Abstract
Production of the structural and enzymatic proteins of type 1 human immunodeficiency virus (HIV-1) is controlled by the rev regulatory gene product. The 116-amino acid Rev protein acts by binding to the Rev response element (RRE), a complex RNA stem–loop structure located within the env gene of HIV. Rev exerts a series of posttranscriptional effects, including the inhibition of viral RNA splicing, the activation of nuclear export of incompletely spliced viral RNAs, and the enhancement of translation of RRE-containing RNAs. Our studies now demonstrate that at least one member of the SR family of splicing factors, SF2/ASF, specifically binds to a subregion of the RRE in vitro in a Rev-dependent manner. Furthermore, expression of high levels of SF2/ASF inhibits Rev function and impairs HIV replication in vivo. Both the in vitro binding of SF2/ASF to the Rev/RRE complex and the in vivo inhibition of Rev action by SF2/ASF are abrogated by mutation of the N-terminal RNA recognition motif but are not affected by mutation of the C-terminal arginine–serine-rich domain. These findings suggest that Rev inhibition of HIV splicing likely involves recruitment of the essential splicing factor SF2/ASF to the Rev/RRE complex. However, these inhibitory effects of Rev on viral RNA splicing are apparently overcome by augmenting the intracellular levels of SF2/ASF expression.
Rev is an essential regulatory protein of the type 1 human immunodeficiency virus (HIV-1) that controls production of proteins required for the assembly of infectious virions (1–5). The 9-kb full-length genomic transcript of HIV undergoes a complex series of posttranscriptional splicing events resulting in the generation of more than 20 mRNAs that encode 9 different viral proteins (for review see refs. 6–8). The resultant mRNAs can be grouped in three size classes: the 2-kb viral RNAs are fully spliced and encode the regulatory proteins of HIV, including Rev, Tat, and Nef, whereas the 9- and 4-kb mRNAs encode the structural, enzymatic, and ancillary viral proteins, including Gag, Pol, Vif, Vpu, Vpr, and Env. The appearance of the 4- and 9-kb classes of mRNAs in the cytoplasm is regulated by the Rev protein (1–5). Rev binds specifically to the Rev response element (RRE), a complex 220-nucleotide RNA stem–loop structure located in the env coding sequence. This binding leads to effective cytoplasmic expression of the singly spliced (4 kb) and unspliced (9 kb) viral mRNAs, thus allowing the transition from early-stage regulatory protein synthesis to late-stage structural and enzymatic viral protein production (2, 6, 7). In the absence of Rev, these incompletely spliced RRE-containing transcripts are retained in the nucleus where they are either completely spliced or degraded (6, 7). Various mechanisms have been suggested to explain the action of Rev, including inhibition of viral mRNA splicing, activation of viral RNA transport from the nucleus to the cytoplasm, and enhanced translation of RRE-containing viral RNAs (1, 6–8). These mechanisms are not mutually exclusive, and Rev may exert effects at multiple levels.
HIV transcripts are inherently inefficient splicing substrates that are also rather unstable in the nuclei of T lymphocytes (2, 7). These viral RNAs contain cis-acting elements termed cis-repressor sequences or inhibitory sequences, which may be responsible for instability, nuclear retention, and/or inefficient translation (1, 2, 7, 9). Rev appears to overcome the effects of these cis-repressor/inhibitory sequence elements. Nuclear retention of the incompletely spliced viral mRNAs could involve cellular factors involved in pre-mRNA splicing (10–12). In this regard, Rev has been shown to inhibit spliceosome assembly and splicing of RRE-containing mRNAs in vitro (13, 14). These findings raised the possibility that Rev might interact directly or indirectly with specific splicing factors. Here we report that the RRE specifically binds the serine–arginine-rich (SR) protein splicing factor SF2/ASF in a Rev-dependent manner. We further show that exogenous expression of SF2/ASF inhibits Rev action and that both the in vitro binding and in vivo inhibitory effects of SF2/ASF are dependent on the RNA binding but not the RS domain of this splicing factor.
MATERIALS AND METHODS
Protein Purification.
HeLa nuclear extracts and 65–90% NH42SO4-saturated precipitates were generated as described (15, 16). SF2/ASF was purified from this fraction by preparative SDS/PAGE and renatured by the method of Hager and Burgess (17).
RNA Gel Mobility Shifts.
RNA electrophoretic mobility shift assays (REMSAs) were performed essentially as described (18). Radiolabeled HIV-1 RRE RNAs were transcribed in vitro using T7 polymerase and wild-type and mutant RRE plasmid templates linearized with Asp-718 (19). Recombinant Rev protein (a generous gift from P. Wingfield, National Institutes of Health, Bethesda) was added to binding reactions and incubated on ice for 10 min before the addition of SF2/ASF. The binding reactions were analyzed on native 5% (acrylamide/bisacrylamide ratio, 29:1) polyacrylamide gels, electrophoresed at 4°C in a Tris-borate buffer system.
Cells and Transfections.
CV1 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and transfected using LipofectAMINE reagent (GIBCO/BRL) according to the manufacturer’s recommendations.
Plasmids.
SF2/ASF wild-type and mutant plasmids were constructed by Klenow treatment of NdeI–BamHI fragments from pET9c-SF2, pET9c-RT, and pET9c-FF-DD plasmids (20) followed by subcloning into the SmaI site of pCMV-4. The SRp20 and SRp35 expression plasmids were constructed by subcloning the EcoRI fragments from SRp20JG and SRp35JG (21) into the EcoRI site of pCMV-4Δ. The SRp40 expression plasmid was constructed by subcloning the EcoRI–NotI fragment from SRp40JG (21) into the corresponding sites of pCMV-4. The pDM128 and pDM128 Rex-RE plasmids were kindly provided by T. Parslow (University of California, San Francisco). The full-length, infectious HIV-1 clone R7/3 was as described (8).
Chloramphenicol Acetyltransferase (CAT) and p24 Gag Assays.
CAT assays were performed 48 hr after transfection as described (22). COS cells were transfected with a replication- competent HIV-1 proviral clone, R7/3 (8), either alone or in the presence of various amounts of sense or antisense pSF2/ASF expression plasmid. Cell culture supernatants were assayed 72 hr later for p24 Gag protein using a p24 antigen capture ELISA kit (Coulter).
Reverse Transcription (RT) PCR.
For RNA RT-PCR analysis, COS cells were transfected as described with 1 μg of pgTAT plasmid (23) pcRev and 0–400 ng of pSF2/ASF. Total RNA was isolated with the RNeasy Total RNA kit (Qiagen, Chatsworth, CA). Total RNA (200 ng) was analyzed with the cDNA Cycle Kit (Invitrogen) and Taq DNA polymerase (Promega). The oligonucleotide primers used to detect spliced and unspliced forms of tat mRNA were ATGGAGCCAGTAGATCCTAGA as the exon 1 forward primer and AGGCATGTGTGGCCCAAACAT as the exon 2 reverse primer.
RESULTS
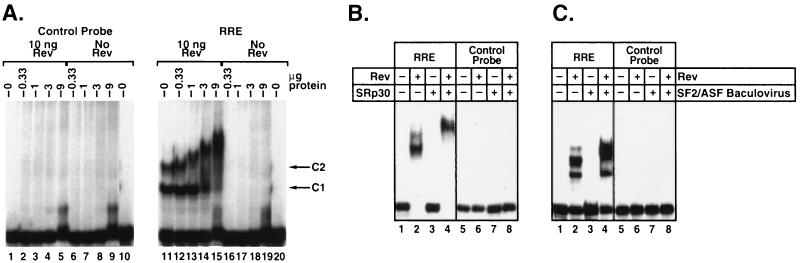
As a first step to identifying cellular proteins that may participate in Rev action, HeLa cell nuclei were fractionated using the ammonium sulfate purification procedure (16). These nuclear fractions were then screened by REMSA for factors that specifically interact with the RRE complex only in the presence of Rev. The RNA probe used in these studies corresponded to the high-affinity Rev-binding domain (7, 18, 24) containing the IIa, IIb, IIc, and III stem loops of the RRE (25, 26) (see Fig. 2A). When Rev was mixed with the RRE probe, two complexes (termed C1 and C2) with different electrophoretic mobilities were detected (Fig. 1A, lane 11) (18, 25). Addition of the precipitate from the 65–90% ammonium sulfate-saturated fraction produced an incremental decrease in the migration of both of these complexes, perhaps involving the binding of more than one protein (Fig. 1A, lanes 12–15). However, this RNA-binding activity was not observed when the ammonium sulfate precipitate was mixed with the wild-type RRE probe in the absence of Rev (Fig. 1A, lanes 16–20) or with a control “base-switched” (BS) RRE probe (19) in the presence or absence of Rev (Fig. 1A, lanes 1–10). This BS probe preserves the overall stem–loop structure of the RRE but alters the primary base order (27).
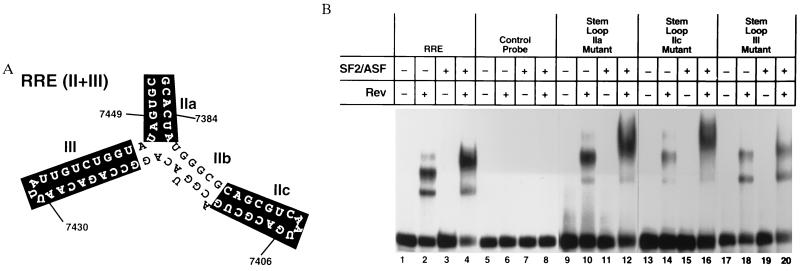
Figure 2.
(A) Structure of the HIV RRE probe used in the RNA-binding experiments. The solid boxes correspond to the sites of the BS mutations tested for SF2/ASF-binding activity in the presence and absence of Rev. The RNA sequences indicated by the boxed regions are as follows: BSIIa, CGUAAU and AUUACG; BSIIc, GUCGCAGUUACUGCGAC; and BSIII UGGUCUGUUAAUAA CAGACCA. The high-affinity Rev-binding site is located in the IIb portion of this structure. Heterologous anchor sequences are also indicated. (B) REMSA using wild-type and mutant RRE probes. Binding reactions were performed with the wild-type or mutant RRE probes as indicated in the presence and absence of Rev and recombinant baculovirus-derived SF2/ASF.
Figure 1.
SF2/ASF binds to the HIV Rev/RRE complex. (A) REMSA autoradiograms are shown in which various amounts of the precipitate from a 65–90% NH42SO4-saturated fraction of HeLa cell nuclear extract were added to Rev-binding reactions performed with 32P-radiolabeled sense and RRE (Right) and control (Left) RNA probes (18). The C1 and C2 complexes formed by Rev binding to the RRE are shown. (B) REMSA demonstrating specific binding of the Rev/RRE complex to SF2/ASF renatured after purification from HeLa cell nuclei by preparative SDS/PAGE (17). In lanes 3 and 4, 300 ng of SF2/ASF was used in reactions. (C) REMSA with 900 ng of recombinant SF2/ASF purified from baculovirus-infected Sf9 cells (21). Binding reactions were performed as described in Materials and Methods.
Analysis of the precipitate from the 65–90% ammonium sulfate-saturated fraction by silver staining demonstrated the presence of several bands as expected. However, proteins with apparent molecular masses of ≈20, 30, 40, 55, and 70–75 kDa were particularly prominent; these sizes were suggestive for the SR family of splicing factors (16). The presence of SR splicing factors in this fraction was demonstrated by marked reactivity with the anti-SR protein monoclonal antibody mAb-104 (16). One major band recognized by mAb-104 displayed an apparent molecular mass of 33 kDa, which was consistent with the sizes of three SR proteins SF2/ASF, 9G8, and SC35 (16, 28) The presence of SF2/ASF in this band was confirmed using the SF2/ASF-specific mAb-96 (data not shown). Several of the SR proteins that reacted with mAb-104 were subsequently purified by SDS/PAGE, renatured (17), and tested in the REMSA. Addition of the band containing SF2/ASF (and perhaps SC35 and 9G8) decreased the migration of the Rev/RRE complexes (Fig. 1B, lane 4). However, this fraction, like the crude precipitate, did not detectably bind to the RRE in the absence of Rev (lane 3) or to a control RNA probe in the presence or absence of Rev (Fig. 1B, lanes 7 and 8). To confirm this result, recombinant SF2/ASF and SC35 were expressed in baculovirus-infected Sf9 cells and tested in the REMSA (Fig. 1C). The recombinant SF2/ASF also produced a decrease in the electrophoretic mobility of the Rev/RRE complexes (Fig. 1C, lane 4) but did not bind to the RRE in the absence of Rev (Fig. 1C, lane 3). In contrast, recombinant SC35 bound weakly to the RRE in the presence or absence of Rev (data not shown). Addition of recombinant 9G8 or SRp40 did not alter the mobility of the Rev/RRE complex or bind to the RRE alone (data not shown).
SF2/ASF is an RNA-binding protein that is likely to interact directly with the RRE in a Rev-dependent manner. To determine the effect of RRE mutations on SF2/ASF binding to the Rev/RRE complex, the indicated BS RRE mutations were tested in the REMSA (Fig. 2A.) The binding of Rev to BS mutations in region IIa, IIc and III was very similar to binding to the wild-type RRE (Fig. 2B, lane 2 versus lanes 10, 14, and 18). In contrast, whereas SF2/ASF binding in the presence of Rev with the BSIIa and BSIIc mutants was similar to that obtained with the wild-type RRE probe (Fig. 2B, lane 4 versus lanes 12 and 16), SF2/ASF binding to the BSIII mutant was greatly reduced (Fig. 2B, lane 20). These findings suggest that stem–loop III may function as an SF2/ASF-binding site on the RRE. Of note, previous structure/function studies of the RRE have demonstrated that stem–loop III is essential for maximal Rev response (27). Specifically, the BSIII mutation results in a 50% reduction in Rev responsiveness (27). In addition, Rev binding appears to be a prerequisite for SF2/ASF binding at this site (Fig. 2B, lanes 11 and 15 versus lanes 12 and 16).
SF2/ASF, SC35, and other SR proteins are not only essential splicing factors, they also participate in selecting alternative splice sites (29, 30). SF2/ASF binds to oligonucleotides containing a 5′ splice site (31) and also enhances U1 small nuclear ribonucleoprotein (snRNP) binding to pre-mRNAs (32). SF2/ASF and SC35 also bind to the U1-70K protein of U1-snRNP (21, 32). In addition, SF2/ASF and SC35 interact with the 35-kDa subunit of U2-snRNP auxiliary factor, U2AF35, which in turn is complexed with the RNA-binding subunit of U2AF (U2AF65) (21). The simultaneous binding of these SR proteins to U1-70K at the 5′ splice site and U2AF35 at the 3′ splice site suggests that one function of SF2/ASF and SC35 is to bridge interactions between the two splice sites allowing subsequent recruitment of additional snRNPs to the complex (21).
One attractive mechanism to explain the inhibitory effects of Rev on HIV splicing would be that the binding of SF2/ASF to the Rev/RRE complex interferes with the normal bridging function of SF2/ASF. If this is the case, overexpression of exogenous SF2/ASF might restore this bridging function and antagonize the effect of Rev. To test this hypothesis, the SF2/ASF cDNA was cloned into the pCMV4 eukaryotic expression plasmid and cotransfected into CV1 cells together with the pcRev and the Rev-dependent CAT-reporter plasmid pDM128 derived from the env region of HIV (Fig. 3A) (33). Transcripts produced by pDM128 contain a single intron containing the HIV-1 RRE and the CAT coding sequence, which is excised when it is spliced (33). Cells transfected with pDM128 alone express only spliced transcripts and yield minimal levels of CAT activity; however, when Rev is cotransfected with pDM128, high levels of CAT enzyme are expressed (Fig. 3B). Transfection of increasing amounts of the pSF2/ASF in the presence of Rev significantly inhibited Rev function as evidenced by diminished CAT expression (Fig. 3B).
Figure 3.
Transient expression SF2/ASF in CV1 cells inhibits HIV Rev function. (A) Diagram of the pDM128 plasmid used in these studies. (B) CV1 cells were transfected with the Rev-responsive pDM128 CAT reporter plasmid in the presence and absence of Rev and various amounts of pSF2/ASF expression plasmid. DNA concentration was held constant by the addition of pCMV4 parental plasmid. Forty-eight hours after transfection, the cells were lysed and CAT assays were performed. (C) CV1 cells were transfected with the pDM128–RexRE reporter plasmid in which HIV-1 RRE was replaced by the HTLV-I RexRE. An HTLV-I Rex expression plasmid pcRex was transfected instead of HIV-1 pcRev. (D and E) CV1 cells were transfected with pDM128, HIV-1 Rev, and various amounts of two different SF2/ASF substitution mutants including SF2/ASF-FF-DD altered in the RNP-1 region of the RRM and the SF2-RT, where all serines in the RS domain were replaced with threonine (20). (F) CV1 cells cotransfected with pDM128, HIV-1 Rev, and various amounts of a second SR protein expression vector pSRp40. Stable expression of SF2/ASF FF-DD, SF2/ASF RT, and SRp40 was confirmed by immunoblotting of transfected CV-1 cell lysates.
To explore further the specificity of these inhibitory effects of SF2/ASF, its effect on human T-lymphotropic virus type I (HTLV-I) Rex action via the HTLV-I Rex response element (RexRE) was evaluated. The Rex/RexRE system of HTLV-I is functionally homologous to the Rev/RRE system; indeed, Rex can function through the RRE as well as the RexRE (34). Addition of SF2/ASF produced only minimal inhibitory effects on Rex action via the RexRE (Fig. 3C). This lack of significant inhibition on the RexRE-containing plasmid further supports specificity in the observed SF2/ASF inhibition of Rev action via the RRE.
SF2/ASF contains two distinct functional domains that are required for its constitutive splicing activity in vitro. These domains include the N-terminal RNA recognition motif (RRM) and the C-terminal RS domain (20). Two mutants of SF2/ASF selectively altered in each of these domains were next tested for inhibitory effects on Rev function in the pDM128 CAT expression assay. The first mutant, FF-DD, corresponds to a dual aspartic acid substitution for the phenylalanine residues located at positions 56 and 58 in the RNP-1 submotif of the RRM (20). This mutant did not inhibit expression of the Rev-dependent CAT activity (Fig. 3D). However, immunoblotting confirmed high-level expression of this mutant protein in these transfected cells (data not shown). The second mutant, RT, corresponds to an RS domain mutant in which all of the serine residues have been substituted with threonine (20). Cotransfection of this RT mutant produced inhibitory effects on Rev function indistinguishable from those found with wild-type SF2/ASF (Fig. 3E). These results suggest that the RS domain is dispensable for SF2/ASF-mediated inhibition of Rev-dependent CAT expression. Given the conservative nature of the threonine for serine mutation in the RS domain, a third SF2/ASF mutant was prepared, which lacked the entire RS domain (Δ RS). Cotransfection of this deletion mutant also produced inhibitory effects on Rev function identical to those of the wild-type and RT mutants of SF2/ASF (data not shown). These findings confirm that the RRM, but not the RS, domain of SF2/ASF is required for the inhibition of Rev-dependent gene expression.
To explore whether other SR proteins exerted similar inhibitory effects on Rev action, SRp40 was tested in the pDM128 assay. Like SF2/ASF, SRp40 can restore in vitro splicing activity to splicing-deficient extracts (S100) prepared from HeLa cells (16, 35). However, in contrast to SF2/ASF, SRp40 exerted essentially no inhibitory effects on Rev action in vivo (Fig. 3E) and failed to assemble with the Rev/RRE complex in vitro (data not shown). These findings indicate that not all SR proteins can counter Rev function and assemble with the Rev/RRE complex.
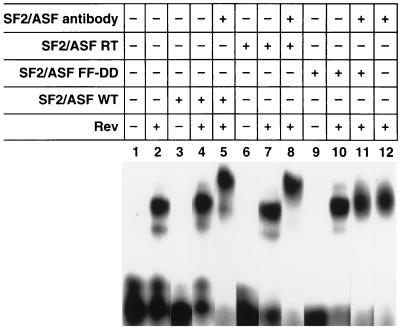
The lack of inhibitory activity seen with the SF2/ASF FF-DD mutant in the Rev-dependent pDM128 CAT assay could reflect the inability of this mutant to bind to the Rev/RRE complex. To address this possibility, wild-type SF2/ASF and the FF-DD and RT mutants were cloned into recombinant baculovirus expression plasmids and expressed in Sf9 cells. REMSA was subsequently performed using purified recombinant polyhistidine-tagged wild-type and mutant SF2/ASF FF-DD, and RT proteins prepared by nickel (II)–nitrolotriacetic acid–Sepharose affinity chromatography. These assays were performed as described in Fig. 1, and in addition, antibody specific for SF2/ASF was added to confirm the presence of wild-type or mutant SF2/ASF proteins in the retarded Rev/RRE complex. The addition of both wild-type SF2/ASF and the RT mutant to the Rev/RRE-binding reactions decreased the mobility of the Rev/RRE complexes (Fig. 4, lanes 4 and 7), and the addition of antibody to SF2/ASF further retarded the complex (Fig. 4, lanes 5 and 8). In contrast, the addition of the SF2/ASF FF-DD mutant protein alone or with antibody to SF2/ASF to Rev/RRE-binding reactions did not affect the mobility of the complex, (Fig. 4, lanes 10 and 11), suggesting that the FF-DD mutant cannot assemble with the Rev/RRE complex.
Figure 4.
The SF2/ASF FF-DD mutant does not assemble with the Rev/RRE complex in vitro. Wild-type SF2/ASF and the FF-DD and RT mutants of SF2/ASF were cloned into recombinant baculovirus expression plasmids and expressed in Sf9 cells. REMSA was performed using these recombinant polyhistidine-tagged proteins after purification on nickel affinity columns. As indicated, REMSA was performed in the presence and absence of Rev and wild-type or mutant SF2/ASF and antibody to SF2/ASF.
To investigate whether the inhibitory effects of SF2/ASF on Rev function were exerted at the level of pre-mRNA splicing, the pgTAT plasmid (23) was transfected into COS cells in the presence and absence of vectors encoding Rev and SF2/ASF. RNA was harvested from these cells, and RT-PCR analysis was performed with primers specific for the first and second coding exons of tat. Analysis of the RNA isolated from cells transfected with pgTAT alone revealed a 258-bp PCR product representing the spliced form of tat mRNA (Fig. 5, lane 1). In the presence of Rev, an additional 2588-bp PCR product was detected that corresponds to the unspliced tat pre-mRNA. When increasing amounts of pSF2/ASF were cotransfected with the pgTAT and pcRev plasmids, the unspliced form of tat pre-mRNA progressively decreased in a dose-dependent manner (Fig. 5, lanes 3–5). Due to efficient and nonlinear amplification of the smaller spliced product, it was not possible to assess whether the spliced mRNA correspondingly increased as the unspliced pre-mRNA declined. These results indicate that SF2/ASF inhibits Rev function at the level of viral RNA splicing.
Figure 5.
The effect of transient overexpression of SF2/ASF on tat pre-mRNA splicing. COS cells were transfected with the pgTAT plasmid in the presence and absence of expression vectors encoding Rev and SF2/ASF (23). Total cellular RNA was prepared 48 hr after transfection and analyzed by RT-PCR with primers specific for the first and second coding exons of tat. In the absence of Rev, the pre-mRNA undergoes complete splicing, which produces a two-exon form of tat RNA that is amplified as a 258-bp RT-PCR product (lane 1). Rev supplied in trans results in the appearance of the unspliced tat pre-mRNA, which is amplified as a 2588-bp RT-PCR product (lane 2). The unspliced pre-mRNA diminished as increasing amounts of the SF2/ASF expression plasmid were transfected (lanes 3–5).
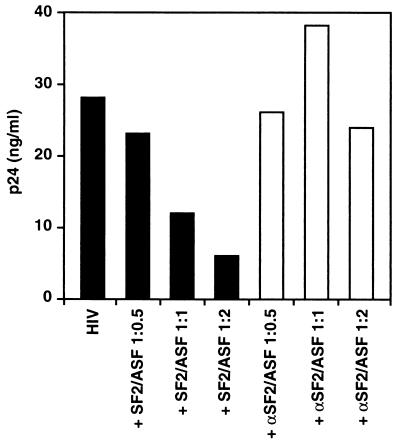
Finally, the effects of exogenous SF2/ASF expression on the replication of HIV-1 were examined. Viral replication was assayed by measuring the accumulation of p24 Gag protein in the cell culture supernatants. As shown in Fig. 6, SF2/ASF induced dose-related inhibition of viral replication in COS cells transfected with a replication-competent R7/3 provirus (8). In contrast, no inhibitory effects were observed after cotransfection of these cells with an antisense SF2/ASF expression vector. These results thus indicate that the inhibitory effects of SF2/ASF observed on Rev-dependent reporter plasmids also extends to the replication of HIV.
Figure 6.
Coexpression of SF2/ASF inhibits HIV-1 replication. COS cells were transfected with a replication-competent HIV-1 proviral clone, R7/3 (8), either alone or in the presence of various amounts of plasmids encoding sense (SF2/ASF) or antisense (αSF2/ASF) (ratio shown depicts amount of R7/3 provirus transfected versus SF2/ASF expression plasmid). DNA concentrations were kept constant in all transfections by addition of the control parental plasmid pCMV4. Cell culture supernatants were assayed 72 hr after transfection for p24 Gag protein using a p24 antigen capture ELISA kit (Coulter).
DISCUSSION
In these studies we have demonstrated that SF2/ASF can physically associate with the RRE in vitro when Rev is bound and that overexpression of exogenous SF2/ASF produces dose-related inhibition of Rev function and HIV replication in vivo. SF2/ASF appears to bind within the stem–loop III region of the RRE, which is distinct from the high-affinity Rev-binding site located in the stem–loop IIb region of this element. Of note, a prior study (36) has suggested that Rev binding to the RRE produces a conformational change in the stem–loop III region leading to diminished cleavage by the double-strand-specific RNase CV probe. The apparent increase in single strandedness within this stem–loop III region is consistent with the single-stranded RNA-binding properties of the highly conserved RNA-binding domain recently described for the U1A spliceosomal protein (37). Our studies have further shown that mutation of the RNP-1 domain present in SF2/ASF inhibits its assembly with the Rev/RRE complex in vitro and blocks the inhibitory effects of SF2/ASF on Rev function in vivo. In contrast, mutation of the RS domain of SF2/ASF does not alter either its ability to assemble with the Rev/RRE complex in vitro or its inhibition of Rev action in vivo.
Previous studies of in vitro splicing of RRE-containing mRNAs revealed that Rev, or a peptide derived from the basic RNA-binding domain of Rev (Rev 34–50) (13, 14), inhibits splicing. This inhibition appeared to occur at a step in the splicing reaction after formation of the “commitment complex.” In mammalian cells, pre-mRNAs are committed to splicing in the pre-spliceosomal complex E, which corresponds to U1-snRNP bound to the 5′ splice donor site (38). Formation of this commitment complex allows a pre-mRNA to be preferentially spliced even when an excess of competitor RNA is added (39). Of note, SR proteins have been shown to participate with U1-snRNP in promoting this first sequence-specific recognition event leading to the formation of the E complex (32, 40). However, SR proteins also appear to mediate subsequent interactions between components of the E complex and possibly the A complex, which additionally contains U2-snRNP bound near the 3′ splice site in the pre-mRNA. Specifically, the SR proteins likely bridge interactions between the 5′ and 3′ splice sites (21, 41). It is possible that Rev/RRE binding to SF2/ASF inhibits the formation of the A complex, which corresponds to pre-mRNA bound to the U1- and U2-snRNPs but still lacking the U4/U6·U5-snRNP complex (14). The binding of SF2/ASF by the Rev/RRE complex may interfere with these bridging functions. In addition, SF2/ASF may recruit U1 (or other snRNPs) to bind to the RRE, possibly resulting in the formation of a transient splicing inhibitory complex. The resulting inhibition of spliceosome assembly would then allow Rev-mediated nuclear export to occur. How overexpression of wild-type or mutant versions of SF2/ASF that are either lacking or altered in the RS protein interaction domain can overcome the effects of Rev is not yet understood, and the mechanisms may be distinct. It is possible that SF2/ASF RS domain substitution or deletion mutants displace endogenous SF2/ASF bound to the Rev/RRE complex thereby acting as transdominant inhibitors of SF2/ASF and inhibiting Rev function. We note, however, that a possibly related situation has been observed for SF2/ASF effects on the simian virus 40 early pre-mRNA splicing in vitro (29) and in vivo (42). In vitro, high concentrations of SF2/ASF result in use of the proximal 5′ splice site resulting in the production of small t mRNA (29); however, overexpression of SF2/ASF in vivo results in the inhibition of small t splicing by unknown mechanisms (42). The lack of requirement of the RS domain for SF2/ASF inhibition of Rev function in vivo was surprising, particularly since this domain has been implicated in protein–protein interactions necessary for constitutive splicing reactions. However, prior studies have shown that the SR domain is not required in splice site switching assays (20, 43).
During the course of our studies, precedent for the involvement of SF2/ASF in the regulation of alternative splicing in other viral systems has emerged. For example, Rous sarcoma virus contains a negative regulator of splicing that inhibits env and src splicing (44, 45); SF2/ASF binds specifically to this negative regulator of splicing, and this RNA–protein interaction is believed to mediate the observed inhibition of splicing (46). A second example involves the equine infectious anemia virus, which contains an exonic enhancer element in a bicistronic mRNA encoding Tat and Rev. This exonic enhancer has also been shown to interact with SF2/ASF (47). Rev binding to this exonic enhancer results in exon skipping and the induction of an mRNA encoding Tat only (47). The adenovirus L1 transcription unit is a third example of an alternatively spliced pre-mRNA that contains one 5′ splice site that can be joined to one of two different 3′ splice sites to produce either 52,55K or IIIa mRNAs (48). Like the complex retroviruses, adenoviruses display early and late stages of viral gene expression. During early gene expression, the 52,55K 3′ splice site is used efficiently due to the presence of an intronic splicing repressor element (3RE) that interacts specifically with SF2/ASF (48), and like the situation with Rev binding to the RRE, results in inhibition of A complex formation (14). Of note, when this element is moved to the second IIIa exon, it no longer represses splicing but rather enhances it (48). These examples suggest that SF2/ASF can exert both a positive and negative influence on alternative splicing in mammalian cells, and that its action is determined by the location of the SF2/ASF-binding site identified in the pre-mRNA. The conditional Rev-dependent SF2/ASF-binding site in the RRE would correspond to a negative element in the HIV system promoting an inhibition of splicing. Intriguingly, the RRE and SF2/ASF-binding site is also appropriately positioned in the exon of env to perhaps act as a splicing enhancer for env mRNA production. Both unspliced genomic mRNA and singly spliced env mRNA expression are controlled by Rev and both are needed for the assembly of infectious virions. However, whether SF2/ASF exerts such a “dual effect” in the HIV system remains unknown.
These results suggest that SF2/ASF interacts with the Rev/RRE complex to inhibit viral RNA splicing, thus enhancing the expression of the unspliced and singly spliced viral mRNAs needed for the propagation of viral infection. Additional effects of Rev are likely to occur at the level of facilitating viral RNA transport or translation. These transport effects could involve the specific interplay of other cellular factors, such as the nucleoporin-like Rab/Rip proteins (49–51) or eIF 5A (52, 53), with the leucine-rich activation domain of Rev (7), which functions as a nuclear export signal (54, 55). Mutation of this leucine-rich domain results in the formation of potent dominant–negative inhibitors of Rev (56), which already have shown promise in early clinical trials (57). In this regard, manipulation of the intracellular levels of SF2/ASF might produce additive or synergistic inhibitory effects with these Rev-dominant negative inhibitors.
Acknowledgments
We thank Adrian R. Krainer for the pT7A.A-p32, pET9c-SF2, pET9c-RT, and pET9c-FF-DD plasmids and SF2 antibody; Tris Parslow for critical review of the manuscript and the pDM128 and pDM128 RexRE plasmids; P. Wingfield for the Rev protein; A. Zahler for the mAb-104 antibody; K. Lynch for the baculovirus SF2/ASF; D. Brust for constructing the pSF2/ASF plasmid; and Gary Howard and Stephen Ordway for editorial assistance. D.M.P. was supported by a Public Health Service Grant 5T32 AI077334-07. We also wish to acknowledge core support from the University of California, San Francisco, Center for AIDS Research.
Footnotes
Abbreviations: RRE, Rev response element; SR, serine–arginine rich; REMSA, RNA electrophoretic mobility shift assay; CAT, chloramphenicol acetyltransferase; RT, reverse transcription; BS, base switched; snRNP, small nuclear ribonucleoprotein; HTLV-I, human T-lymphotropic virus type I; RexRE, Rex response element; RRM, RNA recognition motif.
References
- 1.Arrigo S J, Chen I S. Genes Dev. 1991;5:808–819. doi: 10.1101/gad.5.5.808. [DOI] [PubMed] [Google Scholar]
- 2.Felber B K, Hadzopoulou C M, Cladaras C, Copeland T, Pavlakis G N. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S Y, Byrn R, Groopman J, Baltimore D. J Virol. 1989;63:3708–3713. doi: 10.1128/jvi.63.9.3708-3713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knight D M, Flomerfelt F A, Ghrayeb J. Science. 1987;236:837–840. doi: 10.1126/science.3033827. [DOI] [PubMed] [Google Scholar]
- 5.Malim M H, Hauber J, Le S Y, Maizel J V, Cullen B R. Nature (London) 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 6.Emerman M, Vazeux R, Peden K. Cell. 1989;57:1155–1165. doi: 10.1016/0092-8674(89)90053-6. [DOI] [PubMed] [Google Scholar]
- 7.Malim M H, Bohnlein S, Hauber J, Cullen B R. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 8.Feinberg M B, Jarrett R F, Aldovini A, Gallo R C, Wong S F. Cell. 1986;46:807–817. doi: 10.1016/0092-8674(86)90062-0. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber B K, Pavlakis G N. J Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruskin B, Zamore P D, Green M R. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- 11.Seraphin B, Rosbash M. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Blanco M, Jamison S F, Sharp P A. Genes Dev. 1989;3:1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- 13.Kjems J, Frankel A D, Sharp P A. Cell. 1991;67:169–178. doi: 10.1016/0092-8674(91)90580-r. [DOI] [PubMed] [Google Scholar]
- 14.Kjems J, Sharp P A. J Virol. 1993;67:4769–4776. doi: 10.1128/jvi.67.8.4769-4776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dignam J D. Methods Enzymol. 1990;182:194–203. doi: 10.1016/0076-6879(90)82017-v. [DOI] [PubMed] [Google Scholar]
- 16.Zahler A M, Lane W S, Stolk J A, Roth M B. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 17.Hager D A, Burgess R R. Anal Biochem. 1980;109:76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- 18.Cook K S, Fisk G J, Hauber J, Usman N, Daly T J, Rusche J R. Nucleic Acids Res. 1991;19:1577–1583. doi: 10.1093/nar/19.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powell D M, Zhang M J, Konings D A, Wingfield P T, Stahl S J, Dayton E T, Dayton A I. J Acquired Immune Defic Syndr Hum Retrovirol. 1995;10:317–323. [PubMed] [Google Scholar]
- 20.Caceres J F, Krainer A R. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J Y, Maniatis T. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 22.Beraud C, Sun S C, Ganchi P, Ballard D W, Greene W C. Mol Cell Biol. 1994;14:1374–1382. doi: 10.1128/mcb.14.2.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malim M H, Hauber J, Fenrick R, Cullen B R. Nature (London) 1988;335:181–183. doi: 10.1038/335181a0. [DOI] [PubMed] [Google Scholar]
- 24.Huang X J, Hope T J, Bond B L, McDonald D, Grahl K, Parslow T G. J Virol. 1991;65:2131–2134. doi: 10.1128/jvi.65.4.2131-2134.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malim M H, Cullen B R. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 26.Tiley L S, Malim M H, Tewary H K, Stockley P G, Cullen B R. Proc Natl Acad Sci USA. 1992;89:758–762. doi: 10.1073/pnas.89.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dayton E T, Konings D A, Powell D M, Shapiro B A, Butini L, Maizel J V, Dayton A I. J Virol. 1992;66:1139–1151. doi: 10.1128/jvi.66.2.1139-1151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavaloc Y, Popielarz M, Fuchs J P, Gattoni R, Stevenin J. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge H, Manley J L. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 30.Krainer A R, Conway G C, Kozak D. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 31.Zuo P, Manley J L. Proc Natl Acad Sci USA. 1994;91:3363–3367. doi: 10.1073/pnas.91.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohtz J D, Jamison S F, Will C L, Zuo P, Luhrmann R, Garcia B M, Manley J L. Nature (London) 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 33.Hope T J, Huang X J, McDonald D, Parslow T G. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rimsky L, Hauber J, Dukovich M, Malim M H, Langlois A, Cullen B R, Greene W C. Nature (London) 1988;335:738–740. doi: 10.1038/335738a0. [DOI] [PubMed] [Google Scholar]
- 35.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 36.Kjems J, Brown M, Chang D D, Sharp P A. Proc Natl Acad Sci USA. 1991;88:683–687. doi: 10.1073/pnas.88.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oubridge C, Ito N, Evans P R, Teo C H, Nagal K. Nature (London) 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 38.Michaud S, Reed R. Genes Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- 39.Fu X D. Nature (London) 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 40.Staknis D, Reed R. Mol Cell Biol. 1994;14:2994–3005. doi: 10.1128/mcb.14.5.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu X D, Maniatis T. Proc Natl Acad Sci USA. 1992;89:1725–1729. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Manley J L. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 43.Zuo P, Manley J L. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arrigo S, Beemon K. Mol Cell Biol. 1988;8:4858–4867. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McNally M T, Gontarek R R, Beemon K. Virology. 1991;185:99–108. doi: 10.1016/0042-6822(91)90758-4. [DOI] [PubMed] [Google Scholar]
- 46.McNally L M, McNally M T. J Virol. 1996;70:1163–1172. doi: 10.1128/jvi.70.2.1163-1172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gontarek R R, Derse D. Mol Cell Biol. 1996;16:2325–2331. doi: 10.1128/mcb.16.5.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanopka A, Muhlemann O, Akusjarvi G. Nature (London) 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 49.Bogerd H P, Fridell R A, Madore S, Cullen B R. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 50.Fritz C C, Zapp M L, Green M R. Nature (London) 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 51.Stutz F, Neville M, Rosbash M. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 52.Bevec D, Jaksche H, Oft M, Wohl T, Himmelspach M, Pacher A, Schebesta M, Koettnitz K, Dobrovnik M, Csonga R, Lottspeich F, Hauber J. Science. 1996;271:1858–1860. doi: 10.1126/science.271.5257.1858. [DOI] [PubMed] [Google Scholar]
- 53.Ruhl M, Himmelspach M, Bahr G M, Hammerschmid F, Jaksche H, Wolff B, Aschauer H, Farrington G K, Probst H, Bevec D, Hauber J. J Cell Biol. 1993;123:1309–1320. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 55.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 56.Malim M H, McCarn D F, Tiley L S, Cullen B R. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woffendin C, Ranga U, Yang Z, Xu L, Nabel G J. Proc Natl Acad Sci USA. 1996;93:2889–2894. doi: 10.1073/pnas.93.7.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]