Abstract
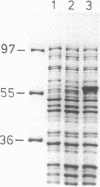
The Staphylococcus xylosus gene scrB, encoding a sucrase, has been isolated from a genomic library of S. xylosus constructed in Escherichia coli. The gene was detected by its ability to confer utilization of the glucose and fructose residues of raffinose in an E. coli strain that is not able to metabolize galactose. It was found to reside within a 1.8-kb DNA fragment, the nucleotide sequence of which was determined. One large open reading frame, which is preceded by a ribosome binding site, is encoded on the fragment. Its deduced amino acid sequence yields a protein with a molecular mass of 57.377 kDa which shows significant homology with bacterial sucrose-6-phosphate hydrolases and sucrases. Overexpression of scrB in E. coli by the bacteriophage T7 polymerase promoter system resulted in the production of a protein with an apparent molecular mass of 58 kDa. Disruption of the scrB gene in the S. xylosus genome rendered S. xylosus unable to utilize sucrose. Thus, the ScrB sucrase is essential for sucrose metabolism in S. xylosus.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnaud M., Vary P., Zagorec M., Klier A., Debarbouille M., Postma P., Rapoport G. Regulation of the sacPA operon of Bacillus subtilis: identification of phosphotransferase system components involved in SacT activity. J Bacteriol. 1992 May;174(10):3161–3170. doi: 10.1128/jb.174.10.3161-3170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslanidis C., Schmid K., Schmitt R. Nucleotide sequences and operon structure of plasmid-borne genes mediating uptake and utilization of raffinose in Escherichia coli. J Bacteriol. 1989 Dec;171(12):6753–6763. doi: 10.1128/jb.171.12.6753-6763.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulkemeyer P., Ebner R., Heilenmann G., Jahreis K., Schmid K., Wrieden S., Lengeler J. W. Molecular analysis of two fructokinases involved in sucrose metabolism of enteric bacteria. Mol Microbiol. 1991 Dec;5(12):2913–2922. doi: 10.1111/j.1365-2958.1991.tb01851.x. [DOI] [PubMed] [Google Scholar]
- Blatch G. L., Scholle R. R., Woods D. R. Nucleotide sequence and analysis of the Vibrio alginolyticus sucrose uptake-encoding region. Gene. 1990 Oct 30;95(1):17–23. doi: 10.1016/0378-1119(90)90408-j. [DOI] [PubMed] [Google Scholar]
- Blatch G. L., Woods D. R. Nucleotide sequence and analysis of the Vibrio alginolyticus scr repressor-encoding gene (scrR). Gene. 1991 May 15;101(1):45–50. doi: 10.1016/0378-1119(91)90222-w. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brückner R. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene. 1992 Dec 1;122(1):187–192. doi: 10.1016/0378-1119(92)90048-t. [DOI] [PubMed] [Google Scholar]
- Chen Y. Y., LeBlanc D. J. Genetic analysis of scrA and scrB from Streptococcus sobrinus 6715. Infect Immun. 1992 Sep;60(9):3739–3746. doi: 10.1128/iai.60.9.3739-3746.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner R., Lengeler J. W. DNA sequence of the gene scrA encoding the sucrose transport protein EnzymeII(Scr) of the phosphotransferase system from enteric bacteria: homology of the EnzymeII(Scr) and EnzymeII(Bgl) proteins. Mol Microbiol. 1988 Jan;2(1):9–17. [PubMed] [Google Scholar]
- Fischer R., von Strandmann R. P., Hengstenberg W. Mannitol-specific phosphoenolpyruvate-dependent phosphotransferase system of Enterococcus faecalis: molecular cloning and nucleotide sequences of the enzyme IIIMtl gene and the mannitol-1-phosphate dehydrogenase gene, expression in Escherichia coli, and comparison of the gene products with similar enzymes. J Bacteriol. 1991 Jun;173(12):3709–3715. doi: 10.1128/jb.173.12.3709-3715.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet A., Arnaud M., Klier A., Rapoport G. Bacillus subtilis sucrose-specific enzyme II of the phosphotransferase system: expression in Escherichia coli and homology to enzymes II from enteric bacteria. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8773–8777. doi: 10.1073/pnas.84.24.8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet A., Klier A., Rapoport G. Nucleotide sequence of the sucrase gene of Bacillus subtilis. Gene. 1986;45(2):221–225. doi: 10.1016/0378-1119(86)90258-1. [DOI] [PubMed] [Google Scholar]
- Gunasekaran P., Karunakaran T., Cami B., Mukundan A. G., Preziosi L., Baratti J. Cloning and sequencing of the sacA gene: characterization of a sucrase from Zymomonas mobilis. J Bacteriol. 1990 Dec;172(12):6727–6735. doi: 10.1128/jb.172.12.6727-6735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz F., Zabielski J., Philipson L., Lindberg M. DNA homology between the arsenate resistance plasmid pSX267 from Staphylococcus xylosus and the penicillinase plasmid pI258 from Staphylococcus aureus. Plasmid. 1983 Mar;9(2):126–137. doi: 10.1016/0147-619x(83)90015-x. [DOI] [PubMed] [Google Scholar]
- Hardesty C., Colón G., Ferran C., DiRienzo J. M. Deletion analysis of sucrose metabolic genes from a Salmonella plasmid cloned in Escherichia coli K12. Plasmid. 1987 Sep;18(2):142–155. doi: 10.1016/0147-619x(87)90042-4. [DOI] [PubMed] [Google Scholar]
- Hardesty C., Ferran C., DiRienzo J. M. Plasmid-mediated sucrose metabolism in Escherichia coli: characterization of scrY, the structural gene for a phosphoenolpyruvate-dependent sucrose phosphotransferase system outer membrane porin. J Bacteriol. 1991 Jan;173(2):449–456. doi: 10.1128/jb.173.2.449-456.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda O., Kato C., Kuramitsu H. K. Nucleotide sequence of the Streptococcus mutans gtfD gene encoding the glucosyltransferase-S enzyme. J Gen Microbiol. 1990 Oct;136(10):2099–2105. doi: 10.1099/00221287-136-10-2099. [DOI] [PubMed] [Google Scholar]
- Kunst F., Pascal M., Lefesant J. A., Walle J., Dedonder R. Purification and some properties of an endocellular sucrase from a constitutive mutant of Bacillus subtilis Marburg 168. Eur J Biochem. 1974 Mar 1;42(2):611–620. doi: 10.1111/j.1432-1033.1974.tb03376.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lepesant J. A., Kunst F., Lepesant-Kejzlarová J., Dedonder R. Chromosomal location of mutations affecting sucrose metabolism in Bacillus subtilis Marburg. Mol Gen Genet. 1972;118(2):135–160. doi: 10.1007/BF00267084. [DOI] [PubMed] [Google Scholar]
- MORSE M. L., ALIRE M. L. An agar medium indicating acid production. J Bacteriol. 1958 Sep;76(3):270–271. doi: 10.1128/jb.76.3.270-271.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin I., Débarbouillé M., Ferrari E., Klier A., Rapoport G. Characterization of the levanase gene of Bacillus subtilis which shows homology to yeast invertase. Mol Gen Genet. 1987 Jun;208(1-2):177–184. doi: 10.1007/BF00330439. [DOI] [PubMed] [Google Scholar]
- Munro C., Michalek S. M., Macrina F. L. Cariogenicity of Streptococcus mutans V403 glucosyltransferase and fructosyltransferase mutants constructed by allelic exchange. Infect Immun. 1991 Jul;59(7):2316–2323. doi: 10.1128/iai.59.7.2316-2323.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Murphy E., Gryczan T. J., Baron E., Edelman I. Penicillinase plasmids of Staphylococcus aureus: restriction-deletion maps. Plasmid. 1979 Jan;2(1):109–129. doi: 10.1016/0147-619x(79)90010-6. [DOI] [PubMed] [Google Scholar]
- Postma P. W., Lengeler J. W. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol Rev. 1985 Sep;49(3):232–269. doi: 10.1128/mr.49.3.232-269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch P. J., De Vos W. M. Characterization of the novel nisin-sucrose conjugative transposon Tn5276 and its insertion in Lactococcus lactis. J Bacteriol. 1992 Feb;174(4):1280–1287. doi: 10.1128/jb.174.4.1280-1287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robeson J. P., Barletta R. G., Curtiss R., 3rd Expression of a Streptococcus mutans glucosyltransferase gene in Escherichia coli. J Bacteriol. 1983 Jan;153(1):211–221. doi: 10.1128/jb.153.1.211-221.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Kuramitsu H. K. Sequence analysis of the Streptococcus mutans scrB gene. Infect Immun. 1988 Aug;56(8):1956–1960. doi: 10.1128/iai.56.8.1956-1960.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Poy F., Jacobson G. R., Kuramitsu H. K. Characterization and sequence analysis of the scrA gene encoding enzyme IIScr of the Streptococcus mutans phosphoenolpyruvate-dependent sucrose phosphotransferase system. J Bacteriol. 1989 Jan;171(1):263–271. doi: 10.1128/jb.171.1.263-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid K., Ebner R., Altenbuchner J., Schmitt R., Lengeler J. W. Plasmid-mediated sucrose metabolism in Escherichia coli K12: mapping of the scr genes of pUR400. Mol Microbiol. 1988 Jan;2(1):1–8. doi: 10.1111/j.1365-2958.1988.tb00001.x. [DOI] [PubMed] [Google Scholar]
- Schmid K., Ebner R., Jahreis K., Lengeler J. W., Titgemeyer F. A sugar-specific porin, ScrY, is involved in sucrose uptake in enteric bacteria. Mol Microbiol. 1991 Apr;5(4):941–950. doi: 10.1111/j.1365-2958.1991.tb00769.x. [DOI] [PubMed] [Google Scholar]
- Schmid K., Schupfner M., Schmitt R. Plasmid-mediated uptake and metabolism of sucrose by Escherichia coli K-12. J Bacteriol. 1982 Jul;151(1):68–76. doi: 10.1128/jb.151.1.68-76.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle R. R., Robb S. M., Robb F. T., Woods D. R. Nucleotide sequence and analysis of the Vibrio alginolyticus sucrase gene (scrB). Gene. 1989 Aug 1;80(1):49–56. doi: 10.1016/0378-1119(89)90249-7. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroza T., Kuramitsu H. K. Sequence analysis of the Streptococcus mutans fructosyltransferase gene and flanking regions. J Bacteriol. 1988 Feb;170(2):810–816. doi: 10.1128/jb.170.2.810-816.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroza T., Ueda S., Kuramitsu H. K. Sequence analysis of the gtfB gene from Streptococcus mutans. J Bacteriol. 1987 Sep;169(9):4263–4270. doi: 10.1128/jb.169.9.4263-4270.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizemore C., Buchner E., Rygus T., Witke C., Götz F., Hillen W. Organization, promoter analysis and transcriptional regulation of the Staphylococcus xylosus xylose utilization operon. Mol Gen Genet. 1991 Jul;227(3):377–384. doi: 10.1007/BF00273926. [DOI] [PubMed] [Google Scholar]
- Sprenger G. A., Lengeler J. W. Analysis of sucrose catabolism in Klebsiella pneumoniae and in Scr+ derivatives of Escherichia coli K12. J Gen Microbiol. 1988 Jun;134(6):1635–1644. doi: 10.1099/00221287-134-6-1635. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Le Coq D., Aymerich S., Gonzy-Tréboul G., Gay P. The DNA sequence of the gene for the secreted Bacillus subtilis enzyme levansucrase and its genetic control sites. Mol Gen Genet. 1985;200(2):220–228. doi: 10.1007/BF00425427. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Le Coq D., Aymerich S. Induction of saccharolytic enzymes by sucrose in Bacillus subtilis: evidence for two partially interchangeable regulatory pathways. J Bacteriol. 1989 Mar;171(3):1519–1523. doi: 10.1128/jb.171.3.1519-1523.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer J. M., Chassy B. M., Krichevsky M. I. Sucrose metabolism by Streptococcus mutans, SL-I. Biochim Biophys Acta. 1971 Feb 28;261(2):379–387. doi: 10.1016/0304-4165(72)90062-1. [DOI] [PubMed] [Google Scholar]
- Ueda S., Shiroza T., Kuramitsu H. K. Sequence analysis of the gtfC gene from Streptococcus mutans GS-5. Gene. 1988 Sep 15;69(1):101–109. doi: 10.1016/0378-1119(88)90382-4. [DOI] [PubMed] [Google Scholar]
- Wehmeier U., Sprenger G. A., Lengeler J. W. The use of lambda plac-Mu hybrid phages in Klebsiella pneumoniae and the isolation of stable Hfr strains. Mol Gen Genet. 1989 Feb;215(3):529–536. doi: 10.1007/BF00427052. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]