Abstract
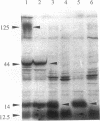
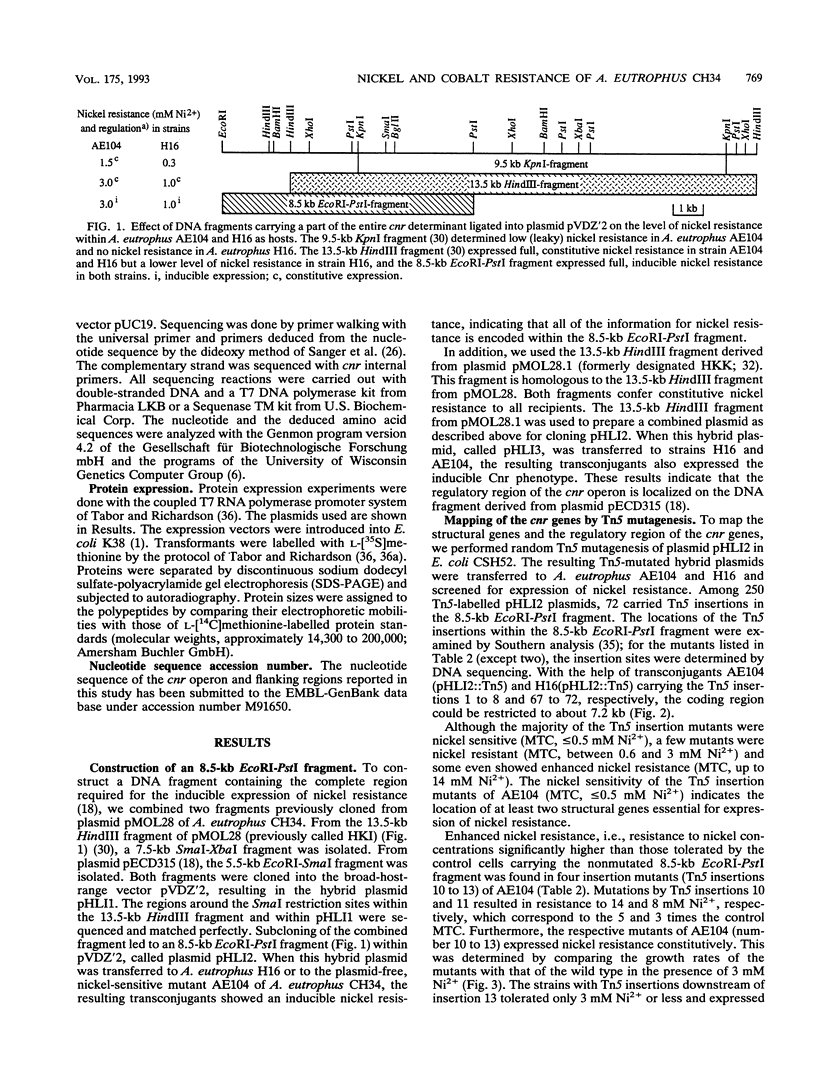
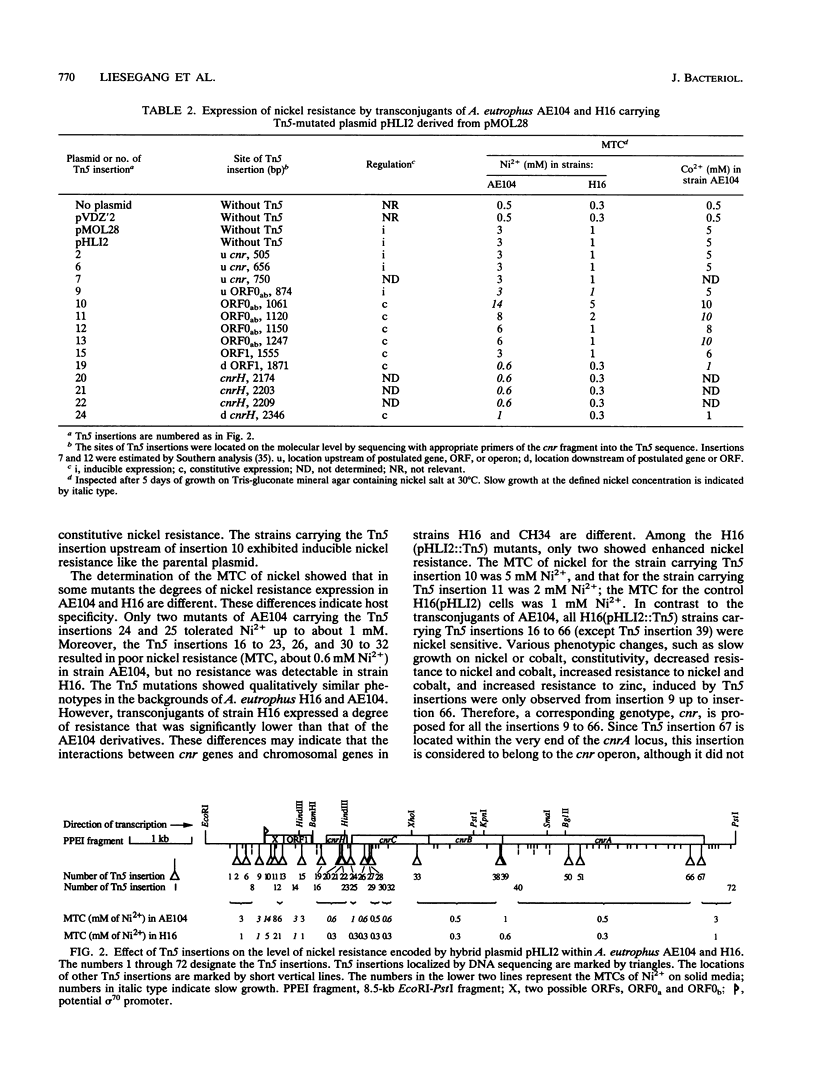
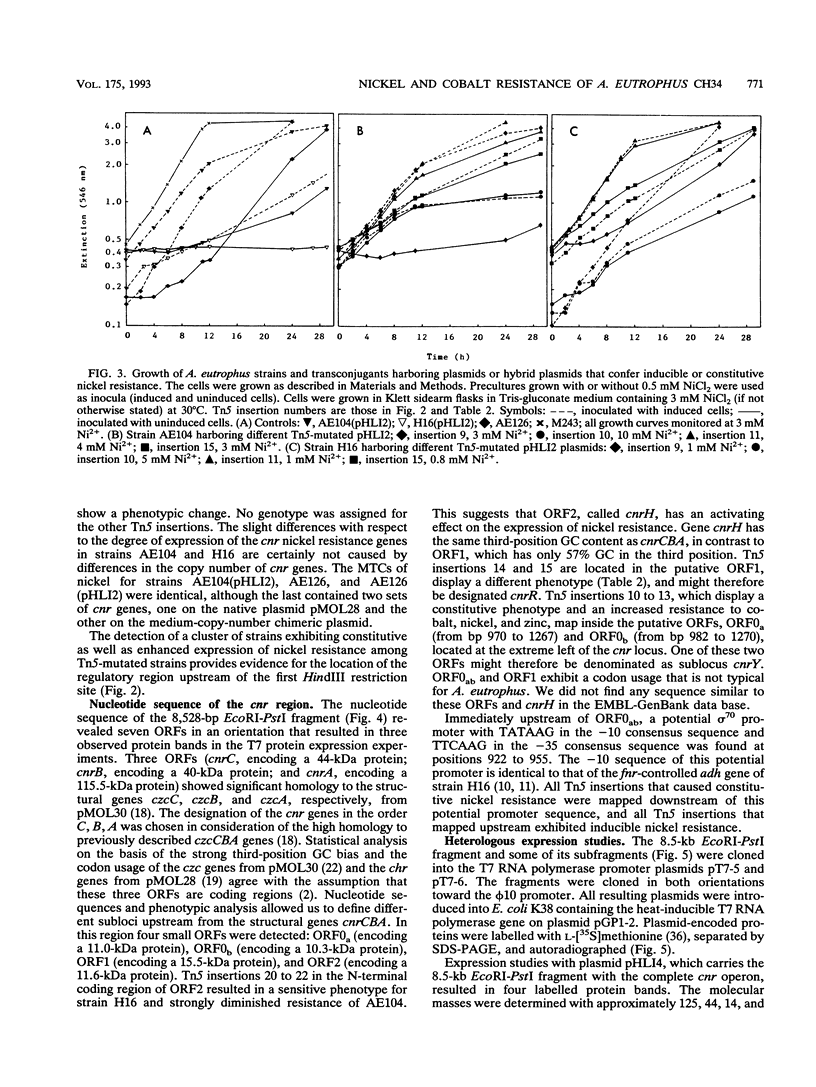
From pMOL28, one of the two heavy metal resistance plasmids of Alcaligenes eutrophus strain CH34, we cloned an EcoRI-PstI fragment into plasmid pVDZ'2. This hybrid plasmid conferred inducible nickel and cobalt resistance (cnr) in two distinct plasmid-free A. eutrophus hosts, strains AE104 and H16. Resistances were not expressed in Escherichia coli. The nucleotide sequence of the 8.5-kb EcoRI-PstI fragment (8,528 bp) revealed seven open reading frames; two of these, cnrB and cnrA, were assigned with respect to size and location to polypeptides expressed in E. coli under the control of the bacteriophage T7 promoter. The genes cnrC (44 kDa), cnrB (40 kDa), and cnrA (115.5 kDa) are probably structural genes; the gene loci cnrH (11.6 kDa), cnrR (tentatively assigned to open reading frame 1 [ORF]; 15.5 kDa), and cnrY (tentatively assigned to ORF0ab; ORF0a, 11.0 kDa; ORF0b, 10.3 kDa) are probably involved in the regulation of expression. ORF0ab and ORF1 exhibit a codon usage that is not typical for A. eutrophus. The 8.5-kb EcoRI-PstI fragment was mapped by Tn5 transposon insertion mutagenesis. Among 72 insertion mutants, the majority were nickel sensitive. The mutations located upstream of cnrC resulted in various phenotypic changes: (i) each mutation in one of the gene loci cnrYRH caused constitutivity, (ii) a mutation in cnrH resulted in different expression of cobalt and nickel resistance in the hosts H16 and AE104, and (iii) mutations in cnrY resulted in two- to fivefold-increased nickel resistance in both hosts. These genes are considered to be involved in the regulation of cnr. Comparison of cnr of pMOL28 with czc of pMOL30, the other large plasmid of CH34, revealed that the structural genes are arranged in the same order and determine proteins of similar molecular weights. The largest protein CnrA shares 46% amino acid similarity with CzcA (the largest protein of the czc operon). The other putative gene products, CnrB and CnrC, share 28 and 30% similarity, respectively, with the corresponding proteins of czc.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames G. F. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs. Membrane, soluble, and periplasmic fractions. J Biol Chem. 1974 Jan 25;249(2):634–644. [PubMed] [Google Scholar]
- Appleyard R K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics. 1954 Jul;39(4):440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Collard J. M., Provoost A., Taghavi S., Mergeay M. A new type of Alcaligenes eutrophus CH34 zinc resistance generated by mutations affecting regulation of the cnr cobalt-nickel resistance system. J Bacteriol. 1993 Feb;175(3):779–784. doi: 10.1128/jb.175.3.779-784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Chandrasekharappa S., Gill J. F., Chatterjee D. K., Chakrabarty A. M. A set of cassettes and improved vectors for genetic and biochemical characterization of Pseudomonas genes. Gene. 1987;57(1):61–72. doi: 10.1016/0378-1119(87)90177-6. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diels L., Mergeay M. DNA probe-mediated detection of resistant bacteria from soils highly polluted by heavy metals. Appl Environ Microbiol. 1990 May;56(5):1485–1491. doi: 10.1128/aem.56.5.1485-1491.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrossek D., Steinbüchel A., Schlegel H. G. Alcohol dehydrogenase gene from Alcaligenes eutrophus: subcloning, heterologous expression in Escherichia coli, sequencing, and location of Tn5 insertions. J Bacteriol. 1988 Nov;170(11):5248–5256. doi: 10.1128/jb.170.11.5248-5256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Mergeay M., Houba C., Gerits J. Extrachromosomal inheritance controlling resistance to cadmium, cobalt, copper and zinc ions: evidence from curing in a Pseudomonas [proceedings]. Arch Int Physiol Biochim. 1978 May;86(2):440–442. [PubMed] [Google Scholar]
- Mergeay M., Nies D., Schlegel H. G., Gerits J., Charles P., Van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985 Apr;162(1):328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergeay M. Towards an understanding of the genetics of bacterial metal resistance. Trends Biotechnol. 1991 Jan;9(1):17–24. doi: 10.1016/0167-7799(91)90007-5. [DOI] [PubMed] [Google Scholar]
- Nies A., Nies D. H., Silver S. Cloning and expression of plasmid genes encoding resistances to chromate and cobalt in Alcaligenes eutrophus. J Bacteriol. 1989 Sep;171(9):5065–5070. doi: 10.1128/jb.171.9.5065-5070.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies A., Nies D. H., Silver S. Nucleotide sequence and expression of a plasmid-encoded chromate resistance determinant from Alcaligenes eutrophus. J Biol Chem. 1990 Apr 5;265(10):5648–5653. [PubMed] [Google Scholar]
- Nies D. H., Nies A., Chu L., Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D. H. Resistance to cadmium, cobalt, zinc, and nickel in microbes. Plasmid. 1992 Jan;27(1):17–28. doi: 10.1016/0147-619x(92)90003-s. [DOI] [PubMed] [Google Scholar]
- Nies D., Mergeay M., Friedrich B., Schlegel H. G. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J Bacteriol. 1987 Oct;169(10):4865–4868. doi: 10.1128/jb.169.10.4865-4868.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen B. P. Recent advances in bacterial ion transport. Annu Rev Microbiol. 1986;40:263–286. doi: 10.1146/annurev.mi.40.100186.001403. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr, Werner P. K., Müller M. Insertion of proteins into bacterial membranes: mechanism, characteristics, and comparisons with the eucaryotic process. Microbiol Rev. 1989 Sep;53(3):333–366. doi: 10.1128/mr.53.3.333-366.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T., Stoppel R. D., Schlegel H. G. High-Level Nickel Resistance in Alcaligenes xylosoxydans 31A and Alcaligenes eutrophus KTO2. Appl Environ Microbiol. 1991 Nov;57(11):3301–3309. doi: 10.1128/aem.57.11.3301-3309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui R. A., Benthin K., Schlegel H. G. Cloning of pMOL28-encoded nickel resistance genes and expression of the genes in Alcaligenes eutrophus and Pseudomonas spp. J Bacteriol. 1989 Sep;171(9):5071–5078. doi: 10.1128/jb.171.9.5071-5078.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui R. A., Schlegel H. G., Meyer M. Inducible and constitutive expression of pMOL28-encoded nickel resistance in Alcaligenes eutrophus N9A. J Bacteriol. 1988 Sep;170(9):4188–4193. doi: 10.1128/jb.170.9.4188-4193.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S., Nucifora G., Chu L., Misra T. K. Bacterial resistance ATPases: primary pumps for exporting toxic cations and anions. Trends Biochem Sci. 1989 Feb;14(2):76–80. doi: 10.1016/0968-0004(89)90048-0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Measurement of DNA length by gel electrophoresis. Anal Biochem. 1979 Dec;100(2):319–323. doi: 10.1016/0003-2697(79)90235-5. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WALDE E. [Studies on growth and synthesis of stored substance by Hydrogenomonas]. Arch Mikrobiol. 1962;43:109–137. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]