Abstract
In budding yeast, TEL1 encodes a protein closely related to ATM. Xrs2 is an Nbs1 homolog and forms a complex with Mre11 and Rad50. We show here that Tel1 associ ates with double-strand breaks (DSBs) through a mechanism dependent on the C terminus of Xrs2. Although Xrs2 is required for the DNA degradation at DSBs, the C-terminal Xrs2 truncation does not affect the degradation. Tel1 and the C terminus of Xrs2 are similarly involved in cell survival and Rad53 phosphorylation after DNA damage. Our findings suggest that the Tel1 association with DNA lesions is required for the activation of DNA damage responses.
Keywords: ATM, Tel1, Xrs2, double-strand break
To ensure accurate transmission of genetic information, cells have evolved surveillance mechanisms that monitor genomic integrity and activate a variety of DNA damage responses, including cell-cycle arrest and alterations in gene expression (Zhou and Elledge 2000). DNA damage responses are initiated through the ATM family proteins, which are large protein kinases with similarity in the catalytic domain to phosphatidylinositol 3-kinases (Zhou and Elledge 2000). This family includes ATM (mammals), ATR (mammals), Mec1(Saccharomyces cerevisiae), Tel1(S. cerevisiae), and Rad3 (Schizosaccharomyces pombe). Each of these proteins falls into two subgroups based on homology; ATM is related most closely to Tel1, whereas ATR is more closely related to Mec1and Rad3.
In the budding yeast S. cerevisiae, Mec1plays a critical role in the DNA damage response pathway, whereas Tel1plays a minor role (Morrow et al. 1995; Sanchez et al. 1996). Mec1physically interacts with Lcd1(also called Ddc2 and Pie1), a protein that exhibits limited homology to the mammalian ATRIP and S. pombe Rad26 protein (Edwards et al. 1999; Paciotti et al. 2000; Rouse and Jackson 2000; Cortez et al. 2001; Wakayama et al. 2001). Both Mec1and Lcd1are recruited to sites of DNA damage, suggesting that the Mec1–Lcd1 complex may act as a primary sensor of DNA damage (Kondo et al. 2001; Melo et al. 2001; Rouse and Jackson 2002). RAD53 encodes a protein kinase related to the mammalian Chk2 protein (Zhou and Elledge 2000). Rad53 functions downstream of Mec1and Tel1, and is activated and phosphorylated after DNA damage (Sanchez et al. 1996; Sun et al. 1996; Usui et al. 2001; Nakada et al. 2003). Recent observations suggest that Rad9 interacts physically with Rad53 to modulate its kinase activity (Gilbert et al. 2001; Schwartz et al. 2002). Activation of the Rad53 pathway results in cell-cycle arrest and transcriptional induction of genes required for damage repair.
The Mre11 complex consists of two highly conserved proteins, Mre11 and Rad50, and a third protein, either Xrs2 in budding yeast or Nbs1in mammals (D'Amours and Jackson 2002). The Mre11 complex has been most extensively characterized in budding yeast, and it is implicated in two different repair processes of double-strand breaks (DSBs): homologous recombination and nonhomologous end joining. In addition to DSB processing, the Mre11 complex is required for mediating the cellular response to DNA damage. Studies of human cells have demonstrated that ATM and the Mre11 complex constitute a DNA damage response pathway (Petrini 2000). ATM phosphorylates Nbs1in response to ionizing radiation (IR), and this Nbs1phosphorylation is important for IR-induced cell-cycle arrest in S phase (Lim et al. 2000; Wu et al. 2000; Zhao et al. 2000). Consistently, phenotypes of cells defective for Mre11 or Nbs1function are very similar to those of ATM-deficient cells with respect to the IR-induced cell-cycle arrest (Petrini 2000). In budding yeast, MRE11, RAD50, and XRS2 are required for activation of the Rad53 pathway following DNA damage (Grenon et al. 2001; Usui et al. 2001). Recent evidence suggests that Tel1 and the Mre11 complex are functionally overlapped in response to DNA damage (D'Amours and Jackson 2001; Usui et al. 2001). Cells with disruption in MRE11, RAD50, or XRS2 are sensitive to DNA damage. Although the tel1Δ mutation by itself does not confer sensitivity to DNA damage, it does cause increased sensitivity in a mec1 background (Morrow et al. 1995; Sanchez et al. 1996). Similarly, mre11Δ and xrs2Δ mutations exacerbate the DNA damage sensitivity of mec1 mutants (D'Amours and Jackson 2001; Usui et al. 2001). Sae2 (also called Com1) is required for some processes involving the Mre11 complex in budding yeast (Rattray et al. 2001; Lobachev et al. 2002). Interestingly, sae2Δ mutations enhance Tel1-mediated Rad53 phosphorylation after DNA damage, possibly because sae2Δ mutations accumulate DNA lesions to activate Tel1, and this enhancement requires the Mre11 complex (Usui et al. 2001). Moreover, Mre11 and Xrs2 are phosphorylated after DNA damage in a Tel1-dependent manner (D'Amours and Jackson 2001; Usui et al. 2001). Altogether, these results indicate that the functions of ATM/Tel1and the Mre11complex in the cellular DNA damage response are conserved between mammals and budding yeast.
In this study, we detected the interaction between Tel1and the C-terminal Xrs2 in a two-hybrid system, and investigated their genetic relationship. We show that the C terminus of Xrs2 is essential for the Tel1-mediated response to DNA damage. We also show that Tel1associates with DSBs, and that its association is dependent on the C terminus of Xrs2. Our results provide the molecular basis for a model in which Xrs2 recruits Tel1to DSBs, thereby participating in the surveillance of DNA damage.
Results and Discussion
To understand the mechanism by which Tel1regulates DNA damage responses, we searched for proteins interacting with Tel1in a two-hybrid screen. We isolated two positive clones (data not shown), both of which contained XRS2 fused to the transcriptional activation domain. Xrs2 consists of 854 amino acid residues, and both positive clones contained the C-terminal regions of Xrs2, one corresponding to amino acids 693–854 and the other to amino acids 474–854.
To examine the function of the Xrs2 C terminus, we constructed a truncated mutation, xrs2-11, which deletes the C-terminal 162 amino acids. Because xrs2Δ mutants are sensitive to DNA damage, we examined the sensitivity of the truncated xrs2-11 mutants to phleomycin and UV light (Fig. 1A,B; Supplementary Fig. 1). Phleomycin is a bleomycin-related antibiotic that induces DSBs in DNA (Moore 1988). UV-induced DNA lesions are possibly converted to DSBs during the cell-cycle progression (Galli and Schiestl 1999). We found that xrs2-11 mutants were as resistant to phleomycin and UV light as wild-type cells. Similar to xrs2-11 mutants, tel1Δ mutants were resistant to phleomycin and UV light.
Figure 1.
Effect of the C-terminal deletion of Xrs2 on DNA sensitivity and the complex formation. (A,B) Sensitivity to phleomycin (A) and UV light (B). Serial dilutions of cultures were spotted on YEPD media without or with phleomycin. Cultures spotted on YEPD media were irradiated with UV light. All of the strains contain the sml1Δ mutation that suppresses the lethality of mec1 mutants (Zhao et al. 1998). Strains used were wild-type (KSC1560), xrs2Δ (KSC1562), xrs2-11 (KSC1563), tel1Δ (KSC1661), mec1-81 (KSC1662), mec1-81 tel1Δ (KSC1564), mec1-81 xrs2-11 (KSC1565), and mec1-81 tel1Δ xrs2-11 (KSC1566). (C) Interaction between Tel1and Xrs2. Cells were untreated (–) or treated with 50 μg/mL phleomycin for 1h (+). Extracts were prepared from cells and immunoprecipitated with anti-HA antibodies. Immunoprecipitates (IP) and whole extracts were subjected to immunoblotting analysis. Strains used were XRS2-myc (KSC1904), xrs2-11-myc (KSC1905), TEL1-HA (KSC1785), TEL1-HA XRS2-myc (KSC1906), and TEL1-HA xrs2-11-myc (KSC1907). (D) Interaction of Xrs2 with Mre11 and Rad50. Extracts were prepared from XRS2-HA (KSC1744), xrs2-11-HA (KSC1869), or untagged cells (KSC1516), and immunoprecipitated with anti-HA antibodies. IPs and whole extracts were subjected to immunoblotting analysis. Phosphatase treatment diminished the slower migrating forms of Xrs2-HA and Xrs2-11-HA (data not shown), suggesting that Xrs2 and Xrs2-11 are phosphorylated proteins.
Although the tel1Δ mutation by itself does not confer sensitivity to DNA damage, it does exacerbate the sensitivity of mec1 mutants to DNA damage (Morrow et al. 1995; Sanchez et al. 1996). We therefore asked whether the xrs2-11 mutation has a similar effect in a mec1 background (Fig. 1A,B; Supplementary Fig. 1). Because the mec1Δ tel1Δ double mutation results in a loss of cell proliferation (Chan et al. 2001), we used a weak allele of mec1, mec1-81, to assess sensitivity to DNA damage. mec1-81 single mutants are sensitive to phleomycin and UV light, and this sensitivity was increased by introduction of either the tel1Δ or xrs2-11 mutation to the same extent. Moreover, the DNA damage sensitivity of these mec1-81 tel1Δ or mec1-81 xrs2-11 double mutants was very similar to that of the mec1-81 tel1Δ xrs2-11 triple mutants.
We then examined whether Tel1and Xrs2 interact physically and whether their interaction is dependent on the C terminus of Xrs2, by coimmunoprecipitation experiments (Fig. 1C). We generated strains containing HA-tagged TEL1 and/or myc-tagged XRS2 or xrs2-11 alleles. After cells were treated or untreated with phleomycin, extracts were prepared from the cells and subjected to immunoprecipitation with anti-HA antibodies. Immunoprecipitates were then analyzed by immunoblotting with anti-HA and anti-myc antibodies. Xrs2-myc was coprecipitated with Tel1-HA in cells expressing both the tagged Tel1 and Xrs2 proteins, and the amount of coprecipitated Xrs2-myc was increased after phleomycin treatment. However, Xrs2-11-myc was not detected in the immunocomplex with Tel1-HA. These results indicate that the C-terminal Xrs2 region mediates the Tel1–Xrs2 interaction in vivo. The Xrs2–Tel1interaction remained intact after DNase I treatment, suggesting that its interaction is not bridged by DNA fragments (data not shown).
Because Xrs2 forms a complex with Mre11 and Rad50, we examined whether the xrs2-11 mutation affects its complex formation (Fig. 1D). Extracts prepared from XRS2-HA, xrs2-11-HA, or untagged cells were subjected to immunoprecipitation with anti-HA antibodies, and immunoprecipitates were analyzed on immunoblots with anti-HA, anti-Mre11, or anti-Rad50 antibodies. Mre11 and Rad50 were detected similarly in immunoprecipitates from XRS2-HA and xrs2-11-HA cells, but not from untagged cells. Expression levels of Xrs2 proteins were not different in XRS2-HA and xrs2-11-HA cells (Fig. 1D). Thus, the xrs2-11 mutation does not affect the Mre11–Rad50–Xrs2 complex formation. Together, these results indicate that Tel1and the C terminus of Xrs2 function similarly in response to DNA damage.
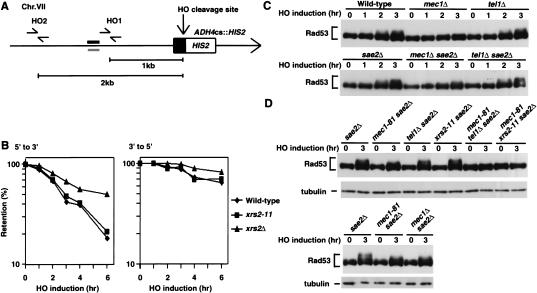
DSB ends are degraded primarily by 5′-to-3′ exonuclease activity, producing long 3′-ended single-stranded DNA (ssDNA) tails (White and Haber 1990), and efficient degradation is dependent on the Mre11 complex function (Ivanov et al. 1994; Lee et al. 1998). We then investigated whether the xrs2-11 mutation affects the degradation rate of DNA ends. To examine the degradation, we used MATa-inc ADH4cs strains, in which the MAT locus is replaced with MATa-inc, a mutation of the HO cleavage site (Sweetser et al. 1994), and a separate cleavage site (HOcs) is introduced in the ADH4 locus (Fig. 2A). In these strains, the HO-induced DSB at the ADH4 locus is not repaired by homologous recombination. Irreparable HO-induced DSBs activate Rad53 kinase at G2/M phase but not at G1(Pellicioli et al. 2001). We then examined the degradation rate following HO expression from the galactose-inducible GAL10 promoter in G2/M-arrested cells. Cells carrying the GAL-HO plasmid were grown initially in sucrose to repress HO expression, and then transferred to medium containing nocodazole to arrest at G2/M. After arrest, galactose was added to induce HO expression. Cells were collected at various time intervals to prepare genomic DNA. Purified DNA was fixed to a membrane and probed with strand-specific sequences near the cleavage site at the ADH4 locus (Fig. 2A). As reported (Ivanov et al. 1994), the rate of 5′-to-3′ degradation of the DNA ends was decreased in xrs2Δ mutants compared to wild-type cells (Fig. 2B). In contrast, no decrease in the rate of 5′-to-3′ degradation was observed in xrs2-11 mutants. Moreover, the xrs2-11 mutation had relatively little effect on the rate of 3′-to-5′ degradation (Fig. 2B). Thus, the C-terminal truncation of Xrs2 does not impair the DNA degradation at the DSB.
Figure 2.
Effect of the C-terminal deletion of Xrs2 on the cellular response to DSBs. (A) A single HO restriction site at the ADH4 locus in MATa-inc cells. A solid box indicates a DNA fragment containing an HO cleavage site, which is marked with HIS2. The black and gray bars indicate probes to examine the rate of degradation of the DSB ends. The primer pairs HO1and HO2 were designed to amplify regions near the cleavage site on the ADH4 locus by PCR for the experiments described in Figures 3 and 4. An arrow represents telomere. The MATa locus was replaced with the MATa-inc allele in which the HO endonuclease does not generate a DSB. (B) Degradation of the HO-induced DSB ends. Wild-type (KSC1516), xrs2Δ (KSC1620), and xrs2-11 (KSC1621) cells carrying YCpA-GAL-HO were grown in sucrose and treated with nocodazole. After arrest at G2/M, the culture was incubated with galactose to induce HO expression. At the indicated time points, aliquots were harvested for DNA preparation. Purified DNAs were fixed to a membrane and probed with 32P-labeled oligonucleotides, each complementary to a 5′-to-3′-degrading or 3′-to-5′-degrading strand as in A. (C) Rad53 phosphorylation after HO expression. Cells carrying YCpT-RAD53-HA and YCpA-GAL-HO were grown in sucrose and treated with nocodazole. After arrest at G2/M, the culture was incubated with galactose to induce HO expression, while part of the culture was maintained in sucrose to repress HO expression. At the indicated time points, aliquots were harvested for immunoblotting analysis. Cells used were wild-type (KSC1560), mec1Δ (KSC1561), tel1Δ (KSC1661), sae2Δ (KSC1567), mec1Δ sae2Δ (KSC1701), and tel1Δ sae2Δ (KSC1594). (D) Effect of the tel1Δ and xrs2-11 mutations on the HO-induced Rad53 phosphorylation. Cells were analyzed as in C to detect Rad53 phosphorylation. Tubulin was detected as a loading control. Cells used were sae2Δ (KSC1567), mec1-81 sae2Δ (KSC1775), tel1Δ sae2Δ (KSC1594), xrs2-11 sae2Δ (KSC1595), mec1-81 xrs2-11 sae2Δ (KSC1597), mec1-81 tel1Δ sae2Δ (KSC1596), and mec1Δ sae2Δ (KSC1701).
Rad53 is activated and phosphorylated after DNA damage (Sanchez et al. 1996; Sun et al. 1996). Although Mec1plays a major role in the DNA damage-induced Rad53 phosphorylation, Tel1contributes to its phosphorylation (Sanchez et al. 1996; Usui et al. 2001; Nakada et al. 2003). We then asked whether Tel1and the C terminus of Xrs2 are similarly involved in the HO-induced Rad53 phosphorylation at G2/M using MATa-inc ADH4cs strains. We first analyzed the status of Rad53 phosphorylation after HO expression in mec1Δ and tel1Δ mutants (Fig. 2C). Cells expressing the Rad53-HA protein were transformed with the GAL-HO plasmid, and were grown as above to induce HO expression. Cells were subsequently collected at various times and analyzed on immunoblots to monitor the modification of Rad53. Rad53 phosphorylation became apparent 2 h after incubation with galactose in wild-type cells. Similarly, Rad53 became phosphorylated after HO expression in tel1Δ cells. In contrast, no apparent phosphorylation was detected in mec1Δ cells. It was thus difficult to detect the Tel1-dependent Rad53 phosphorylation after HO expression. Because sae2 mutations enhance the Tel1-mediated responses (Usui et al. 2001), we next examined the HO-induced Rad53 phosphorylation in cells carrying an sae2Δ mutation allele (Fig. 2C). The Rad53 phosphorylation status was not significantly altered in sae2Δ and tel1Δ sae2Δ cells compared to wild-type SAE2 and tel1Δ SAE2 cells, respectively. However, in mec1Δ sae2Δ cells, Rad53 phosphorylation became detectable following HO expression, although it is relatively weak (Fig. 2C,D). The Rad53 phosphorylation observed in mec1Δ sae2Δ cells is likely dependent on Tel1, because Rad53 phosphorylation was detectable in mec1-81 sae2Δ cells, but not in mec1-81 tel1Δ sae2Δ cells (Fig. 2D). Thus, the HO-induced Rad53 phosphorylation is dependent on both Mec1and Tel1in the absence of Sae2, suggesting that Mec1and Tel1act in parallel to phosphorylate Rad53 (Usui et al. 2001). We further assessed the effect of the xrs2-11 mutation on the HO-induced Rad53 phosphorylation in sae2Δ strains. The xrs2-11 mutation behaved similar to the tel1Δ mutation; the HO-induced Rad53 phosphorylation was abolished in mec1-81 xrs2-11 sae2Δ cells, but was observed in xrs2-11 sae2Δ cells (Fig. 2D). Consistently, Rad53 kinase activity was increased in mec1-81 sae2Δ cells after HO expression, and this increase was abolished by the introduction of the tel1Δ or xrs2-11 mutation (Supplementary Fig. 2). These results indicate that the tel1Δ and xrs2-11 mutations confer the same defect in the HO-induced Rad53 phosphorylation in the absence of Sae2, further suggesting that the C terminus of Xrs2 is functionally similar to Tel1in mediating DNA damage responses.
We then investigated whether Tel1associates with sites near the HO-induced DSB in sae2Δ strains (Fig. 3A). Cells expressing an HA-tagged Tel1protein were transformed with the GAL-HO plasmid and grown as above to induce HO expression. Cells were then collected at various time intervals and subjected to immunoprecipitation by anti-HA antibodies. Coprecipitated DNA was extracted and amplified by polymerase chain reaction (PCR) using either primer sets (HO1and HO2) corresponding to regions near the HO-cleavage site at the ADH4 locus on chromosome VII or primers for the SMC2 locus containing no cleavage site on chromosome VI (Fig. 2A). PCRs with the primer pairs HO1and HO2 amplify regions 1.0 kb and 2.0 kb distant from the cleavage site, respectively. Interaction of Tel1with the ADH4 locus was detected in sae2Δ cells after HO expression (Fig. 3A). In contrast, no interaction was detected in cells lacking the cleavage site at the ADH4 locus (data not shown). The Tel1interaction is specific to the ADH4 locus, because the PCR-amplified fragment from the SMC2 locus did not increase after HO expression (Fig. 3A). These results indicate that Tel1associates with sites near the HO-induced DSB in sae2Δ cells.
Figure 3.
Association of Tel1with sites near the HO-induced DSB. (A) The Tel1association with DSBs in SAE2 and sae2Δ cells. Cells expressing Tel1-HA (KSC1785 and KSC1786) were transformed with the YCpA-GAL-HO plasmid. Transformed cells were grown in sucrose and treated with nocodazole. After arrest at G2/M, the culture was incubated with galactose to induce HO expression, while part of the culture was maintained in sucrose to repress HO expression. Aliquots of cells were collected at the indicated times and subjected to chromatin immunoprecipitation. PCR was done with the primers for the ADH4 locus shown schematically in Figure 2A and for the control SMC2 locus. PCR products from the respective input extracts are shown in parallel. (B) Effect of the sae2Δ mutation on Tel1 association with DSBs in mec1Δ mutants. Cells expressing Tel1-HA (KSC1873, KSC1874 and KSC1875) were analyzed as in A. (C) Effect of xrs2Δ and xrs2-11 mutations on Tel1association with the DSBs. Cells expressing Tel1-HA (KSC1785, KSC1881 and KSC1882) were analyzed as in the top two panels of B. Immunoprecipitates were also subjected to immunoblotting analysis with anti-HA antibodies (bottom).
We next compared Tel1association with DSBs in SAE2 and sae2Δ cells. Tel1associated with the HO-induced DSB irrespective of the SAE2 or sae2Δ allele (Fig. 3A), although Tel1does not apparently contribute to the HO-induced Rad53 phosphorylation in the presence of SAE2 (see Fig. 2). Because Tel1is clearly involved in Rad53 phosphorylation in mec1Δ sae2Δ mutants, but not in mec1Δ mutants (see Fig. 2), we compared Tel1 association with DSBs in mec1Δ and mec1Δ sae2Δ cells. Tel1associated with the DSB in mec1Δ cells as well as in wild-type cells (Fig. 3B). Again, the sae2Δ mutation did not affect the Tel1association; Tel1associated with the DSB in mec1Δ cells as efficiently as in mec1Δ sae2Δ cells (Fig. 3B). These results suggest that association of Tel1 with DSBs does not necessarily result in activation of the Rad53 pathway.
Because Tel1is functionally similar to the C terminus of Xrs2, we asked whether the Tel1association with DSBs is dependent on XRS2 (Fig. 3C). We found that Tel1 did not associate with the HO-induced DSB in xrs2Δ mutants. Moreover, the Tel1association was not detected in xrs2-11 mutants. Immunoblotting analysis showed that the same amount of Tel1-HA was immunoprecipitated from these cell extracts. Neither xrs2Δ nor xrs2-11 mutation affected the expression level or cellular localization of Tel1(Supplementary Fig. 3). Thus, the C-terminal Xrs2 function is required for the Tel1 association with DSBs.
We also examined whether Xrs2 is recruited to sites near the HO-induced DSB in cells arrested with nocodazole at G2/M (Fig. 4A). Cells expressing an HA-tagged Xrs2 protein were subjected to chromatin immunoprecipitation, and precipitated DNA was amplified by PCR as described above. The Xrs2 interaction with the ADH4 locus became detectable after HO expression (Fig. 4A). This interaction was not detected in cells lacking the cleavage site at the ADH4 locus (data not shown). Thus, Xrs2 associates with the HO-induced DSB. The Xrs2 association was unaffected by introduction of the tel1Δ mutation (Fig. 4B), indicating that Xrs2 is recruited to DSBs independently of Tel1. No apparent change in the Xrs2 association was observed in sae2Δ or mec1Δ cells (Fig. 4B). Consistent with the finding that the C-terminal truncation does not impair the function of Xrs2 in DSB degradation, the Xrs2-11 mutant protein associated with the DSB similar to the wild-type Xrs2 protein (Fig. 4C). Altogether, these results indicate that Tel1associates with DSBs in a mechanism dependent on the C-terminal function of Xrs2.
Figure 4.
Association of Xrs2 with sites near the HO-induced DSB. (A) Time course of the Xrs2 association with DSBs. Cells expressing Xrs2-HA (KSC1744) were analyzed as in Figure 3A. (B) Effect of mec1Δ, tel1Δ and sae2Δ mutations on the Xrs2 association with DSBs. Cells expressing Xrs2-HA were analyzed as in Figure 3B. Cells used were wild-type (KSC1744), tel1Δ (KSC1799), sae2Δ (KSC1798), sml1Δ (KSC1863), and mec1Δ sml1Δ (KSC1864). (C) Effect of the C-terminal truncation on the Xrs2 association with DSBs. XRS2-HA (KSC1744) and xrs2-11-HA (KSC1869) cells were analyzed as in B.
In this study we show that Tel1and the C terminus of Xrs2 are similarly involved in cell survival and Rad53 phosphorylation after DNA damage, and that Tel1associates with DSBs in a manner dependent on the C-terminal Xrs2 region. These results are consistent with a previous model in which Tel1and the Mre11complex constitute a DNA damage response pathway (D'Amours and Jackson 2001; Usui et al. 2001), and suggest that the Tel1association is essential for Tel1-mediated DNA damage responses. We also show that Tel1and Xrs2 interact physically through the C terminus of Xrs2. This Tel1–Xrs2 interaction is increased after phleomycin treatment, suggesting that the presence of DSBs could alter their interaction status. The DNA ends at DSBs are degraded in an XRS2-dependent manner, accumulating ssDNA. However, the C-terminal Xrs2 truncation does not affect the DNA degradation at DSBs, implying that the accumulation of ssDNA per se does not facilitate the Tel1association with DSBs. Tel1associates with DSBs, but its association does not necessarily correlate with activation of the Rad53 pathway. Mutations in SAE2 cause a defect in DSB repair (Rattray et al. 2001; Lobachev et al. 2002), but enhance the Tel1-mediated Rad53 phosphorylation after DNA damage (Usui et al. 2001). After association with DSBs, Tel1 might monitor DSB processing to activate the Rad53 pathway. In mammals, ATM and Nbs1function in the same pathway, and both localize to DSBs (Mirzoeva and Petrini 2001; Perkins et al. 2002; Bakkenist and Kastan 2003; Lukas et al. 2003). However, it has not yet been determined whether Nbs1regulates the association of ATM with DSBs. Because ATM/Tel1and the Mre11complex are highly conserved, it is possible that ATM associates with DSBs through a similar mechanism.
Materials and methods
Strains and plasmids
All yeast strains used are isogenic and are listed in Supplementary Table 1. Construction of strains and plasmids is described in the Supplemental Material.
Two-hybrid screening
Yeast two-hybrid screening of an S. cerevisiae genomic library was carried out using pBD-TEL1as bait as described (Wakayama et al. 2001).
Immunoblotting and immunoprecipitation
Immunoblotting and immunoprecipitation were performed as described (Wakayama et al. 2001). To examine the HO-induced Rad53 phosphorylation, cells carrying YCpA-GAL-HO and YCpT-RAD53-HA were grown in selective medium containing sucrose, and then arrested with 15 μg/mL of nocodazole for 120 min at G2/M. To induce HO expression, galactose was added to the culture to 2% at a final concentration. After nocodazole treatment, microscopic examination revealed that 85% of cells were large-budded, and FACS analysis predicted that 80% of cells contained the G2/M content of DNA. This G2/M arrest profile remained similar after HO expression. Antibodies against Mre11 and Rad50 were described previously (Raymond and Kleckner 1993; Usui et al. 2001).
Measurement of DSB processing
Purified DNA was fixed to a positively charged nylon membrane, and probed with oligonucleotides complementary to DNA strands near the HO restriction site at the ADH4 locus as described (Lee et al. 1998). The sequences of oligonucleotide probes to examine the 5′-to-3′ or 3′-to-5′ degradation are listed in the Supplemental Material. Fixed DNAs were also probed with a DNA fragment containing the TDH3 promoter.
Chromatin immunoprecipitation
The chromatin immunoprecipitation assay was performed essentially as described (Kondo et al. 2001), except for the use of Protein A sepharose. Each lot of Protein A sepharose was tested, confirming no specific PCR amplification with untagged strains. Immunoprecipitation of cross-linked DNA was performed after sonication using anti-HA (12CA5) antibodies. The DNA in the soluble fraction had an average size of 700 base pairs (bp) and a range between 200 and 1400 bp. DNA was also purified from the whole cell extract (designated “input”). The PCR reaction was performed in a nonsaturating condition, in which the rate of PCR amplification is proportional to the concentration of substrate and cycling. The sequences of PCR primers are listed in the Supplemental Material.
Acknowledgments
We thank N. Kleckner, J. Nickoloff, T. Ogawa, K. Ohta, J. Petrini, E. Schiebel, A. Shinohara, M. Shinohara, and T. Usui for materials. Thiswork was supported by Grants-in-Aid from the Ministry of Education and Science of Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1099003.
References
- Bakkenist, C.J. and Kastan, M.B. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499–506. [DOI] [PubMed] [Google Scholar]
- Chan, S.W., Chang, J., Prescott, J., and Blackburn, E.H. 2001. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 11: 1240–1250. [DOI] [PubMed] [Google Scholar]
- Cortez, D., Guntuku, S., Qin, J., and Elledge, S.J. 2001. ATR and ATRIP: Partners in checkpoint signaling. Science 294: 1713–1716. [DOI] [PubMed] [Google Scholar]
- D'Amours, D. and Jackson, S.P. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes & Dev. 15: 2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. The Mre11 complex: At the crossroads of DNA repair and checkpoint signalling. Nature Rev. Mol. Cell Biol. 3: 317–327. [DOI] [PubMed] [Google Scholar]
- Edwards, R.J., Bentley, N.J., and Carr, A.M. 1999. A Rad3–Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell Biol. 1: 393–398. [DOI] [PubMed] [Google Scholar]
- Galli, A. and Schiestl, R.H. 1999. Cell division transforms mutagenic lesions into deletion-recombinagenic lesions in yeast cells. Mutat. Res. 429: 13–26. [DOI] [PubMed] [Google Scholar]
- Gilbert, C.S., Green, C.M., and Lowndes, N.F. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8: 129–136. [DOI] [PubMed] [Google Scholar]
- Grenon, M., Gilbert, C., and Lowndes, N.F. 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3: 844–847. [DOI] [PubMed] [Google Scholar]
- Ivanov, E.L., Sugawara, N., White, C.I., Fabre, F., and Haber, J.E. 1994. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 3414–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, T., Wakayama, T., Naiki, T., Matsumoto, K., and Sugimoto, K. 2001. Recruitment of Mec1and Ddc1checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294: 867–870. [DOI] [PubMed] [Google Scholar]
- Lee, S.E., Moore, J.K., Holmes, A., Umezu, K., Kolodner, R.D., and Haber, J.E. 1998. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409. [DOI] [PubMed] [Google Scholar]
- Lim, D.S., Kim, S.T., Xu, B., Maser, R.S., Lin, J., Petrini, J.H., and Kastan, M.B. 2000. ATM phosphorylates p95/nbs1in an S-phase checkpoint pathway. Nature 404: 613–617. [DOI] [PubMed] [Google Scholar]
- Lobachev, K.S., Gordenin, D.A., and Resnick, M.A. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193. [DOI] [PubMed] [Google Scholar]
- Lukas, C., Falck, J., Bartkova, J., Bartek, J., and Lukas, J. 2003. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 5: 225–260. [DOI] [PubMed] [Google Scholar]
- Melo, J.A., Cohen, J., and Toczyski, D.P. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes & Dev. 21: 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva, O.K. and Petrini, J.H. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol. 21: 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, C.W. 1988. Internucleosomal cleavage and chromosomal degradation by bleomycin and phleomycin in yeast. Cancer Res. 23: 6837–6843. [PubMed] [Google Scholar]
- Morrow, D.M., Tagle, D.A., Shiloh, Y., Collins, F.S., and Hieter, P. 1995. TEL1 an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82: 831–840. [DOI] [PubMed] [Google Scholar]
- Nakada, D., Shimomura, T., Matsumoto, K., and Sugimoto, K. 2003. The ATM-related Tel1protein of Saccharomyces cerevisiae controls a checkpoint response following phleomycin treatment. Nucleic Acids Res. 31: 1715–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti, V., Clerici, M., Lucchini, G., and Longhese, M.P. 2000. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes & Dev. 14: 2046–2059. [PMC free article] [PubMed] [Google Scholar]
- Pellicioli, A., Lee, S.E., Lucca, C., Foiani, M., and Haber, J.E. 2001. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell 7: 293–300. [DOI] [PubMed] [Google Scholar]
- Perkins, E.J., Nair, A., Cowley, D.O., Dyke, T.V., Chang, Y., and Ramsden, D.A. 2002. Sensing of intermediates in V(D)J recombination by ATM. Genes & Dev. 16: 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini, J.H. 2000. The Mre11 complex and ATM: Collaborating to navigate S phase. Curr. Opin. Cell Biol. 12: 293–296. [DOI] [PubMed] [Google Scholar]
- Rattray, A.J., McGill, C.B., Shafer, B.K., and Strathern, J.N. 2001. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: A role for SAE2/COM1. Genetics 158: 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, W.E. and Kleckner, N. 1993. RAD50 protein of S. cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 21: 3851–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, J. and Jackson, S.P. 2000. LCD1: An essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 19: 5793–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9: 857–869. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., Desany, B.A., Jones, W.J., Liu, Q., Wang, B., and Elledge, S.J. 1996. Regulation of RAD53 by the ATM-like kinase MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271: 357–360. [DOI] [PubMed] [Google Scholar]
- Schwartz, M.F., Duong, J.K., Sun, Z., Morrow, J.S., Pradhan, D., and Stern, D.F. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9: 1055–1065. [DOI] [PubMed] [Google Scholar]
- Sun, Z., Fay, D.S., Marini, F., Foiani, M., and Stern, D.F. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes & Dev. 10: 395–406. [DOI] [PubMed] [Google Scholar]
- Sweetser, D.B., Hough, H., Whelden, J.F., Arbuckle, M., and Nickoloff, J.A. 1994. Fine-resolution mapping of spontaneous and double-strand break-induced gene conversion tracts in Saccharomyces cerevisiae reveals reversible mitotic conversion polarity. Mol. Cell. Biol. 14: 3863–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui, T., Ogawa, H., and Petrini, J.H. 2001. A DNA damage response pathway controlled by Tel1and the Mre11complex. Mol. Cell 7: 1255–1266. [DOI] [PubMed] [Google Scholar]
- Wakayama, T., Kondo, T., Ando, S., Matsumoto, K., and Sugimoto, K. 2001. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, C.I. and Haber, J.E. 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9: 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., Ranganathan, V., Weisman, D.S., Heine, W.F., Ciccone, D.N., O'Neill, T.B., Crick, K.E., Pierce, K.A., Lane, W.S., Rathbun, G., et al. 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405: 477–482. [DOI] [PubMed] [Google Scholar]
- Zhao, S., Weng, Y.C., Yuan, S.S., Lin, Y.T., Hsu, H.C., Lin, S.C., Gerbino, E., Song, M.H., Zdzienicka, M.Z., Gatti, R.A., et al. 2000. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature 405: 473–477. [DOI] [PubMed] [Google Scholar]
- Zhao, X., Muller, E.G.D., and Rothstein, R. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pool. Mol. Cell 2: 329–340. [DOI] [PubMed] [Google Scholar]
- Zhou, B.-B.S. and Elledge, S.J. 2000. The DNA damage response: Putting checkpoints in perspective. Nature 408: 433–439. [DOI] [PubMed] [Google Scholar]