Subtle changes in the levels of kinase activity in vivo may also have pronounced biological effects that are not seen using conventional in vitro kinase assays. These factors could contribute to normal phosphorylation of some substrates but the inability to phosphorylate others. These points emphasize the need to evaluate the phosphorylation status of substrates in vivo, as has been performed in this report.
In light of recent observations that forkhead transcription factors are involved in G2/M transcriptional control in mammalian cells, it will be interesting to determine if the pathways identified in this report are also conserved (Alvarez et al. 2001).
Materials and methods
Strains, media, reagents, and yeast genetic protocols
All yeast strains were derivatives of W303 (MATa or MATα ho ade2-1 trp1-1 can1-100 leu2-3, 112 his3-11, 15 ura3-1 ssd1) except where otherwise indicated and are listed in Table 1. Yeast strains were grown in YEP medium or the appropriate selective medium as described previously (Dalton and Treisman 1992; Maher et al. 1995; Kumar et al. 2000). Single-copy integrations were confirmed by Southern blot or PCR analysis. Gene disruptions, yeast transformations, and other yeast techniques were by standard methods (Guthrie and Fink 1991). Liquid culture and qualitative plate-based assays used to determine β-galactosidase activity were as described (Dalton and Treisman 1992; Maher et al. 1995).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303 1a | MATaho ade2-1 trp1-1 can1-100 leu2-3 his3-11 ura3-52 ssd1 | Rodney Rothstein |
| US262 | MATandd1::LEU2 pGAL-NDD1 | Loy et al. 1999 |
| MK146 | MATandd1::LEU2 pGAL-NDD1 Δfkh1::TRP1 fkh2::HIS3 | Koranda et al. 2000 |
| MK465 | MATandd1::LEU2 pGAL-NDD1 FKH1 fkh2::HIS3 | Koranda et al. 2000 |
| S272 | MATacdc28-1n | Surana et al. 1991 |
| S287 | MATabar1 ura3::CLB2uas-ubiYLacZ | Maher et al. 1995 |
| S315 | MATa, clb1 Δ clb2-VI clb3::TRP1 clb4::HIS3 ura3::CLB2uas-ubiYLacZ | Amon et al. 1993 |
| S466 | MATabar1 fkh1::LEU2 ura3::CLB2uas-ubiYLacZ | Kumar et al. 2000 |
| S508 | MATabar1 fkh2::LEU2 ura3::CLB2uas-ubiYLacZ | This study |
| S599 | US262 pADH.NDD1 | This study |
| S600 | S599 fkh2::fkh26myc-TRP1 | This study |
| S601 | S599 fkh2::fkh2ΔC.6myc-TRP1 | This study |
| S602 | S599 fkh2::fkh2Δ6Cdk.6myc-TRP1 | This study |
| S610 | MATafkh1::TRP1 fkh2::HIS3 | This study |
| S618 | S466 fkh2::fkh2ΔC-TRP1 | This study |
| S619 | S618 ndd1::HIS3 | This study |
Plasmid constructions and generation of yeast strains
Except where specified, all constructs have been described previously (Maher et al. 1995; Kumar et al. 2000). Strains expressing 6myc epitope-tagged Fkh2p, Fkh2pΔC, and Fkh2pΔ6Cdk were generated by integration of tagged open reading frame-containing fragments at the FKH2 locus (Kumar et al. 2000). The FKH2 open reading frame and its derivatives were epitope-tagged by inserting a 6myc tag into a NotI site immediately before the STOP codon in YIplac204 (Gietz and Sugino 1988). Fkh2pΔC is Fkh2p amino acids 1–584.6myc and Fkh2pΔ6Cdk is full-length Fkh2p but carries alanine substitutions for serine or threonine residues at positions 589, 683, 697, 759, 771, and 833. These were integrated at the FKH2 locus in strain S508 (see Table 1).
To create a DNA construct to disrupt the NDD1 gene, the entire ORF of NDD1 was amplified by PCR, subcloned into pBS.KS+ (pBS.NDD1) into which a 1.7-kb HIS3 fragment was inserted between EcoRV and MunI sites (blunt). This construct was digested with EcoRI and cotransformed into recipient strains with pADH.NDD1, a plasmid expressing full length Ndd1p from the ADH1 promoter in pVT100-U (Vernet et al. 1987; 2μ, URA3). URA+ transformants were then replica plated onto histidine-selective plates and gene disruption confirmed by PCR analysis. Curing ndd1::HIS3 strains of pADH.NDD1 on five fluoroorotic acid (5-FOA) plates did not give rise to any viable cells except when FKH2 was also disrupted. Triple HA-tagged mutant or wild-type versions of Ndd1p (Ndd1p3HA) were expressed from an ADH1-promoter in YCplac22 (pT.NDD13HA; CEN, ARS1, TRP1) or, from a derivative of pADH.NDD1 that had a triple HA tag inserted immediately before the STOP codon (pADH.NDD13HA). The construct used for recombination to generate 6myc-tagged NDD1, at its own locus, was created by inserting a 6myc tag into a NotI site engineered immediately before the STOP codon in pBS.NDD1. The construct was linearized and cotransformed with pADH.NDD1. No growth or morphological defects were observed in the tagged strain thus confirming functionality.
A GALDBD–FKH2 fusion gene (G.FKH2HA) was constructed by inserting an 867-bp NcoI–BamHI cut PCR fragment into NcoI–BamHI cut pAS2 (URA3, 2 μ; Clontech): 5′-CC.ATG. GCC.. .AAT.ACC (FKH2 codon 291).TAG. GGA.TCC-3′. G.FKH2R87A.HA is identical to G.FKH2HA except that codon 87 was changed from arginine to alanine. GAL1UAS–LacZ, CYC1UAS–LacZ, RP39UAS–LacZ and ΔUAS–LacZ reporter plasmids (URA3, 2 μ) have been described previously (Dalton and Treisman 1992; Maher et al. 1995).
Fkh1p–Fkh2p fusion proteins were as follows: 1/2p-1, Fkh1p1–484–Fkh2p574–862; 1/2p-2, Fkh1p1–303–Fkh2p336–862; 1/2p-3, Fkh1p1–303–Fkh2p347–444–Fkh1p402–484; 1/2p-4, Fkh1p1–76–Fkh2p83–152–Fkh1p142–484. Hybrid genes were inserted into a YCplac33-based expression vector under control of the GAL1 promoter (CEN ARS1 URA3). Constructs were all evaluated in the Δfkh1 Δfkh2 strain to confirm that they were functional.
Northern analysis, cell synchronization, immunoprecipitations, kinase assays, and ChIP assays
Northern blot analysis, probe preparation, and α-factor synchronizations have been described previously (Dalton and Whit-bread 1995; Maher et al. 1995; Kumar et al. 2000). Whole-cell lysates were prepared as described previously by the bead beat method (Maher et al. 1995; Kumar et al. 2000).
All steps prior to and including immunoprecipitations were performed on ice or at 4°C. Cells were grown to midlog phase, collected by centrifugation, and resuspended in an equal volume of lysis buffer (50 mM Hepes at pH 7.9, 250 mM NaCl, 0.5 mM EDTA, 10% glycerol, 0.1% NP40, 1 mM DTT, 50 μM chymostatin, 25 μM leupeptin, 0.2 μM aprotinin, 0.5 mM PMSF, 50 μM TLCK, 100 μM TPCK, 0.4 mM NaF, 0.4 mM NaVO4) to which an equal volume of chilled, acid-washed glass beads was added. Cells were lysed in a bead-beater as previously described (Maher et al. 1995; Kumar et al. 2000) and the lysate clarified by centrifugation at 15,000 × g for 10 min. Immunoprecipitations were performed as described (Harlow and Lane 1988). Treatment of immune complexes with λ-phosphatase (NEB) and purified cdc2–cyclin B complexes (NEB) was as described previously (Hendrickson et al. 1996). Chromatin immunoprecipitation assays were performed essentially as described (Tanaka et al. 1997; Kumar et al. 2000).
Antibodies
Monoclonal antibodies raised against glyceraldehyde phosphate dehydrogenase (Gapdh) were provided by Trevor Lithgow (University of Melbourne). Polyclonal phospho-Ndd1pT319 (pNdd1T319) antibodies were generated using a phosphorylated peptide (CILQRpTPLRS-NH2) to immunize rabbits. The peptide was synthesized with a free cysteine at its N terminus for coupling to SulphoLink Coupling Gel (Pierce) as per the manufacturer's instructions. Phosphospecific antibodies were purified by first depleting antibodies that recognized the unphosphorylated peptide, followed by purification on a column to which the phosphorylated peptide was coupled. SulfoLink Coupling gel (5 mL of 50% slurry) was washed three times in coupling buffer (50 mM Tris-HCl at pH 8.5, 5 mM EDTA) and resuspended in a final volume of 8 mL. Five milligrams of peptide was added to the gel, mixed gently for 60 min at 4°C and uncoupled peptide removed by four additional washes in coupling buffer before being finally resuspended in 8 mL coupling buffer. Uncoupled site on the beads were blocked by the addition of 2-mercaptoethanol to 50 mM for 60 min at 4°C. The gel was washed three times in 8 mL coupling buffer, three times in 1 M NaCl and then four times in PBS. Up to 25 mL of crude sera was mixed with the unphosphorylated peptide column for 2 h at 4°C. This was then poured into a 10-mL EconoColumn (Bio-Rad) and the gel washed with 10 column volumes of PBS. If necessary, the flow-through was reapplied to the nonphospho column to further deplete antibodies recognizing nonphosphopeptide (monitored by ELISA assay). Antibodies in the flow-through were then applied to a second column to which phosphorylated peptide had been coupled. After loading of the flow-through to this column, the gel was washed with 10 column volumes of PBS and bound antibodies eluted with 7 mL 0.2 M glycine at pH 2.5. Each 500 μL fraction was collected into 100 μL of 2 M Tris-HCl at pH 7.5. Enrichment for phosphospecific antibodies was assayed by ELISA against phospho and nonphosphorylated peptide (Harlow and Lane 1988).
Acknowledgments
We thank Gustav Ammerer, Kim Nasmyth, and Uttam Surana for yeast strains and Jörg Heierhorst and Elaine Stead for review of the manuscript. This work was funded by the Australian Research Council through the Center for Molecular Genetics of Development.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1074103.
References
- Althoefer H., Schleiffer, A., Wassmann, K., Nordheim, A., and Ammerer A. 1995. Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 5917–5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez B., Martinez, A.C., Burgering, B.M., and Carrera, A.C. 2001. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature. 413: 744–747. [DOI] [PubMed] [Google Scholar]
- Amon A. 1997. Regulation of B-type cyclin proteolysis by Cdc28-associated kinases in budding yeast. EMBO J. 16: 2693–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A., Tyers, M., Futcher, B., and Nasmyth, K. 1993. Mechanisms that make the yeast cell cycle clock tick: G2 cyclins repress G1 cyclins. Cell 74: 993–1007. [DOI] [PubMed] [Google Scholar]
- Breeden L.L. 2000. Cyclin transcription. Timing is everything. Curr. Biol. 10: R586–R588. [DOI] [PubMed] [Google Scholar]
- Dalton S. and Treisman, R. 1992. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell 68: 597–612. [DOI] [PubMed] [Google Scholar]
- Dalton S., and Whitbread, L. 1995. Cell cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA replication in budding yeast. Proc. Natl. Acad. Sci. 92: 2514–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D. and Jackson, S.P. 2002. The FHA domain. FEBS Lett. 513: 58–66. [DOI] [PubMed] [Google Scholar]
- Durocher D., Taylor, I.A., Sarbassova, D., Haire, L.F., Westcott, S.L., Jackson, S.P., Smerdon, S.J., and Yaffe, M.B. 2000. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signalling mechanisms. Mol. Cell 6: 1169–1182. [DOI] [PubMed] [Google Scholar]
- Futcher B. 2002. Transcriptional regulatory networks and the yeast cell cycle. Curr. Opin. Cell Biol. 14: 676–683. [DOI] [PubMed] [Google Scholar]
- Guthrie C. and Fink, G. 1991. Guide to yeast genetics and molecular biology. Meth. Enzymol. Academic press, Vol. 194. [PubMed]
- Gietz R.D. and Sugino, A. 1988. New yeast—Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 30: 527–534. [DOI] [PubMed] [Google Scholar]
- Hammet A., Pike, B.L., Mitchelhill, K.I., Teh, T., Kobe, B., House, C.M., Kemp, B.E., and Heierhorst, J. 2000. FHA domain boundaries of the dun1p and rad53p cell cycle checkpoint kinases. FEBS Lett. 471: 141–146. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane, D. 1988. Antibodies: A laboratory manual. Cold Spring Harbor Laboratory Press, New York.
- Hendrickson M., Madine, M., Dalton, S., and Gautier, J. 1996. Phosphorylation of MCM4 by cdc2 protein kinase inhibits the activity of the minichromosome maintenance complex. Proc. Natl. Acad. Sci. 93: 12223–12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K. and Bucher, P. 1995. The FHA domain: A putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem. Sci. 20: 347–349. [DOI] [PubMed] [Google Scholar]
- Hollenhorst P.C., Bose, M.E., Mielke, M.R., Muller, U., and Fox, C.A. 2000. Forkhead genes in transcriptional silencing, cell morphology and the cell cycle: Overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics 154: 1533–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst P.C., Pietz, G., and Fox, C.A. 2002. Mechanisms controlling differential promoter-occupancy by the yeast forkhead Fkh1p and Fkh2p: Implications for regulating the cell cycle and differentiation. Genes & Dev. 15: 2445–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P. and Tyers, M. 2000. The fork'ed path to mitosis. Gen. Biol. 1: 1022.1–1022.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koranda M., Schleiffer, A., Endler, L., and Ammerer, G. 2000. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature 406: 94–98. [DOI] [PubMed] [Google Scholar]
- Kumar R., Reynolds, D., Shevchenko, A., Shevchenko, A., Goldstone, S.D., and Dalton, S. 2000. Forkhead transcription factors, Fkh1p and Fkh1p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 10: 896–906. [DOI] [PubMed] [Google Scholar]
- Lew D.J. and Reed, S.I. 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120: 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lee, G., Van Doren, S.R., and Walker, J.C. 2000. The FHA domain mediates phosphoprotein interactions. J. Cell. Sci. 113: 4143–4149. [DOI] [PubMed] [Google Scholar]
- Liao H., Byeon, I.J., and Tsai, M.D. 1999. Structure and function of a new phosphopeptide-binding domain the FHA2 of Rad53. J. Mol. Biol. 294: 1041–1049. [DOI] [PubMed] [Google Scholar]
- Loy C.J., Lydall, D., and Surana, U. 1999. NDD1, a high-dosage suppressor of cdc28-1n, is essential for expression of a subset of late-S-phase-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 3312–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D., Ammerer, G., and Nasmyth, K. 1991. A new role for MCM1 in yeast: Cell cycle regulation of SWI5 transcription. Genes & Dev. 5: 2405–2419. [DOI] [PubMed] [Google Scholar]
- Maher M., Cong, F., Kindelberger, D., Nasmyth, K., and Dalton, S. 1995. Cell cycle-regulated transcription of the CLB2 gene is dependent on Mcm1 and a ternary complex factor. Mol. Cell. Biol. 15: 3129–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pic A., Lim, F.-L., Ross, S.J., Veal, E.A., Johnson, A.L., Sultan, M.R.A., West, A.G., Johnston, L.H., Sharrocks, A.D., and Morgan, B.A. 2000. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 19: 3750–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike B.L., Hammet, A., and Heierhorst, J. 2001. Role of N-terminal forkhead-associated domain in the cell cycle checkpoint of the Rad53 kinase. J. Biol. Chem. 276: 14019–14026. [DOI] [PubMed] [Google Scholar]
- Spellman P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, R., Eisen, M.B., Brown, P.O., Botstein, D., and Futcher, B. 1998. Comprehensive identification of cell cycle regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9: 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., Coic, E., Zhou, Z., Durrens, P., and Haber, J.E. 2002. Saccharomyces forkhead protein Fkh1 regulates donor preference during mating-type switching through the recombination enhancer. Genes & Dev. 16: 2085–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U., Robitsch, H., Price, C., Schuster, T., Fitch, I., Futcher, A.B., and Nasmyth, K. 1991. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell 65: 145–161. [DOI] [PubMed] [Google Scholar]
- Surana U., Amon, A., Dowzer, C., McGrew, J., Byers, B., and Nasmyth, K. 1993. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 12: 1969–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Knapp, D., and Nasmyth, K. 1997. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90: 649–660. [DOI] [PubMed] [Google Scholar]
- Vernet T., Dignard, D., and Thomas, D.Y. 1987. A family of yeast expression vectors containing the phage f1 intergenic region. Gene 52: 225–233. [DOI] [PubMed] [Google Scholar]
- Zhu G., Spellman, P.T., Volpe, T., Brown, P.O., Botstein, D., and Futcher, B. 2000. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406: 90–94. [DOI] [PubMed] [Google Scholar]