Abstract
Food-ingested foreign DNA is not completely degraded in the gastrointestinal tract of mice. Phage M13mp18 DNA as a test molecule devoid of homology to mouse DNA was pipette-fed to or added to the food supply of mice. The fate of this foreign DNA in the animals was followed by several methods. In 84 animals, fragments of M13mp18 DNA were detected in the contents of the small intestine, the cecum (until 18 h), the large intestine, or the feces. In 254 animals, M13mp18 DNA fragments of up to 976 bp were found in blood 2–8 h after feeding. In buffer-fed control animals, M13mp18 DNA could not be detected. M13mp18 DNA fragments were traced by PCR in peripheral leukocytes and located by fluorescent in situ hybridization in about 1 of 1000 white cells between 2 and 8 h, and in spleen or liver cells up to 24 h after feeding, but not later. M13mp18 DNA could be traced by fluorescent in situ hybridization in the columnar epithelial cells, in the leukocytes in Peyer’s patches of the cecum wall, in liver cells, and in B cells, T cells, and macrophages from spleen. These findings suggest transport of foreign DNA through the intestinal wall and Peyer’s patches to peripheral blood leukocytes and into several organs. Upon extended feeding, M13mp18 DNA could be recloned from total spleen DNA into a λ vector. Among about 2.5 × 107 λ plaques, one plaque was isolated that contained a 1299 nucleotide pair fragment (nt 4736–6034) of sequence-identified M13mp18 DNA. This fragment was covalently linked to an 80 nt DNA segment with 70% homology to the mouse IgE receptor gene. The DNA from another λ plaque also contained mouse DNA, bacterial DNA, and rearranged λ DNA. Two additional plaques contained M13mp18 DNA fragments of at least 641 (nt 2660–3300) or 794 (nt 4640–5433) nucleotide pairs. The medical and evolutionary implications of these observations may be considerable.
Uptake and fate of foreign DNA ingested with the daily food supply in the gastrointestinal tract (GI) of mammals have hardly been investigated. The epithelial lining of the GI tract presents a huge surface for contacts with foreign DNA and/or DNA–protein complexes. We have shown that, after each meal, this surface is exposed for hours to DNA fragments of very different sources (1). In mice, about 1–2% of orally ingested phage M13mp18 DNA persist transiently as fragments between 1 and 7 h after feeding in the gut and feces. The bulk of these fragments ranges in size from 100 to 400 bp, with some extending to about 1700 bp. A very small proportion (≤0.1%) of the orally administered M13mp18 DNA has been detected between 2 and 8 h after feeding in the animals’ blood stream (1).
This project originated in the late 1980s as an extension of studies on the fate of adenovirus DNA in cell culture (refs. 2 and 3 and J. Schröer and W.D., unpublished work) and in adenovirus-transformed cells or in adenovirus type 12-induced hamster tumor cells (4, 5). The epithelial lining of the GI tract has been considered akin to a monolayer culture of mammalian cells exposed to foreign DNA. A pathway of foreign DNA from the contents of the murine GI tract through epithelial cells and Peyer patches in the intestinal wall has been investigated leading to peripheral white blood cells, to the spleen and liver. Under certain experimental conditions, the administered foreign (M13) DNA has been recloned from total spleen DNA where it has been found covalently linked to mouse DNA.
MATERIALS AND METHODS
Basic Experiment.
Most techniques employed were described previously (1). In all experiments described here, linearized or circular double-stranded M13mp18 DNA was orally administered to 3–6 (12)-month-old C57BL/6 male or female mice. At various times after feeding, mice were anesthetized with ether, the abdominal fur was thoroughly washed, and one set of instruments was used to open the peritoneal cavity, meticulously avoiding damage to the abdominal organs. A second set of surgical instruments was subsequently used to open the thoracic cavity by cutting the diaphragm. Finally, the still palpitating heart was punctured with a disposable 0.9-mm gauge needle, and blood (about 0.5–1 ml) was drawn with a syringe. Animal experiments were performed in accordance with permit 23.203.2K13 16/95 from the local state administration. Control analyses on abdominal fur washing fluids performed by using the dot blot hybridization and PCR methods revealed no evidence for detectable fur contamination.
Isolation of Peripheral Blood Leukocytes by Ficoll Gradient Centrifugation.
Freshly drawn blood was diluted with RPMI 1640 medium to a total volume of 3 ml. A volume of 3 ml Ficoll-Paque (Pharmacia, 5.7 g Ficoll 400 in 100 ml) was centrifuged at 1000 × g for 30 s in a 10-ml plastic tube. Subsequently, the blood was poured into the tube. The mixture was centrifuged at 1000 × g for 10 min at room temperature. Fractions were collected from the top with a Pasteur pipette (plasma and leukocytes) or with a syringe (erythrocytes). Leukocytes were washed twice with phosphate-buffered saline (PBS) (6).
Fractionation of Spleen Cells on Magnetic Bead Columns Loaded with Ferro-Magnetically Tagged Antibodies to Specific Surface Proteins (7).
At 18 h after feeding mice 50 μg of M13mp18 DNA, spleen cells were passed over magnetic bead columns loaded with specific monoclonal antibodies against CD4 on T helper cells (8), CD8 on cytotoxic T cells (9), CD45 on B cells (10), and CD11 on macrophages (11). These cell populations were found to be 78%, 91%, 99%, and 88% pure, respectively, by FACS scanning in a FACSort sorter (Becton Dickinson) using the cellquest computer program. DNA was extracted by standard methods from individual cell fractions.
Quantitation of Foreign DNA.
This method was as described in the legend to Fig. 1.
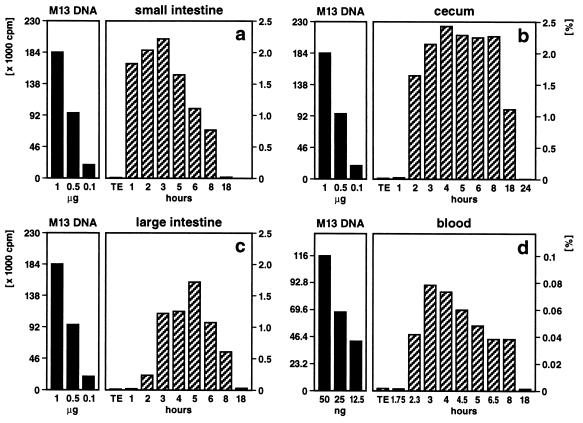
Figure 1.
Quantitation of M13mp18 foreign DNA detectable by dot blot hybridization in different segments of the intestine (a–c) or in peripheral blood (d). Fifty micrograms of M13mp18 DNA (19) (7250 bp) was pipette-fed to 2–6 (12)-month-old mice. As indicated, at times after feeding, the DNA was extracted (1) from the contents of the small intestine (a), the cecum (b), the large intestine (c), or from blood (d). The persisting M13mp18 DNA was identified by dot blot hybridization (1). Aliquot portions of the extracted DNA were fixed on a positively charged Qiagen nylon membrane and hybridized to 32P-labeled (20) M13mp18 DNA followed by autoradiography. In reconstitution experiments, amounts of M13mp18 DNA ranging from 5 ng to 1 μg were also bound to the membranes. Subsequently, autoradiographically localized dots were excised, and the amounts of 32P label membrane bound in these spots were determined by Cerenkov counting. Considering the fraction of the total reextracted DNA from gut segments or blood applied to the dot blot matrix, percentages (scales on right) of the 50 μg of orally administered M13mp18 test DNA were calculated (hatched bars). The scales in cpm and the solid bars on the left referred to the actual 32P Cerenkov cpm for reference M13mp18 DNA.
Histological Sections.
Organs were removed and immediately fixed in 4% formalin for 12 h at room temperature. Subsequently, the samples were embedded in Paraplast using an automated device (Tissue Tek VIP, Vogel) according to a standard schedule. Organs were sectioned at 2–5 μm thickness in a MICROM rotation microtom. Sections were spread on 55°C water, transferred to precleaned glass slides, and dried at 58°C for 30 min.
Fluorescent in Situ Hybridization (FISH) (12–15).
Sections were treated with xylol (10 min), ethanol (100%, 95%, 90%, 80%, 70%, 50%, and 30%, each for 2 min), PBS (5 min), 4% paraformaldehyde in PBS (20 min), PBS (2 × 5 min), proteinase K (20 μg/ml, 35 min), and subsequently dehydrated in a reversed ethanol series. Sections were then air dried. M13mp18 DNA as hybridization probe was nick-translated (16) using biotin 16-dUTP. The hybridized biotinylated probe was detected with fluorescein isothiocyanate-labeled avidin. In 500 μl of hybridization buffer, 2 μg of biotinylated M13mp18 DNA was dissolved, and 20 μl of this solution was incubated per section under a sealed coverslip at 95°C for 6 min for denaturation. After rapid cooling to −20°C for 1 min, sections were held at 42°C for 16–18 h in a humid chamber. Sections were then washed twice in 2 × SSC at 20°C and once in 0.1 × SSC at 42°C. One times SSC is 0.15 M NaCl/15 mM sodium citrate. Photographs of FISH experiments were taken in an Olympus Fluorescence Microscope BHS or a Zeiss Axioscope H with filter systems for multicolor photography.
Recloning of M13mp18 DNA Sequences from Total Spleen DNA.
Three feeding regimens were applied. (i) Mice were fed 50 μg of M13mp18 DNA once, and total spleen DNA was extracted 18 h later. (ii) Mice were fed 50 μg of M13mp18 DNA three times at 24-h intervals, and total spleen DNA was prepared 18 h after the last feeding. (iii) Mice were fed 50 μg of M13mp18 DNA daily for 1 week, and total spleen DNA was prepared 18 h after the last feeding. Total spleen DNA was extracted by the SDS/proteinase K phenol method (17), and subsequently cleaved with BamHI and ligated into the BamHI site in the polycloning segment of the λDASHII vector DNA. After in vitro packaging, plating of λ plaques, plaque transfer to Qiagen (Hilden, Germany) nylon membranes and hybridization to [32P]-nick-translated M13mp18 DNA, one positive clone was identified after feeding regimen ii in about 108 plaques and three positive clones were found after regimen iii in about 2.5 × 107 plaques. Positive plaques were replaqued three times, amplified, and recombinant λ DNA was prepared from CsCl-purified λ phage. The λDASHII vector plaques did not hybridize with M13mp18 DNA. Recombinant λ DNA was analyzed by restriction, nucleotide sequencing (18) and PCR. Recombinant λ DNA was also 32P-labeled by nick-translation and hybridized to restricted M13mp18 DNA or to genomic mouse DNA on Southern blot membranes to ascertain the nature of the recloned DNA sequences (see Fig. 5 b and c).
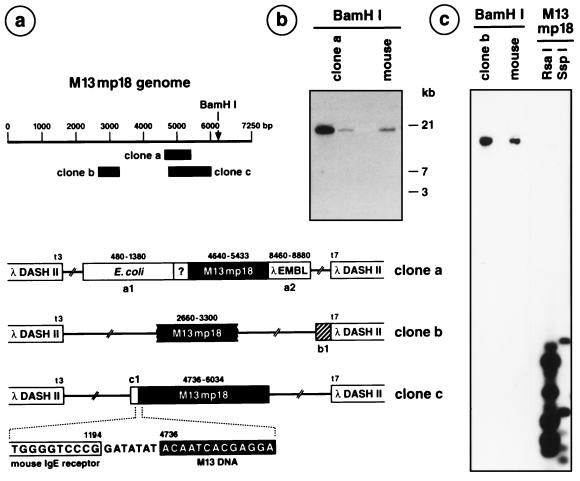
Figure 5.
Recloning of M13mp18 DNA from spleen DNA of mice fed with M13mp18 DNA. In brief, the segments of M13mp18 DNA present in the DNA of the M13mp18 DNA-positive recombinant λ clones were identified by hybridizing the 32P-labeled recombinant λ DNA to RsaI- or SspI-cleaved M13mp18 DNA (e.g., c, right lanes). On the basis of the known nucleotide sequence of M13mp18 DNA (19), primers were selected and the exact M13mp18 DNA represented was determined. By using primers from the known M13mp18 DNA (solid boxes in a), the adjacent nucleotide sequences were determined and identified by computer-aided National Center for Biotechnology Information searches. The λDASHII clone a was derived from feeding experiment ii, the clones b and c from the schedule iii experiment. (a) Map of the linearized M13mp18 genome. The locations of the nucleotide sequence-identified M13mp18 segments present in λDASHII clones a–c are indicated as solid blocks. In clone a, a1 represents the Escherichia coli DNA sequence of an ATP-dependent Clp-protease, the question mark a sequence of unknown, possibly mouse, origin. The segment a2 carries rearranged DNA from the left arm of λ DNA. In clone b, the DNA sequence shown as a hatched segment b1 exhibits homologies to mouse DNA (129/sv Clora cell 10 kDa, mCC10 gene). (b) Clone a DNA was 32P-labeled by nick-translation and hybridized to BamHI-restricted clone a DNA or genomic mouse DNA on a Southern blot under stringent hybridization (68°C, 2 × SSC) and wash conditions (68°C, 2 × 15 min with 2 × SSC, 0.1% SDS; 2 × 25 min with 0.2 × SSC, 0.1% SDS). (c) A similar experiment for clone b DNA, which is also shown to hybridize to m13mp18 DNA and to genomic mouse DNA. The 80 bp DNA segment in c1 was 70% homologous to the mouse IgE receptor gene.
RESULTS AND DISCUSSION
Quantitation of Food-Ingested Foreign DNA in Different Organ Systems.
Fragments of orally administered M13mp18 DNA were recovered from the feces (37 animals), different parts of the gut (42 animals), the blood (254 animals), or spleen and liver (42 animals). After gastric passage, >95% of the M13mp18 DNA was lost. Fragments of M13mp18 DNA between 100 and ≈1700 bp in size were recovered reproducibly from the feces or from the contents of different intestinal segments. M13mp18 DNA fragments between 194 and 976 bp were identified by PCR in the blood of 254 DNA-fed animals. From organs of TE buffer-fed control animals, M13mp18 sequences were never isolated. By comparison to the results of reconstitution experiments it was determined that the small intestine contained about 2.2–0.7% (1–8 h after feeding), the cecum 2.4–1.1% (2–18 h), the large intestine 0.2–1.7% (2–8 h), and blood about 0.04–0.08% (2.3–8 h) of the M13mp18 DNA orally administered (Fig. 1).
Blood was obtained by heart puncture from buffer- or from M13mp18 DNA-fed mice at 2–8 h after mock-feeding or feeding, respectively. Blood cells were fractionated by centrifugation of freshly drawn blood on a Ficoll gradient. The fraction of peripheral leukocytes contained a mixture of granulocytes, monocytes, and lymphocytes as shown by Giemsa staining (data not shown). The DNA was extracted from leukocytes by standard protocols (17, 21) and tested for the presence of M13mp18 DNA fragments by PCR (22) using synthetic oligodeoxyribonucleotide primers (map positions on M13mp18 DNA, Fig. 2c). The results of PCR analyses and Southern blot transfers (23, 24) with leukocyte-extracted DNA from 25 animals fed with M13mp18 DNA were similar to those shown in Fig. 2a and revealed authentic M13mp18 DNA in leukocytes at 4 h after feeding. DNA extracted from the plasma, erythrocyte, or wash fractions, or from the leukocytes of five buffer-fed control animals did not yield M13mp18 DNA-specific signals. The white cell population from spleen was fractionated by the magnetic bead method at 18 h after feeding. The results of PCR and hybridization analyses revealed M13mp18 DNA in DNA from cytotoxic T cells, B cells, and macrophages (Fig. 2a).
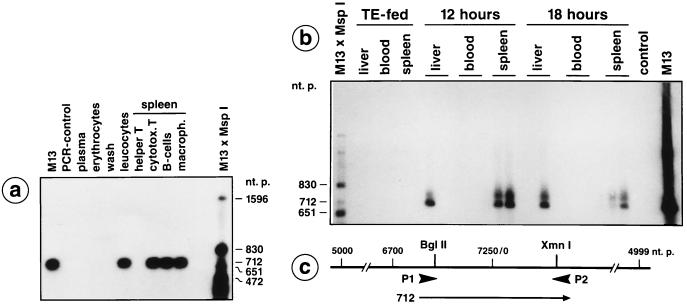
Figure 2.
Detection of M13mp18 DNA fragments in peripheral blood leukocytes, in liver, and in different spleen cells by PCR. (a) A male C57BL/6 mouse was pipette-fed 50 μg of M13mp18 DNA. At 4 h after feeding, blood was drawn by heart puncture and the plasma, erythrocyte, and leukocyte fractions were prepared by Ficoll gradient centrifugation. The leukocytes were washed twice in Tris-saline or PBS and incubated for 2 min in 0.15 M NaCl, 10 mM Tris·HCl (pH 7.5), 2 mM MgCl2, 1% Nonidet P-40. The thus liberated nuclei were sedimented, resuspended in 10 mM Tris·HCl (pH 7.5), 1 mM EDTA, 1% SDS, 1.3 mg/ml proteinase K, and incubated for 2 h at 37°C. Nucleic acids were then prepared by phenol/chloroform extraction. Similarly, the plasma, erythrocyte, and wash fractions were extracted for possibly present DNA. By using the synthetic oligodeoxyribonucleotide primers P1 and P2 designated on the M13mp18 DNA map in c, the indicated M13mp18 DNA segments were amplified as described earlier (1). (a) At 18 h after feeding a different animal, the different spleen cells were fractionated by the magnetic bead method as described. DNA from the designated cell fractions was prepared and PCR analyzed. (b) Detection of M13mp18 DNA in spleen and liver cells by PCR at 12 or 18 h after feeding. DNA samples from TE-fed animals or from blood of animals 12 or 18 h after feeding M13mp18 DNA did not contain M13mp18 DNA. The PCR products were blotted to positively charged membranes, and M13mp18 DNA was identified by hybridization to 32P-labeled M13mp18 DNA and by autoradiography.
We concluded that small amounts of the M13mp18 DNA ingested orally by mice could be recovered as fragments of maximally 712 bp, 2–8 h after feeding, both from the cytoplasm and the nuclei of peripheral leukocytes and up to 18 h after feeding in cytotoxic T cells, B cells, and macrophages from spleen (Fig. 2a). Thus, foreign DNA in peripheral blood and spleen was predominantly present in leukocytes, perhaps because of their defense functions. The leukocytes carrying the foreign DNA had possibly migrated from the Peyer’s patches in the gut wall to the bloodstream and spleen.
FISH.
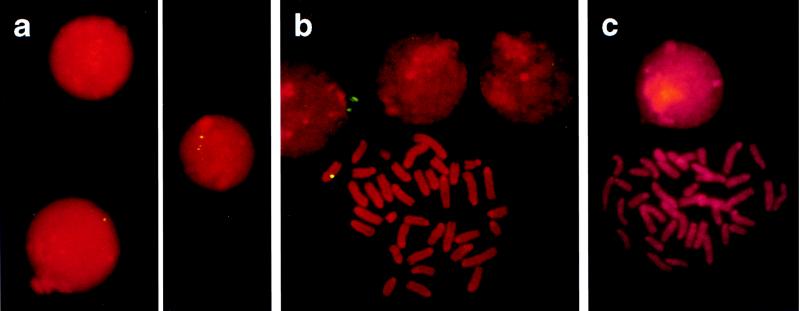
By using the FISH technique (12–15), M13mp18 DNA could be visualized in about 1 of 1000 peripheral leukocytes, between 2 and 8 h after feeding (Fig. 3a). Chromosome squash preparations were made from spleen cells between 12 and 18 h after feeding and 48 h after cultivating them (Fig. 3b). M13mp18 DNA-specific signals were elicited in a small subpopulation of cells or on metaphase chromosomes, albeit frequently only on one of the chromatids (Fig. 3b). In cells prepared from TE buffer-fed animals, such signals were never obtained in nuclei of leukocytes or on chromosomes of spleen cells (Fig. 3c). These data corroborated the PCR results on DNA from isolated leukocytes (Fig. 2a). M13mp18 sequences persisted at least until 18 h after feeding in spleen and liver cells as shown by PCR (Fig. 2b). In minute quantities, fragments of foreign DNA ingested with food could transgress the intestine–blood barrier in mice and could be found in about 0.1% of the peripheral leukocytes up to 8 h after feeding. Foreign DNA persisted in spleen and liver up to 18 h after feeding.
Figure 3.
Ingested foreign DNA detected in peripheral leukocytes and in spleen cells by FISH. (a) Peripheral blood leukocytes from M13mp18 DNA-fed mice were analyzed by the FISH technique using biotinylated M13mp18 DNA as hybridization probe and FITC-tagged avidin for the detection of biotinylated DNA (12–15). (b) Spleen cells from M13mp18 DNA-fed mice were incubated at 37°C for 48 h in RPMI 1640 medium containing 10% fetal calf serum, 2.4 mM glutamine, 10 mM 2-mercaptoethanol, 30 μg/ml lipopolysaccharide, 300 μg/ml penicillin, 100 μg/ml streptomycin. Subsequently, ethidium bromide (10 μg/ml) and colcemid (50 μg/ml) were added, and incubation was continued for 2 h at 37°C. Finally, cells were incubated for 20 min at 37°C in 0.075 KCl, fixed at 4°C in methanol:acetic acid (3:1), and attached to precleaned glass slides. (c) Control with spleen cells from a TE-fed mouse.
Uptake of Foreign DNA Through the Intestinal Wall Mucosa.
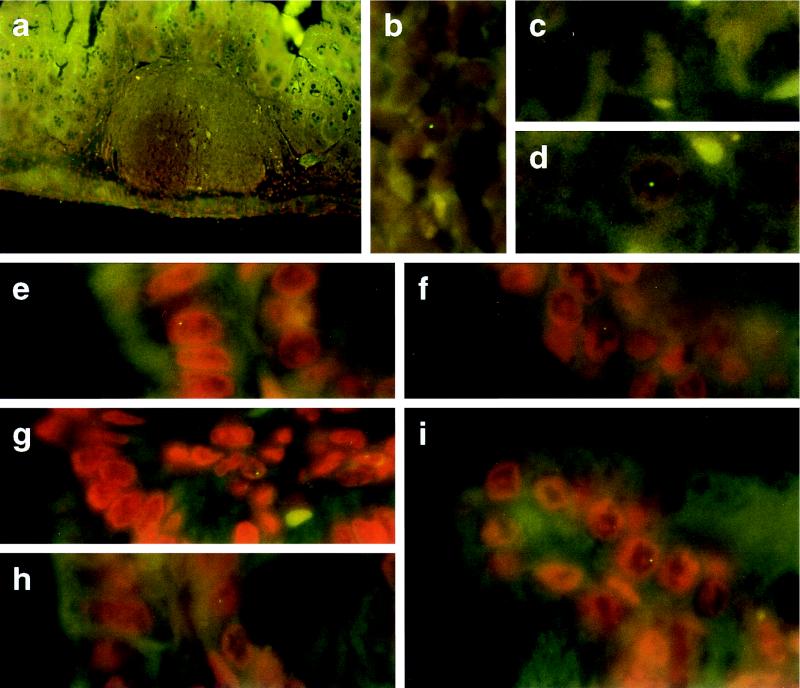
We traced the pathway of the foreign DNA through the intestinal wall by FISH on histological sections of the cecum of mice fed with M13mp18 DNA 3–8 h earlier. M13mp18 DNA-specific signals could be identified in the nuclei of columnar epithelia in the cecum (Fig. 4 e–i) and in leukocytes in Peyer’s patches of the intestinal wall (Fig. 4b); similarly in leukocytes in spleen (Fig. 3b) analyzed 3–12 h after feeding, and in liver (Fig. 4d) analyzed 3–8 h after feeding. The M13mp18 DNA-containing liver cells were frequently close to blood vessels. Controls from TE-fed mice (Fig. 4c) were negative. Between 40 and 60 cecum, liver or spleen sections from a total of 12 different M13mp18 DNA-fed animals, and from 5 TE-fed controls were investigated with similar results. The data implied that foreign DNA was taken up by intestinal wall epithelia, reached leukocytes in the Peyer’s patches, and was then transported into the organism via peripheral blood leukocytes. Foreign DNA was found as late as 18 h after feeding in spleen and liver.
Figure 4.
Histological sections through the cecum wall (a, b, and e–i) or through liver (c and d) from mice that had been fed 50 μg of M13mp18 DNA (a, b, d, and e–i) or TE-buffer (c). Tissue samples were prepared as described. M13mp18 DNA was identified by FISH. Tissues were counterstained in propidium iodide solution (200 ng/ml of H2O). (a and b) Peyer’s patch inside the cecum wall 5 h after feeding; (e–i) cecum epithelia from mice 3–5 h after feeding; (c) liver from a TE-fed control mouse; (d) liver from an animal 6 h after feeding M13mp18 DNA. (a, ×50; b–i, ×1250.)
M13mp18 DNA Segments Could Be Recloned from Spleen DNA of Mice Fed with This Phage DNA.
We had to cope with the detection of minute amounts of foreign DNA. It was therefore necessary to reclone M13mp18 DNA from mouse organ DNA to assess the validity of the results and to determine the state of foreign DNA in the host cells. As described in Materials and Methods, three feeding schedules were employed. The results were as follows: (i) After a singular feeding of 50 μg of M13mp18 DNA, 108 λDASHII clones were screened without success for M13mp18 recombinants. (ii) After three consecutive feedings at daily intervals, one recombinant clone was isolated among 108 λ plaques (Fig. 5, clone a). (iii) Daily feedings for 1 week produced three recombinant clones among 2.5 × 107 λ plaques (Fig. 5, clones b and c). These experiments were performed in a different building at the university to minimize the possibility of inadvertant contaminations with the test M13mp18 DNA.
The M13mp18 DNA in recombinant λ DNA preparations (from feeding schedules ii and iii) was resequenced (18) by using appropriate oligodeoxyribonucleotide primers and an Applied Biosystems A373 DNA sequencer. The M13mp18 DNA segments present in recombinant λ clones had previously been identified by hybridizing the [32P]-labeled recombinant DNA to a mixture of authentic M13mp18 DNA fragments, which had been generated with MspI, RsaI, or SspI and Southern blotted (23, 24). Clone a was isolated through regimen ii and contained the nucleotide 4640–5433 (794 bp) fragment of M13mp18 DNA. The flanking nucleotide sequences were as indicated in Fig. 5a and were identified with the blastn program in a GenBank search. The nucleotide sequence immediately adjacent to nucleotide 4640 of M13mp18 was determined. It represented an unknown sequence not found in any of the gene bank searches (December 1996) and was possibly of mouse origin. Feeding schedule iii resulted in the isolation of three M13mp18 DNA-positive λ clones. Their 32P-labeled DNA hybridized to M13mp18 DNA and to genomic mouse DNA (Fig. 5, clones b and c). The M13mp18 DNA segments recloned in two of the three positive clones were assigned to nucleotides 2660–3300 (Fig. 5, clone b) and 4736–6034 (Fig. 5, clone c), respectively. The three analyzed λ clones contained different M13mp18 DNA segments, which did not encompass the BamHI restriction site used for cloning. This fact further argued against contamination scenarios. In clone c, 80 bp of DNA with 70% homology to the mouse IgE receptor gene (33) was found adjacent to nucleotide 4736 of M13mp18 DNA, linked via a GATATAT sequence of unknown origin (Fig. 5, clone c). The isolation of these recombinant clones supports the conclusion that the authentic M13mp18 DNA administered by feeding can be retrieved from mouse spleen DNA (Fig. 5, clones a–c) and can be covalently linked to mouse DNA (Fig. 5, clone c).
Maximal Longevity of M13mp18 DNA in Mouse Tissues.
After a single feeding episode of 50 μg of M13mp18 DNA to mice, this foreign DNA could be detected by the dot blot (cecum) or PCR method (all other organs) up to 18 h in the contents of the cecum (Fig. 1b), up to 8 h in DNA from peripheral white blood cells, up to 24 h in DNA from spleen and liver (data not shown). DNA was extracted from blood, spleen and liver also at 42 h, 68 h, and 1 week after a single feeding and was found to be devoid of M13mp18 DNA.
Conclusions.
The design for this project was derived from studies on the de novo methylation of foreign (adenovirus) DNA integrated into established mammalian genomes (17, 25–27). The methylation of integrated foreign DNA was considered an ancient cellular defense mechanism against the activity of foreign genes (28). The need for such a defense mechanism has been sought in the constant intake of foreign DNA for which the most likely portal of entry into an organism is its GI tract. The mechanism of foreign DNA uptake by the intestinal wall epithelia is unknown; it could be akin to the incorporation of foreign DNA by cells in culture (3). Peyer’s patch M cells (29), membranous epithelial cells, are a major route by which pathogenic microorganisms like Salmonella typhimurium, Vibrio cholerae, or Shigella, could penetrate the intestinal epithelial barrier (30–32). It will be interesting to investigate whether M cells are also a target for the interaction with macromolecules that have resisted digestion in the GI tract. Apparently, the intestinal tract is not an absolute barrier against the uptake of macromolecules or even of microorganisms.
Orally administered M13mp18 DNA can be recloned from spleen DNA in linkage to DNA with 70% homology to the mouse IgE receptor gene (Fig. 5, clone c). At least in one case, the DNA recloned from spleen also contained bacterial DNA possibly transported from the gut through the intestinal wall by a route akin to M13mp18 test DNA (Fig. 5, clone a). Blood or spleen leukocytes could thus serve as a depository for much foreign DNA regularly penetrating the intestinal mucosa. Linkage of food-ingested foreign DNA to mouse DNA in the spleen invites speculation about general evolutionary consequences and medically relevant implications in terms of mutagenesis and carcinogenesis.
Acknowledgments
We thank Rudi Manz and Andreas Radbruch (Institute of Genetics, Cologne) for introducing R.S. to the magnetic bead technique, Martin Hansmann (Cologne/Frankfurt) for help with histological sections, Herbert Pfister for allowing us to do some of the experiments at the Institute of Virology, Irmgard Hölker for the preparation of synthetic oligodeoxyribonucleotides, Petra Böhm for expert editorial work, and Udo Ringeisen for graphic design. This research was supported by the Deutsche Forschungsgemeinschaft through Grants SFB274-A1 and Do 165/12-1.
Footnotes
Abbreviations: GI, gastrointestinal tract; FISH, fluorescent in situ hybridization.
References
- 1.Schubbert R, Lettmann C, Doerfler W. Mol Gen Genet. 1994;242:495–504. doi: 10.1007/BF00285273. [DOI] [PubMed] [Google Scholar]
- 2.Doerfler W. Proc Natl Acad Sci USA. 1968;60:636–643. doi: 10.1073/pnas.60.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groneberg J, Brown D T, Doerfler W. Virology. 1975;64:115–131. doi: 10.1016/0042-6822(75)90084-7. [DOI] [PubMed] [Google Scholar]
- 4.Doerfler W. Adv Cancer Res. 1995;66:313–344. doi: 10.1016/s0065-230x(08)60259-6. [DOI] [PubMed] [Google Scholar]
- 5.Doerfler W. Biochim Biophys Acta Rev Cancer. 1996;1288(2):F79–F99. doi: 10.1016/0304-419x(96)00024-8. [DOI] [PubMed] [Google Scholar]
- 6.Dulbecco R L, Vogt M. J Exp Med. 1954;99:167–182. doi: 10.1084/jem.99.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miltenyi S, Müller W, Weichel W, Radbruch A. Cytometry. 1990;11:231–239. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 8.Dialnyas D P, Quan Z S, Wall K A, Pierres A, Quintans J, Loken M R, Pierres M, Fitch F W. J Immunol. 1983;131:2445–2451. [PubMed] [Google Scholar]
- 9.Ledbetter J A, Herzenberg L A. Immunol Rev. 1979;47:63–90. doi: 10.1111/j.1600-065x.1979.tb00289.x. [DOI] [PubMed] [Google Scholar]
- 10.Coffmann R L. Immunol Rev. 1982;69:5–23. doi: 10.1111/j.1600-065x.1983.tb00446.x. [DOI] [PubMed] [Google Scholar]
- 11.Springer T, Galfre G, Secher D S, Milstein C. Eur J Immunol. 1978;8:539–551. doi: 10.1002/eji.1830080802. [DOI] [PubMed] [Google Scholar]
- 12.Lichter P, Tang C-J C, Call K, Hermanson G, Evans G A, Housman D, Ward D C. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- 13.Heller H, Kämmer C, Wilgenbus P, Doerfler W. Proc Natl Acad Sci USA. 1995;92:5515–5519. doi: 10.1073/pnas.92.12.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer zu Altenschildesche G, Wilgenbus P, Heller H, Tjia S T, Doerfler W. Chromosoma. 1996;104:341–344. doi: 10.1007/BF00337222. [DOI] [PubMed] [Google Scholar]
- 15.Kessel M, Gruss P. Cell. 1991;67:89–104. doi: 10.1016/0092-8674(91)90574-i. [DOI] [PubMed] [Google Scholar]
- 16.Rigby P W J, Dieckmann M, Rhodes C, Berg P. J Mol Biol. 1977;113:237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- 17.Sutter D, Westphal M, Doerfler W. Cell. 1978;14:569–585. doi: 10.1016/0092-8674(78)90243-x. [DOI] [PubMed] [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Messing J, Vieira J. Gene. 1983;19:269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- 20.Feinberg A P, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 21.Behn-Krappa A, Doerfler W. Hum Mol Genet. 1993;2:993–999. doi: 10.1093/hmg/2.7.993. [DOI] [PubMed] [Google Scholar]
- 22.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 23.Southern E M. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 24.Koetsier P, Schorr J, Doerfler W. BioTechniques. 1993;15:260–262. [PubMed] [Google Scholar]
- 25.Sutter D, Doerfler W. Proc Natl Acad Sci USA. 1980;77:253–256. doi: 10.1073/pnas.77.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doerfler W. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- 27.Orend G, Knoblauch M, Kämmer C, Tjia S T, Schmitz B, Linkwitz A, Meyer zu Altenschildesche G, Maas J, Doerfler W. J Virol. 1995;69:1226–1242. doi: 10.1128/jvi.69.2.1226-1242.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doerfler W. Biol Chem Hoppe-Seyler. 1991;372:557–564. [PubMed] [Google Scholar]
- 29.Owen R L, Jones A L. Gastroenterology. 1974;66:189–203. [PubMed] [Google Scholar]
- 30.Clark M A, Jepson M A, Simmons N L, Hirst B H. Histochem J. 1994;26:271–280. [PubMed] [Google Scholar]
- 31.Jones B, Pascopella L, Falkow S. Curr Opin Immunol. 1995;7:474–478. doi: 10.1016/0952-7915(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 32.Siebers A, Finlay B B. Trends Microbiol. 1996;4:22–29. doi: 10.1016/0966-842x(96)81501-0. [DOI] [PubMed] [Google Scholar]
- 33.Richards M L, Katz D H, Lin F T. J Immunol. 1991;147:1067–1074. [PubMed] [Google Scholar]