Abstract
FliG, FliM, and FliN are three proteins of Salmonella typhimurium that affect the rotation and switching of direction of the flagellar motor. An analysis of mutant alleles of FliM has been described recently (H. Sockett, S. Yamaguchi, M. Kihara, V. M. Irikura, and R. M. Macnab, J. Bacteriol. 174:793-806, 1992). We have now analyzed a large number of mutations in the fliG and fliN genes that are responsible for four different types of defects: failure to assembly flagella (nonflagellate phenotype), failure to rotate flagella (paralyzed phenotype), and failure to display normal chemotaxis as a result of an abnormally high bias to clockwise (CW) or counterclockwise (CCW) rotation (CW-bias and CCW-bias phenotypes, respectively). The null phenotype for fliG, caused by nonsense or frameshift mutations, was nonflagellate. However, a considerable part of the FliG amino acid sequence was not needed for flagellation, with several substantial in-frame deletions preventing motor rotation but not flagellar assembly. Missense mutations in fliG causing paralysis or abnormal switching occurred at a number of positions, almost all within the middle one-third of the gene. CW-bias and CCW-bias mutations tended to segregate into separate subclusters. The null phenotype of fliN is uncertain, since frameshift and nonsense mutations gave in some cases the nonflagellate phenotype and in other cases the paralyzed phenotype; in none of these cases was the phenotype a consequence of polar effects on downstream flagellar genes. Few positions in FliN were found to affect switching: only one gave rise to the CW mutant bias and only four gave rise to the CCW mutant bias. The different properties of the FliM, FliG, and FliN proteins with respect to the processes of assembly, rotation, and switching are discussed.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blair D. F., Berg H. C. Mutations in the MotA protein of Escherichia coli reveal domains critical for proton conduction. J Mol Biol. 1991 Oct 20;221(4):1433–1442. doi: 10.1016/0022-2836(91)90943-z. [DOI] [PubMed] [Google Scholar]
- Blair D. F., Kim D. Y., Berg H. C. Mutant MotB proteins in Escherichia coli. J Bacteriol. 1991 Jul;173(13):4049–4055. doi: 10.1128/jb.173.13.4049-4055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret R. B., Borkovich K. A., Simon M. I. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- Chun S. Y., Parkinson J. S. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988 Jan 15;239(4837):276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- Francis N. R., Irikura V. M., Yamaguchi S., DeRosier D. J., Macnab R. M. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc Natl Acad Sci U S A. 1992 Jul 15;89(14):6304–6308. doi: 10.1073/pnas.89.14.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Aizawa S., Dean G. E., Macnab R. M. Identification of the M-ring protein of the flagellar motor of Salmonella typhimurium. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7483–7487. doi: 10.1073/pnas.84.21.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imae Y., Atsumi T. Na+-driven bacterial flagellar motors. J Bioenerg Biomembr. 1989 Dec;21(6):705–716. doi: 10.1007/BF00762688. [DOI] [PubMed] [Google Scholar]
- Jones C. J., Homma M., Macnab R. M. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989 Jul;171(7):3890–3900. doi: 10.1128/jb.171.7.3890-3900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. J., Macnab R. M. Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J Bacteriol. 1990 Mar;172(3):1327–1339. doi: 10.1128/jb.172.3.1327-1339.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagishi I., Müller V., Williams A. W., Irikura V. M., Macnab R. M. Subdivision of flagellar region III of the Escherichia coli and Salmonella typhimurium chromosomes and identification of two additional flagellar genes. J Gen Microbiol. 1992 Jun;138(6):1051–1065. doi: 10.1099/00221287-138-6-1051. [DOI] [PubMed] [Google Scholar]
- Khan S., Dapice M., Reese T. S. Effects of mot gene expression on the structure of the flagellar motor. J Mol Biol. 1988 Aug 5;202(3):575–584. doi: 10.1016/0022-2836(88)90287-2. [DOI] [PubMed] [Google Scholar]
- Kihara M., Homma M., Kutsukake K., Macnab R. M. Flagellar switch of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989 Jun;171(6):3247–3257. doi: 10.1128/jb.171.6.3247-3257.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R. M. Examination of bacterial flagellation by dark-field microscopy. J Clin Microbiol. 1976 Sep;4(3):258–265. doi: 10.1128/jcm.4.3.258-265.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
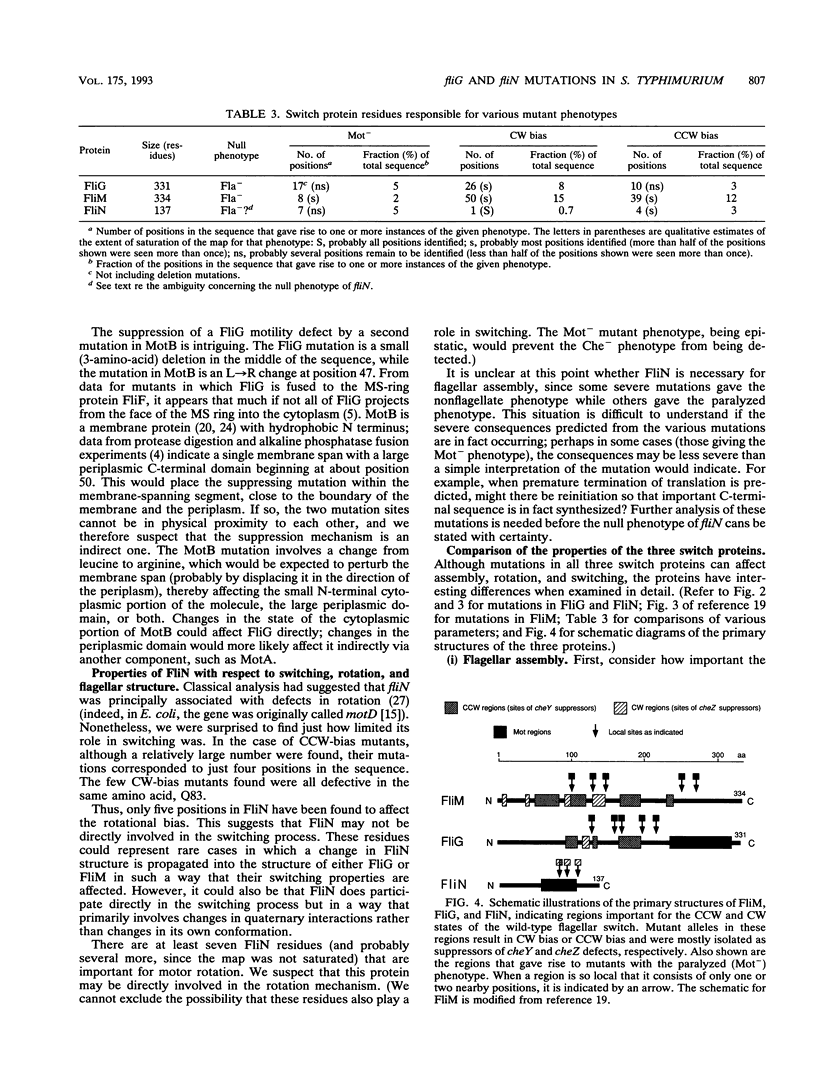
- Magariyama Y., Yamaguchi S., Aizawa S. Genetic and behavioral analysis of flagellar switch mutants of Salmonella typhimurium. J Bacteriol. 1990 Aug;172(8):4359–4369. doi: 10.1128/jb.172.8.4359-4369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Parker S. R., Talbert P. B., Houts S. E. Interactions between chemotaxis genes and flagellar genes in Escherichia coli. J Bacteriol. 1983 Jul;155(1):265–274. doi: 10.1128/jb.155.1.265-274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid S., Matsumura P., Eisenbach M. Restoration of flagellar clockwise rotation in bacterial envelopes by insertion of the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7157–7161. doi: 10.1073/pnas.83.19.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman S. J., Meyers M., Volz K., Matsumura P. A chemotactic signaling surface on CheY defined by suppressors of flagellar switch mutations. J Bacteriol. 1992 Oct;174(19):6247–6255. doi: 10.1128/jb.174.19.6247-6255.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sockett H., Yamaguchi S., Kihara M., Irikura V. M., Macnab R. M. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J Bacteriol. 1992 Feb;174(3):793–806. doi: 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stader J., Matsumura P., Vacante D., Dean G. E., Macnab R. M. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J Bacteriol. 1986 Apr;166(1):244–252. doi: 10.1128/jb.166.1.244-252.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T., Oosawa K., Aizawa S. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J Mol Biol. 1992 Oct 5;227(3):672–677. doi: 10.1016/0022-2836(92)90216-7. [DOI] [PubMed] [Google Scholar]
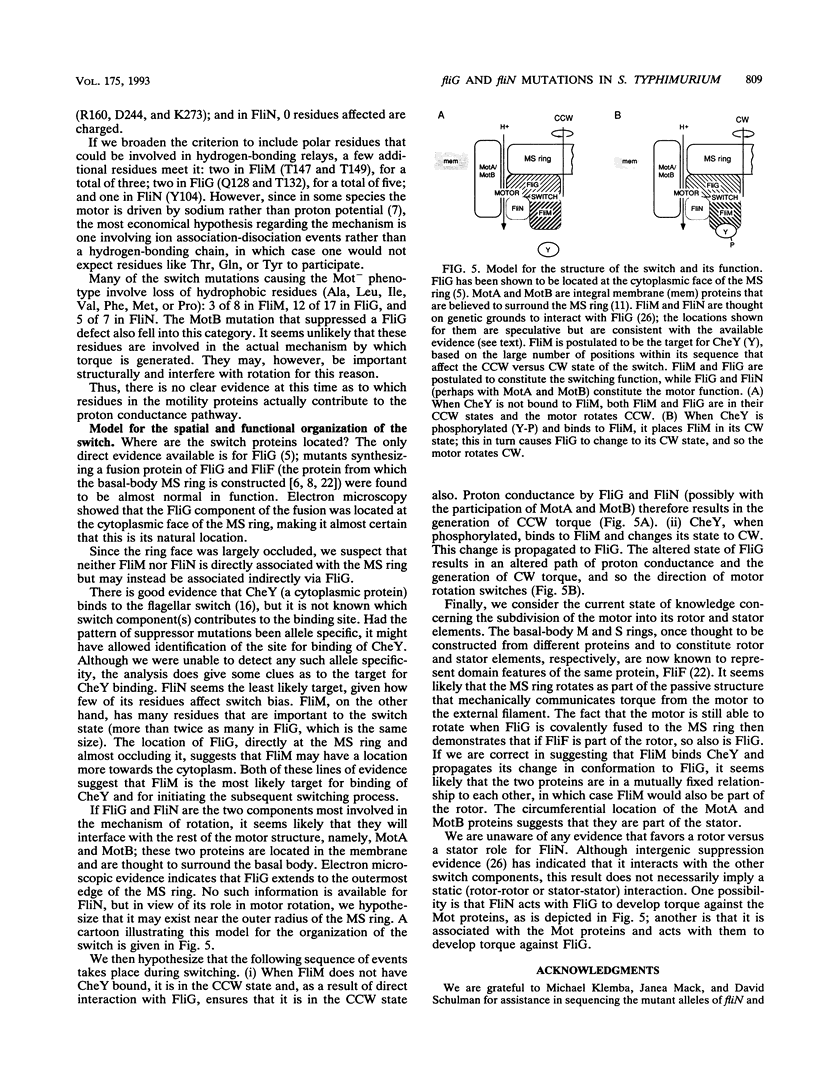
- Vogler A. P., Homma M., Irikura V. M., Macnab R. M. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J Bacteriol. 1991 Jun;173(11):3564–3572. doi: 10.1128/jb.173.11.3564-3572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. L., Macnab R. M. Co-overproduction and localization of the Escherichia coli motility proteins motA and motB. J Bacteriol. 1990 Jul;172(7):3932–3939. doi: 10.1128/jb.172.7.3932-3939.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Aizawa S., Kihara M., Isomura M., Jones C. J., Macnab R. M. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986 Dec;168(3):1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Fujita H., Ishihara A., Aizawa S., Macnab R. M. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol. 1986 Apr;166(1):187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Iino T., Horiguchi T., Ota K. Genetic analysis of fla and mot cistrons closely linked to H1 in Salmonella abortusequi and its derivatives. J Gen Microbiol. 1972 Apr;70(1):59–75. doi: 10.1099/00221287-70-1-59. [DOI] [PubMed] [Google Scholar]


