Abstract
Tuberous sclerosis complex (TSC) is a genetic disease caused by mutation in either TSC1 or TSC2. The TSC1 and TSC2 gene products form a functional complex and inhibit phosphorylation of S6K and 4EBP1. These functions of TSC1/TSC2 are likely mediated by mTOR. Here we report that TSC2 is a GTPase-activating protein (GAP) toward Rheb, a Ras family GTPase. Rheb stimulates phosphorylation of S6K and 4EBP1. This function of Rheb is blocked by rapamycin and dominant-negative mTOR. Rheb stimulates the phosphorylation of mTOR and plays an essential role in regulation of S6K and 4EBP1 in response to nutrients and cellular energy status. Our data demonstrate that Rheb acts downstream of TSC1/TSC2 and upstream of mTOR to regulate cell growth.
Keywords: TSC2, Rheb, mTOR, S6K, GAP, tuberous sclerosis complex
Tuberous sclerosis complex (TSC) is characterized by hamartoma formation in a wide range of tissues and is a genetic disorder caused by mutations in either the TSC1 or the TSC2 gene (Kwiatkowski 2003). The most serious clinical complications are mental retardation, epilepsy, and autism caused by tumor growth in the brain (Gomez 1991). Other symptoms include renal dysfunction, dermatological abnormalities, and heart problems. Heterozygous deletions of either TSC1 or TSC2 produce similar phenotypes and increase tumor incidence particularly in the kidney, whereas homozygous deletion of either gene is embryonic lethal (Au et al. 1998; Kobayashi et al. 2001). Genetic data from Drosophila show that TSC1 and TSC2 negatively regulate cell growth and cell size (Gao and Pan 2001; Potter et al. 2001; Tapon et al. 2001).
Recent studies have established that TSC1/TSC2 inhibits phosphorylation of the ribosomal S6 kinase (S6K) and the eukaryotic initiation factor 4E-binding protein (4EBP1), two key regulators of translation (Goncharova et al. 2002; Inoki et al. 2002; Manning et al. 2002; Tee et al. 2002). Phosphorylation of S6K and 4EBP1 enhances translation (Gingras et al. 1999). How TSC1/TSC2 regulates the phosphorylation of S6K and 4EBP1 is a key question yet to be answered. The mammalian target of rapamycin (mTOR) is directly responsible for phosphorylation of both S6K and 4EBP1 (Brown et al. 1995; Hara et al. 1998). Recent studies have suggested that TSC1/TSC2 acts through mTOR to regulate the phosphorylation of S6K and 4EBP1 (Gao et al. 2002; Inoki et al. 2002; Tee et al. 2002). Consistent with this model is that TSC1 and TSC2 are important for cellular nutrient response, which requires the function of mTOR (Gao et al. 2002). However, it is unclear how TSC1/TSC2 inhibits mTOR activity.
The C-terminal region of TSC2 displays significant homology to the Rap GTPase-activating protein (GAP; The European Chromosome 16 Tuberous Sclerosis Consortium 1993). In fact, GAP activity of TSC2 toward Rap1 and Rab5 had previously been reported (Wienecke et al. 1995; Xiao et al. 1997). However, the reported GAP activity was extremely low, and the functional significance is unclear. Rheb is a small GTPase initially isolated as a Ras homolog enriched in brain (Yamagata et al. 1994) and is widely expressed. Rheb shares higher sequence identity with Ras than with Rho family members. The biological function of mammalian Rheb is unclear. Conflicting studies report that Rheb both inhibits and activates the Raf-MAP kinase pathway (Clark et al. 1997; Yee and Worley 1997). Interestingly, mutation of the Rheb gene in Schizosaccharomyces pombe produces a phenotype similar to nutrient starvation, indicating that Rheb is possibly involved in nutrient signaling (Mach et al. 2000).
In this report, we provide direct biochemical data demonstrating that TSC2 has GAP activity toward Rheb in vitro. Furthermore, we show that TSC2 regulates Rheb-GTP levels in vivo. We show that one of the roles of Rheb is to stimulate phosphorylation of S6K and 4EBP1, two of the best characterized cellular downstream targets of TSC1/TSC2. Both the effector domain and the GTP binding are essential for Rheb function. The ability of Rheb to stimulate S6K phosphorylation requires the function of mTOR, indicating that mTOR acts downstream of Rheb. Consistent with this is that Rheb stimulates the phosphorylation of mTOR on serine residue 2448. Furthermore, our data support that Rheb plays an important role in cellular responses to energy limitation and nutrient starvation. Together, this study provides a model that Rheb is a direct downstream target of TSC2 and acts upstream of mTOR to regulate translation and cell growth.
Results and Discussion
TSC2 stimulates GTP hydrolysis of Rheb
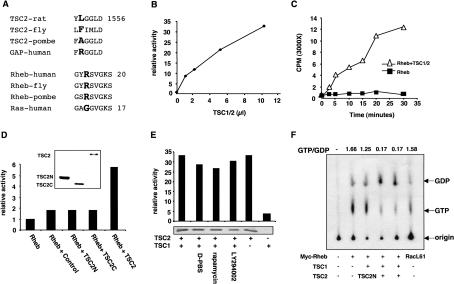
The C-terminal region of TSC2 contains a putative GAP domain with significant homology to RapGAP (The European Chromosome 16 Tuberous Sclerosis Consortium 1993; Scheffzek et al. 1998). However, the precise physiological and biochemical functions of the putative TSC2 GAP domain have not been demonstrated. Interestingly, a high frequency of TSC-associated mutations occurs in the C-terminal putative GAP domain of TSC2, indicating the GAP domain may be important for TSC2 function (Jin et al. 1996; Momose et al. 2002; Kwiatkowski 2003). To study the biochemical functions of TSC2, we expressed and purified the GAP domain of TSC2 in Escherichia coli and tested for GAP activity toward the Ras subfamily GTPases (Ras, Rap, TC21, and Rheb) and the Rho family GTPases (Rac and Cdc42). Our in vitro assays failed to detect significant GAP activity, whereas the positive control of RasGAP1 showed activity toward Ras (data not shown). It is worth noting that the catalytic arginine residue essential for GAP activity in the Rap GAP family is not conserved in TSC2 (Fig. 1A), suggesting that TSC2 may have no GAP activity (Scheffzek et al. 1998). It is also possible that TSC2 has GAP activity, but a different arginine or a completely different catalytic mechanism may be used.
Figure 1.
TSC2 has GAP activity toward Rheb. (A) Rheb and TSC2 are an unusual GTPase and GAP. The catalytic active-site arginine (the residue in bold) in RapGAP is not conserved in TSC2. Rheb contains an arginine residue (the residue in bold) at the position corresponding to codon 12 of Ras, which has a glycine. Numbers indicate positions of the last residues. (B) Immunoprecipitated TSC1/TSC2 stimulates GTP hydrolysis of Rheb. Increasing amounts (in microliters) of immunoprecipitated TSC1/TSC2 were incubated with GST-Rheb at room temperature for 20 min. Release of free 32P-phosphate was measured by radioactive counting. Background activity of a control immunoprecipitation was subtracted. The basal GTPase activity of Rheb was arbitrarily set as 1. (C) Time-dependent GTP hydrolysis of Rheb stimulated by TSC1/TSC2. GTP hydrolysis was determined in the absence (▪) and presence (▵) of TSC1/TSC2. (D) Neither the N-terminal nor the C-terminal region of TSC2 has GAP activity toward Rheb. Two truncated TSC2 constructs (TSC2N, amino acids 1-1007; TSC2C, amino acids 1008-1765) were coexpressed with TSC1 and immunoprecipitated and assayed for GAP activity in vitro. The amount of HA-TSC2 used in this assay is equivalent to 1 μL in B. The expression levels of these proteins were determined by Western blot. (E) TSC2, but not TSC1, has GAP activity. Transfected HEK293 cells were untreated or treated with D-PBS, rapamycin (20 nM), or LY294002 (50 μM) for 30 min as indicated. The relative amount of TSC2 used in the GAP assay was determined by Western blot and is shown below each bar. (F) TSC1/TSC2 decreases the Rheb-GTP levels in vivo. Myc-Rheb was transfected in HEK293 cells and labeled with 32P-phosphate. Myc-Rheb was immunoprecipitated, and the bound nucleotides were eluted and resolved on a cellulose plate. Cotransfection with TSC1 and TSC2 are indicated. Myc-RacL61 was included as a control. The ratio of GTP/GDP was calculated by the formula GTP counts/3 divided by GDP counts/2, and indicated on top of each lane.
To further test for GAP activity of TSC2, we expressed TSC2 together with TSC1 in HEK293 cells. The TSC1/TSC2 complex was purified by immunoprecipitation, and in vitro GAP activity was determined using various small GTPases. We made particular efforts on Rheb because deletion of Rheb in S.pombe produces phenotypes similar to nutrient starvation (Mach et al. 2000). We found that the TSC1/TSC2 complex stimulated the GTP hydrolysis of Rheb (Fig. 1B,C) but not Ras. The GAP activity of TSC2 requires the full-length protein because neither the N-terminal nor the C-terminal region, which contains the GAP homology domain, displayed activity toward Rheb (Fig. 1D). To exclude the possibility that the GAP activity detected in the TSC1/TSC2 immunoprecipitation is due to another coprecipitated GAP, we made TSC2-R1701Q and TSC2-R1703Q mutants, which are found in TSC patients, and detected no GAP activity (data not shown). Therefore, we conclude that TSC2 is an Rheb GAP. The lack of GAP activity of TSC2C could be due to the requirement of residues in the N-terminal region of TSC2 for GAP activity or incorrect folding of TSC2C. The ability of TSC2 to stimulate GTPase activity of Rheb is very interesting because TSC2 does not have the conserved active-site arginine found in other related RapGAPs and Rheb is an unusual GTPase (Fig. 1A). Almost all Ras family GTPases contain a conserved glycine residue at the position equivalent to codon 12 in Ras (Lowy and Willumsen 1993). Mutation of this glycine to almost any residue, including valine found in many cancers, activates Ras family GTPases. The RasV12 mutant is known to be resistant to down-regulation by RasGAP (Trahey and McCormick 1987). Interestingly, Rheb has an arginine residue at the corresponding position codon 12 and has a much lower basal GTPase activity than Ras (data not shown). However, the GTPase activity of Rheb can be effectively stimulated by TSC1/TSC2. Therefore, Rheb and TSC2 are unusual GTPases and GAPs, respectively.
To determine whether TSC1 is required for the GAP activity of TSC2, HA-TSC2 alone was expressed in HEK293 cells and immunoprecipitated. The precipitated TSC2, which may also coimmunoprecipitate a small amount of endogenous TSC1, showed significant GAP activity toward Rheb (Fig. 1E). In contrast, immunoprecipitated TSC1 showed a very low level of GAP activity toward Rheb possibly due to coprecipitation of endogenous TSC2 protein. These observations indicate that TSC1 stabilizes TSC2 in vivo, but is not directly required for GAP activity. TSC1/TSC2 was also immunoprecipitated under various conditions, including treatments with rapamycin, LY294002, and D-PBS, which are all known to inhibit phosphorylation of S6K and 4EBP1. We observed that these treatments did not directly affect the GAP activity of immunoprecipitated TSC1/TSC2 (Fig. 1E).
The effect of TSC2 on Rheb GTP levels was also determined in vivo. The nucleotide-binding status of Rheb was determined by in vivo labeling with 32P-phosphate. Interestingly, Rheb contains a high basal GTP level (Fig. 1F). The high GTP level of wild-type Rheb is consistent with it having an arginine at a position equivalent to codon 12 of Ras (Fig. 1A). Expression of TSC1/TSC2 decreased the GTP/GDP ratio of cotransfected Rheb ∼10-fold (Fig. 1F). As a control, expression of the N-terminal domain of TSC2 had no significant effect on the GTP level of Rheb. These data provide in vivo evidence that TSC2 is an Rheb GAP.
Rheb stimulates the phosphorylation of S6K and 4EBP1
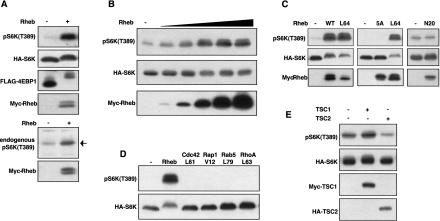
If Rheb is a key downstream target of TSC2, Rheb should stimulate the phosphorylation of S6K and 4EBP1. Expression of the wild-type Rheb induced a dramatic increase in S6K phosphorylation and mobility shift of 4EBP1 (Fig. 2A). Rheb also increased the phosphorylation of endogenous S6K in transfected HEK293 cells. In contrast, Rheb did not stimulate ERK phosphorylation (data not shown). The high activity of wild-type Rheb is consistent with it being predominantly GTP bound (Fig. 1F). Rheb also stimulates S6K phosphorylation in a dose-dependent manner (Fig. 2B).
Figure 2.
Rheb stimulates phosphorylation of S6K and 4EBP1. (A) Rheb stimulates phosphorylation of S6K and 4EBP1. HA-S6K and Flag-4EBP1 were transfected into HEK293 cells in the presence or absence of Myc-Rheb (100 ng). Phosphorylation of S6K was determined by phosphospecific antibody, pS6K(T389), whereas phosphorylation of 4EBP1 was determined by mobility shift (upper panels). Phosphorylation of endogenous S6K was also determined (lower panels). (B) Rheb stimulates phosphorylation of S6K in a dose-dependent manner. HA-S6K (15 ng) was cotransfected with 0, 2, 5, 20, 50, and 100 ng of Myc-Rheb. (C) The effector domain of Rheb is required to stimulate S6K phosphorylation. HA-S6K was cotransfected with wild-type Rheb (100 ng), RhebL64 (40 ng), Rheb-5A (200 ng), and RhebN20 (300 ng). The relative expression of Rheb mutants is also shown. (D) Stimulation of S6K phosphorylation by different GTPases. Constitutively active mutants (100 ng for each) of Cdc42, Rap1, Rab5, RhoA, and the wild-type Rheb (40 ng) were used to stimulate phosphorylation of S6K. (E) TSC2, but not TSC1, inhibits S6K phosphorylation. HA-S6K was coexpressed with either Myc-TSC1 or HA-TSC2. Phosphorylation of S6K was determined.
Five residues between 38 and 42 in the effector domain of Rheb were mutated to alanines to produce Rheb5A. We found that mutation of the effector domain completely abolished the ability of Rheb to stimulate S6K phosphorylation (Fig. 2C). We also tested constitutively active Rheb (RhebL64) and observed that RhebL64 is approximately two to three times more active than the wild type (Fig. 2C). In an attempt to create a dominant-negative mutant, we made an RhebN20. Surprisingly, RhebN20 does not show any dominant-negative effect nor does it stimulate S6K (Fig. 2C). A similar dominant-negative mutant of RasN17 is effective in blocking Ras function through sequestration of the Ras guanine nucleotide exchange factor (GEF; Lowy and Willumsen 1993). This observation suggests that Rheb may not need a GEF because of its high basal GTP level and that GTP binding is required to stimulate S6K phosphorylation.
To determine the specificity of Rheb, several other GTPases were examined, including Cdc42, Rap1, RhoA, and Rab5. Constitutively active mutants of these GTPases fail to stimulate S6K phosphorylation (Fig. 2D). These results demonstrate that Rheb is specific to stimulate S6K phosphorylation. We tested the effect of TSC1 or TSC2 alone on S6K phosphorylation. Expression of TSC2, but not TSC1, significantly decreased the basal phosphorylation of cotransfected S6K (Fig. 3E), consistent with the fact that TSC2 alone has GAP activity. The ability of TSC2 but not TSC1 to inhibit S6K indicates that TSC2 is the active component of the TSC1/TSC2 complex, whereas TSC1 may have regulatory functions, such as localization and stabilization. Furthermore, these observations suggest that inhibition of endogenous Rheb by TSC2 decreases S6K phosphorylation, thus supporting the importance of Rheb in S6K regulation.
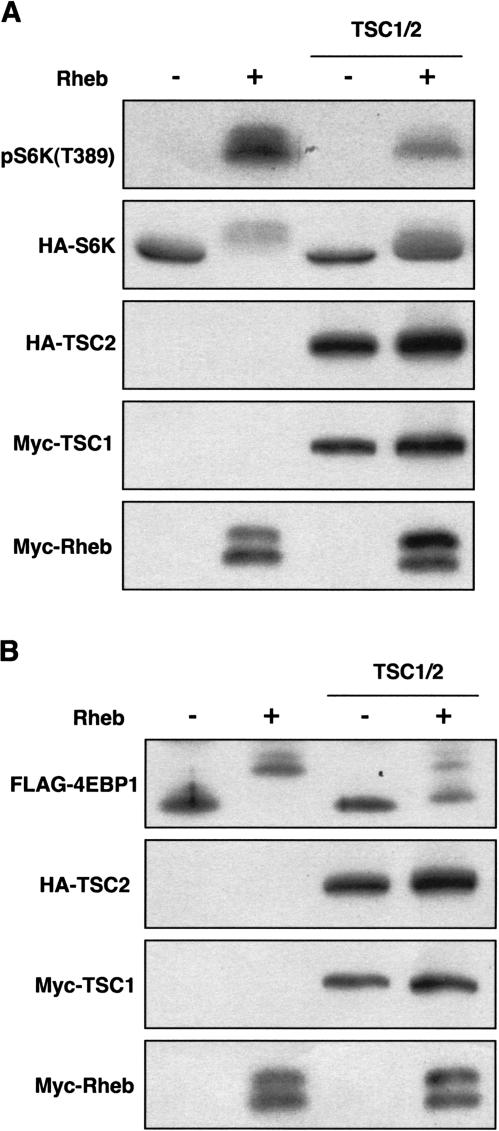
Figure 3.
TSC1/TSC2 inhibits Rheb function in vivo. (A) TSC1/TSC2 inhibits the ability of Rheb to induce S6K phosphorylation. (B) TSC1/TSC2 inhibits the ability of Rheb to induce 4EBP1 phosphorylation.
TSC1/TSC2 inhibits Rheb function
The functional relationship between TSC1/TSC2 and Rheb was examined in vivo. Cotransfection of TSC1/TSC2 suppressed the ability of Rheb to stimulate S6K (Fig. 3A). Similar results were observed with 4EBP1 (Fig. 3B). These data further support the idea that TSC2 acts as a GAP toward Rheb.
mTOR acts downstream of Rheb
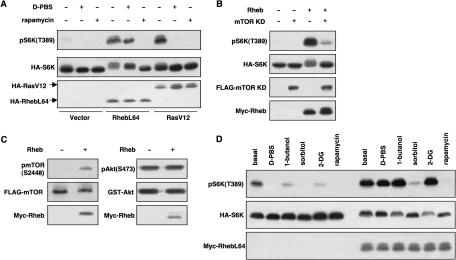
Previous studies have suggested that mTOR acts downstream of TSC2 (Gao et al. 2002; Inoki et al. 2002; Tee et al. 2002). To determine the relationship between Rheb and mTOR, the effect of rapamycin was investigated. Treatment of cells with rapamycin completely blocked the stimulatory effect of Rheb on S6K phosphorylation (Fig. 4A), indicating that Rheb functions upstream of mTOR. To further test the functional relationship between Rheb and mTOR, a kinase-inactive mutant of mTOR was coexpressed with Rheb. We found that the dominant-negative mTOR-KD significantly blocked the phosphorylation of S6K by Rheb (Fig. 4B). We had previously observed that TSC1/TSC2 inhibits the phosphorylation of S2448 of mTOR (Inoki et al. 2002; Kenerson et al. 2002). Coexpression of Rheb caused a significant increase of mTOR S2448 phosphorylation, whereas phosphorylation of Akt was not affected (Fig. 4C), indicating that Rheb activates mTOR.
Figure 4.
Rheb functions upstream of mTOR and is involved in response to various signals. (A) Rheb acts downstream of nutrient signals and upstream of mTOR. HA-S6K was cotransfected with RhebL64 or RasV12 as indicated. Cells were treated with rapamycin or D-PBS for 30 min. Phosphorylation of S6K was determined. (B) Kinase inactive mTOR KD blocks Rheb-induced S6K phosphorylation. (C) Rheb stimulates the phosphorylation of mTOR. Flag-mTOR or GST-Akt was cotransfected with Rheb and immunoprecipitated. (Left) Phosphorylation of mTOR was detected by the anti-phospho mTOR (S2448) antibody. (Right) Phosphorylation of Akt was monitored by the anti-phospho Akt (S473) antibody. (D) Role of Rheb in S6K regulation by various signaling pathways. HA-S6K was transfected alone (left half) or together with RhebL64 (right half). Cells were treated with D-PBS, 1-butanol (0.3%), sorbitol (600 mM), 2-DG (25 mM), or rapamycin (20 nM) for 30 min as indicated.
Rheb is involved in multiple signaling pathways that regulate S6K
Phosphorylation of S6K is regulated by numerous cellular signals including nutrients (Proud 2002). The functional significance of Rheb in nutrient signaling was analyzed. In Rheb-transfected HEK293 cells, nutrient starvation (treatment with D-PBS that contains glucose, but not amino acids) induced a minor dephosphorylation of S6K (Fig. 4A). In contrast, nutrient starvation inhibited S6K phosphorylation in RasV12-transfected cells (Fig. 4A). These results demonstrate that Rheb plays an important role in the nutrient signaling pathway, but Ras is not directly involved in nutrient signaling. We also examined the effect of osmotic stress by sorbitol, energy depletion by 2-deoxyglucose (2-DG), and inhibition of phospholipase D by 1-butanol. These treatments are known to induce S6K dephosphorylation (Fig. 4D; Dennis et al. 2001; Fang et al. 2001; Desai et al. 2002). Rheb significantly blocked the effect of 2-DG and 1-butanol, but had little effect on S6K dephosphorylation induced by sorbitol (Fig. 4D). These data suggest that Rheb acts in the cellular energy-sensing pathway and the phospholipase D pathway, but is not directly involved in the osmotic stress pathway. Consistently, TSC1/TSC2 also plays an important role in regulation of S6K in response to cellular energy depletion (data not shown).
The importance of TSC1/TSC2 in cell growth regulation has been well established (Potter and Xu 2001). The major cellular function of TSC1/TSC2 is to inhibit the phosphorylation of S6K and 4EBP1, likely through mTOR. This study provides a significant advance in the understanding of the molecular mechanism of how TSC1/TSC2 regulates the phosphorylation of S6K and 4EBP-1. Based on both in vitro and in vivo biochemical experiments, we conclude that TSC2 is an Rheb-specific GAP and Rheb is a key downstream target of TSC1/TSC2 (Fig. 5). Rheb is closely related to Ras and is an unusual GTPase in that it is predominantly GTP bound in cells. The biological function of Rheb has not been clearly defined. Our studies support that Rheb promotes cell growth by stimulating phosphorylation of S6K and 4EBP1, but has no role in ERK activation.
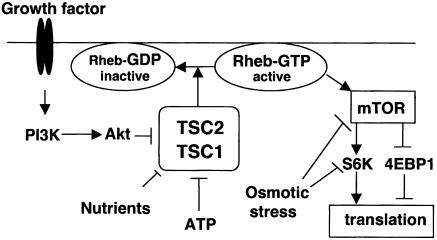
Figure 5.
A proposed model of Rheb functions downstream of TSC1/TSC2 and upstream of mTOR. TSC2 acts as a GAP to inactivate Rheb by directly stimulating GTP hydrolysis. Rheb stimulates mTOR. Nutrient and cellular energy status signals through Rheb, whereas osmotic stress signals independently of Rheb.
Inhibition of Rheb by TSC1/TSC2 is likely to be critical for the function of these two tumor suppressor proteins because Rheb potently regulates the phosphorylation of S6K and 4EBP1. Interestingly, mutation of the Rheb ortholog in S.pombe displays phenotypes characteristic of nutrient starvation (Mach et al. 2000). Our data are consistent with the genetic observations made in S.pombe and support the notion that Rheb plays an essential role in nutrient signaling. S.pombe also contains TSC1- and TSC2-related molecules (Matsumoto et al. 2002). We have also shown that TSC2 is an unusual GAP because it lacks the conserved arginine found in other Rap family GAPs. Yet it has very high and selective GAP activity toward Rheb. Future studies to elucidate the catalytic mechanism of TSC2 on Rheb-GTP hydrolysis will provide new insights into how this unusual GTPase and GAP work.
Our studies present a model in which Rheb is involved in S6K regulation in response to nutrients and energy levels (Fig. 5). In contrast, osmotic stress regulates S6K independently of Rheb. A major function of Rheb is to stimulate mTOR; however, we did not detect a significant association between Rheb and mTOR, suggesting that Rheb may activate mTOR indirectly. Future efforts will focus on how Rheb stimulates mTOR. Our model predicts that TSC2 acts through Rheb to regulate translation (Fig. 5). It suggests that Rheb positively regulates cell growth and plays an important role in the development of TSC. Therefore, inhibition of Rheb could be a potential target for the treatment of TSC and cancer.
Materials and methods
Antibodies and plasmids
Anti-phospho S6K (T389), anti-mTOR, and anti-phospho mTOR (S2448) antibodies were from Cell Signaling Inc., and anti-Myc antibodies were from Santa Cruz Biotechnology. Anti-HA and anti-Flag were purchased from Covance and Sigma, respectively. TSC1 and TSC2 constructs were described previously (Inoki et al. 2002). The HA-tagged S6K1 (α II) construct was generously provided by J. Blenis (Harvard University, Cambridge, MA) and Flag-tagged mTOR and kinase inactive mTOR (mTORKD) were from S. Schreiber (Harvard University, Cambridge, MA). Rab5 is a gift of G. Li (University of Oklahoma, Health Sciences Center, Oklahoma City, OK). All other DNA constructs including Ras, Rheb, Rap1, Rac, Cdc42, RhoA, 4E-BP1, Akt, and ERK2 were laboratory stocks. All mutant constructs of TSC2 and Rheb were created by PCR mutagenesis and verified by DNA sequencing.
Cell culture, transfection, and immunoprecipitation
HEK293 cells were seeded and maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). Transfection was performed using Lipofectamine Reagent (Invitrogen) following the manufacturer's instructions. Cells were lysed in lysis buffer (10 mM Tris-HCl at pH 7.5, 100 mM NaCl, 1% NP-40, 1% Triton X-100, 50 mM NaF, 2 mM EDTA, 1 mM PMSF, 10 μg/mL leupeptin, 10 μg/mL aprotinin) and immunoprecipitated with the indicated antibodies and protein G-Sepharose beads. Immunocomplexes were subjected to SDS-PAGE.
In vitro GTPase assay
GST-Rheb, GST-Ras, GST-Rac, and GST-Rap were expressed in the bacterial strain BL21 and purified using glutathione-Sepharose 4B beads (Sigma) as described. Small GTPases were loaded with 32P-γ-GTP before GAP assays. Here, 10 μg of small GTPase protein bound on glutathione beads was incubated with 50 μCi of 32P-γ-GTP in loading buffer (20 mM Tris at pH 8, 5 mM EDTA, 1 mM DTT, 0.1 mg/mL BSA) in a volume of 20 μL. The loading reaction was stopped by addition of MgCl2 to 10 mM. Then the small GTPase protein was eluted.
HA-TSC2 was transfected to HEK293 cells and immunoprecipitated by anti-HA antibody. The immune complexes were washed three times with wash buffer (20 mM Tris at pH 7.4, 800 mM NaCl, 2 mM EDTA, 1% NP-40) and two times with GAP assay wash buffer (20 mM Tris at pH 8, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT). The final GAP assays included 0.5 μg of 32P-γ-GTP loaded small GTPase, and immunoprecipitated TSC2 from 1 × 107 transfected to HEK293 cells. Reactions were carried out in 40 μL of GAP assay buffer (20 mM Tris at pH 8, 10 mM MgCl2, and 4 mM DTT), at room temperature for 20 min. The reaction was stopped by 300 μL of charcoal buffer (5% charcoal, 20 mM phosphoric acid, 0.6 M HCl), then vortexed for 10 min and spun at 13,000 rpm for 10 min. Then 100 μL of supernatant was taken, and the 32P release was determined by scintillation counting.
In vivo labeling of Rheb
The cells were washed once with phosphate-free DMEM and incubated with 0.5 mCi/mL 32P-orthophosphate (ICN) for 4 h. The cells were lysed with labeling lysis buffer (1% Triton X-100, 50 mM HEPES at pH 7.4, 100 mM NaCl, 5 mM MgCl2, 1 mg/mL BSA, 1 mM DTT, 1 mM PMSF, 10 μg/mL leupeptin, 10 μg/mL aprotinin). Myc-tagged Rheb was immunoprecipitated. The Rheb-bound nucleotides were eluted with elution buffer (2 mM EDTA, 0.2% SDS, 1 mM GDP, 1 mM GTP) at 68°C for 20 min. Then the eluted nucleotides were subjected to thin layer chromatography using PEI cellulose plates (Baker-flex) in 0.75 M KH2PO4 (pH 3.4).
Acknowledgments
We thank Tianqing Zhu for technical assistance. We also thank Weiquan Li and Huira Chong for plasmid construction and Haris G. Vikis and Jennifer Aurandt for critical reading of the manuscript. This work was supported by grants from the NIH and the Walther Cancer Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Corresponding author.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1110003.
References
- Au K.S., Rodriguez, J.A., Finch, J.L., Volcik, K.A., Roach, E.S., Delgado, M.R., Rodriguez Jr., E., and Northrup, H. 1998. Germ-line mutational analysis of the TSC2 gene in 90 tuberous-sclerosis patients. Am.J. Hum.Genet. 62: 286-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.J., Beal, P.A., Keith, C.T., Chen, J., Shin, T.B., and Schreiber, S.L. 1995. Control of p70 S6 kinase by kinase activity of FRAP in vivo. Nature 377: 441-446. [DOI] [PubMed] [Google Scholar]
- Clark G.J., Kinch, M.S., Rogers-Graham, K., Sebti, S.M., Hamilton, A.D., and Der, C.J. 1997. The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. J.Biol.Chem. 272: 10608-10615. [DOI] [PubMed] [Google Scholar]
- Dennis P.B., Jaeschke, A., Saitoh, M., Fowler, B., Kozma, S.C., and Thomas, G. 2001. Mammalian TOR: A homeostatic ATP sensor. Science 294: 1102-1105. [DOI] [PubMed] [Google Scholar]
- Desai B.N., Myers, B.R., and Schreiber, S.L. 2002. FKBP12-rapamycin-associated protein associates with mitochondria and senses osmotic stress via mitochondrial dysfunction. Proc.Natl.Acad.Sci. 99: 4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The European Chromosome 16 Tuberous Sclerosis Consortium. 1993. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell 75: 1305-1315. [DOI] [PubMed] [Google Scholar]
- Fang Y., Vilella-Bach, M., Bachmann, R., Flanigan, A., and Chen, J. 2001. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294: 1942-1945. [DOI] [PubMed] [Google Scholar]
- Gao X. and Pan, D. 2001. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes & Dev. 15: 1383-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Zhang, Y., Arrazola, P., Hino, O., Kobayashi, T., Yeung, R.S., Ru, B., and Pan, D. 2002. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat.Cell Biol. 4: 699-704. [DOI] [PubMed] [Google Scholar]
- Gingras A.C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu.Rev.Biochem. 68: 913-963. [DOI] [PubMed] [Google Scholar]
- Gomez M.R. 1991. Phenotypes of the tuberous sclerosis complex with a revision of diagnostic criteria. Ann.NY Acad.Sci. 615: 1-7. [DOI] [PubMed] [Google Scholar]
- Goncharova E.A., Goncharov, D.A., Eszterhas, A., Hunter, D.S., Glassberg, M.K., Yeung, R.S., Walker, C.L., Noonan, D., Kwiatkowski, D.J., Chou, M.M., et al. 2002. Tuberin regulates p70 S6 kinase activation and ribosomal protein S6 phosphorylation. A role for the TSC2 tumor suppressor gene in pulmonary lymphangioleiomyomatosis (LAM). J.Biol.Chem. 277: 30958-30967. [DOI] [PubMed] [Google Scholar]
- Hara K., Yonezawa, K., Weng, Q.P., Kozlowski, M.T., Belham, C., and Avruch, J. 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol.Chem. 273: 14484-14494. [DOI] [PubMed] [Google Scholar]
- Inoki K., Li, Y., Zhu, T., Wu, J., and Guan, K.L. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat.Cell Biol. 4: 648-657. [DOI] [PubMed] [Google Scholar]
- Jin F., Wienecke, R., Xiao, G.H., Maize Jr., J.C., DeClue, J.E., and Yeung, R.S. 1996. Suppression of tumorigenicity by the wild-type tuberous sclerosis 2 (Tsc2) gene and its C-terminal region. Proc.Natl.Acad. Sci. 93: 9154-9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenerson H.L., Aicher, L.D., True, L.D., and Yeung, R.S. 2002. Activated mammalian target of rapamycin pathway in the pathogenesis of tuberous sclerosis complex renal tumors. Cancer Res. 62: 5645-5650. [PubMed] [Google Scholar]
- Kobayashi T., Minowa, O., Sugitani, Y., Takai, S., Mitani, H., Kobayashi, E., Noda, T., and Hino, O. 2001. A germ-line Tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by Tsc2 mutation in mice. Proc.Natl. Acad.Sci. 98: 8762-8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski D.J. 2003. Tuberous sclerosis: From tubers to mTOR. Ann. Hum.Genet. 67: 87-96. [DOI] [PubMed] [Google Scholar]
- Lowy D.R. and Willumsen, B.M. 1993. Function and regulation of ras. Annu.Rev.Biochem. 62: 851-891. [DOI] [PubMed] [Google Scholar]
- Mach K.E., Furge, K.A., and Albright, C.F. 2000. Loss of Rhb1, a Rheb-related GTPase in fission yeast, causes growth arrest with a terminal phenotype similar to that caused by nitrogen starvation. Genetics 155: 611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning B.D., Tee, A.R., Logsdon, M.N., Blenis, J., and Cantley, L.C. 2002. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol.Cell 10: 151-162. [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Bandyopadhyay, A., Kwiatkowski, D.J., Maitra, U., and Matsumoto, T. 2002. Role of the Tsc1-Tsc2 complex in signaling and transport across the cell membrane in the fission yeast Schizosaccharomyces pombe. Genetics 161: 1053-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose S., Kobayashi, T., Mitani, H., Hirabayashi, M., Ito, K., Ueda, M., Nabeshima, Y., and Hino, O. 2002. Identification of the coding sequences responsible for Tsc2-mediated tumor suppression using a transgenic rat system. Hum.Mol.Genet. 11: 2997-3006. [DOI] [PubMed] [Google Scholar]
- Potter C.J. and Xu, T. 2001. Mechanisms of size control. Curr.Opin. Genet.Dev. 11: 279-286. [DOI] [PubMed] [Google Scholar]
- Potter C.J., Huang, H., and Xu, T. 2001. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 105: 357-368. [DOI] [PubMed] [Google Scholar]
- Proud C.G. 2002. Regulation of mammalian translation factors by nutrients. Eur.J.Biochem. 269: 5338-5349. [DOI] [PubMed] [Google Scholar]
- Scheffzek K., Ahmadian, M.R., and Wittinghofer, A. 1998. GTPase-activating proteins: Helping hands to complement an active site. Trends Biochem.Sci. 23: 257-262. [DOI] [PubMed] [Google Scholar]
- Tapon N., Ito, N., Dickson, B.J., Treisman, J.E., and Hariharan, I.K. 2001. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105: 345-355. [DOI] [PubMed] [Google Scholar]
- Tee A.R., Fingar, D.C., Manning, B.D., Kwiatkowski, D.J., Cantley, L.C., and Blenis, J. 2002. Tuberous sclerosis complex-1 and complex-2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc.Natl.Acad.Sci. 99: 13571-13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahey M. and McCormick, F. 1987. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science 238: 542-545. [DOI] [PubMed] [Google Scholar]
- Wienecke R., Konig, A., and DeClue, J.E. 1995. Identification of tuberin, the tuberous sclerosis-2 product. Tuberin possesses specific Rap1GAP activity. J.Biol.Chem. 270: 16409-16414. [DOI] [PubMed] [Google Scholar]
- Xiao G.H., Shoarinejad, F., Jin, F., Golemis, E.A., and Yeung, R.S. 1997. The tuberous sclerosis 2 gene product, tuberin, functions as a Rab5 GTPase activating protein (GAP) in modulating endocytosis. J.Biol. Chem. 272: 6097-6100. [DOI] [PubMed] [Google Scholar]
- Yamagata K., Sanders, L.K., Kaufmann, W.E., Yee, W., Barnes, C.A., Nathans, D., and Worley, P.F. 1994. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J. Biol.Chem. 269: 16333-16339. [PubMed] [Google Scholar]
- Yee W.M. and Worley, P.F. 1997. Rheb interacts with Raf-1 kinase and may function to integrate growth factor- and protein kinase A-dependent signals. Mol.Cell.Biol. 17: 921-933. [DOI] [PMC free article] [PubMed] [Google Scholar]