Abstract
Bone remodeling is central to maintaining the integrity of the skeletal system, wherein the developed bone is constantly renewed by the balanced action of osteoblastic bone formation and osteoclastic bone resorption. In the present study, we demonstrate a novel function of the Stat1 transcription factor in the regulation of bone remodeling. In the bone of the Stat1-deficient mice, excessive osteoclastogenesis is observed, presumably caused by a loss of negative regulation of osteoclast differentiation by interferon (IFN)-β. However, the bone mass is unexpectedly increased in these mice. This increase is caused by excessive osteoblast differentiation, wherein Stat1 function is independent of IFN signaling. Actually, Stat1 interacts with Runx2 in its latent form in the cytoplasm, thereby inhibiting the nuclear localization of Runx2, an essential transcription factor for osteoblast differentiation. The new function of Stat1 does not require the Tyr 701 that is phosphorylated when Stat1 becomes a transcriptional activator. Our study provides a unique example in which a latent transcription factor attenuates the activity of another transcription factor in the cytoplasm, and reveals a new regulatory mechanism in bone remodeling.
Keywords: Stat1, Runx2, osteoblast, bone remodeling
The integrity of the vertebrate skeletal system is maintained by two distinct regulatory processes: the embryonic developmental and postnatal regulatory processes (Olsen et al. 2000; Karsenty and Wagner 2002). In the former process, the development of the skeletal system depends on the differentiation of chondrocytes, osteoblasts, and osteoclasts, all of which coordinate endochondral and membranous bone formation. On the other hand, in the latter process, referred to as bone remodeling, the bone matrix is constantly degraded by osteoclasts and deposited by osteoblasts (Manolagas 2000). The balanced action of these two cell types is critical for the normal homeostasis of the skeletal system in adults. Therefore, tipping this balance in favor of either cell type sometimes leads to pathological conditions. Excessive bone resorption is seen in autoimmune arthritis, periodontitis, postmenopausal osteoporosis, Paget's disease, and bone tumors, whereas excessive bone formation or defective bone resorption lead to osteosclerosis or osteopetrosis (Rodan and Martin 2000; Takayanagi et al. 2000a; Teitelbaum 2000). Thus, the investigation of the regulatory mechanism of differentiation of these two cell types is important in the understanding of the physiology and pathology of the skeletal system.
Previously, we have shown that both type I and type II interferon systems (i.e., IFN-α/β and IFN-γ systems) are critical for the regulation of the skeletal system by suppressing osteoclastogenesis (Takayanagi et al. 2000b, 2002b). These two IFNs exert their inhibitory functions by distinct mechanisms, as briefly described below (Stark et al. 1998; Taniguchi et al. 2001; Takayanagi et al. 2002c). During osteoclast differentiation, induced by RANKL (receptor activator of NF-κB ligand), the IFN-β gene is induced in osteoclast precursor cells and IFN-β inhibits the differentiation by interfering with the RANKL-induced expression of c-Fos, an essential transcription factor for osteoclastogenesis. This inhibition is dependent on the IFN-activated transcription factor, interferon stimulated gene factor 3 (ISGF3), which consists of activated Stat1, Stat2, and a member of the family of interferon regulatory factors (IRFs), IRF-9. On the other hand, IFN-γ is critical in the T-cell-mediated regulation of osteoclastogenesis. RANKL expression is induced by activated T cells, but RANKL signaling is negatively regulated by IFN-γ, which is also induced by the activated T-cells, thereby balancing RANKL signaling (Kong et al. 1999; Takayanagi et al. 2000b). Stat1 (signal transducer and activator of transcription 1) is required for this IFN-γ signal-mediated inhibition, in which the phosphorylated Stat1 forms a homodimer, termed IFN-γ-activated factor (GAF). Despite the distinct mechanisms of regulating osteoclast differentiation by IFN-β and IFN-γ, Stat1 is involved in both mechanisms, suggesting the integral role of Stat1 in the regulation of bone metabolism.
In this regard, it is interesting that an activating mutation in FGFR3 results in human achondroplasia, one of the most common congenital diseases characterized by dwarfism, and that Stat1 is involved in this pathogenesis (Rousseau et al. 1994; Deng et al. 1996). The skeletal malformation in achondroplasia is caused by the suppression of chondrocyte proliferation by excessive FGF signaling, leading to impaired endochondral ossification. FGF induces the phosphorylation of Stat1, and the suppressive effect of FGF on chondrocyte proliferation is not observed in the skeletal tissue derived from mice lacking Stat1, suggesting that activation of Stat1 is involved in the suppressive effect of FGF on chondrocyte proliferation in endochondral bone formation during embryogenesis (Su et al. 1997; Sahni et al. 1999, 2001). Notwith-standing, it is unknown at present if and how Stat1 is involved in the control of transcriptional program(s) of the skeletal system at the postnatal stage.
The transcription factor Runx2, also called Cbfa1, is a runt family transcription factor, and it plays a central role in the determination of osteoblast differentiation (Ducy et al. 1997, 2000). Targeted disruption of Runx2 results in the complete lack of bone formation by osteoblasts, revealing that Runx2 is essential for both endochondral and membranous bone formation (Komori et al. 1997). The haploinsufficiency of the Runx2 gene leads to cleidocranial dysplasia in humans, and Runx2+/– mice show a similar phenotype characterized by abnormal membranous ossification, suggesting that the expression level of Runx2 is closely related to the promotion of membranous ossification (Otto et al. 1997). Among growth factors involved in osteoblast differentiation, bone morphogenetic protein (BMP) family proteins are most notable, because they have a strong ability to induce ectopic bone formation in vivo (Wang et al. 1990; Yamaguchi et al. 1991), and targeted disruption of Tob, an inhibitory protein of BMP signaling, results in the increased bone mass (Yoshida et al. 2000). Other factors regulating bone formation include Fos family proteins, which function in the context of AP-1. Their stimulatory effect on bone formation is underscored by the phenotype of the transgenic mice of Fra-1 and ΔFosB, similarly exhibiting enhanced bone formation (Jochum et al. 2000; Sabatakos et al. 2000).
In the present study, we first show an unexpected phenotype in the skeletal system of the mice deficient in the Stat1 gene (Stat1–/– mice). The bone of the mutant mice exhibits excessive osteoclastogenesis, which is expected from our previous report demonstrating the critical role of the IFN-activated ISGF3 in negative regulation of osteoclastogenesis. Surprisingly, however, the bone mass is increased in these mice, suggesting a possibility that Stat1 plays a critical role(s) in inhibiting the bone-forming process and that this inhibitory function dominates over the inhibitory function of bone-resorbing process in the context of IFN signaling.
To date, the negative regulatory mechanism of bone formation has been poorly understood. We show here that Stat1 is involved in attenuating the transcriptional activity of Runx2 in a unique manner, that is, by interaction with Runx2 in the cytoplasm. This new function of Stat1 is independent of IFN signaling. We further show that the in vitro differentiation of osteoblasts derived from the Stat1–/– mice is increased, which is accompanied by the up-regulation of the DNA-binding activity of Runx2. Our results thus reveal a novel physiological function of Stat1 in the regulation of osteoblast differentiation in the postnatal stage, and offer an interesting example of an otherwise latent transcription factor in the cytoplasm that has an active and physiologically critical role.
Results
Increased bone mass in the Stat1–/– mice
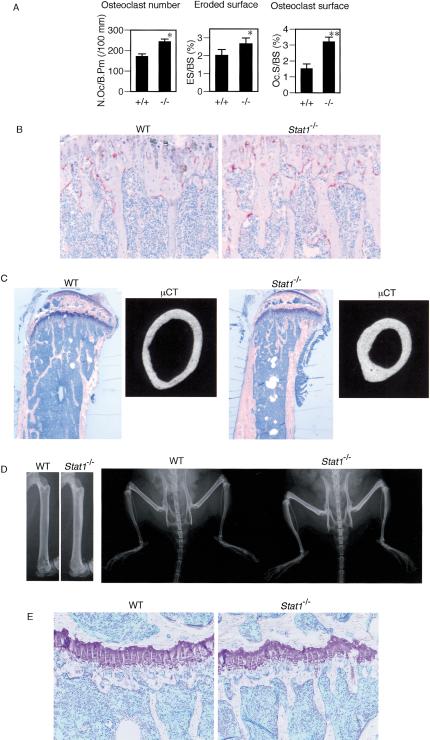
To investigate the physiological function of Stat1 in the skeletal system, we examined the bone phenotype of Stat1–/– mice (Meraz et al. 1996) at the age of 12 wk. We reported previously that both Stat1 and IRF-9 are required for IFN-β-mediated inhibition of osteoclastogenesis, suggesting that Stat1 participates in this inhibition in the context of ISGF3 (Takayanagi et al. 2002b). Consistently, we observed an increased osteoclast number and enhanced osteoclastic bone resorption in the tibia of the Stat1–/– mice (Fig. 1A). Tartrate-resistant acid phosphatase (TRAP) staining of the epiphyseal area of adult mice shows that the Stat1–/– mice have significantly more osteoclasts in vivo than the wild-type (WT) littermates (Fig. 1B), an observation similar to the mice deficient in one of the IFN-α/β receptor components, IFNAR1 (IFNAR1–/– mice; Takayanagi et al. 2002b).
Figure 1.
Increased bone mass in Stat1–/– mice in spite of enhanced osteoclastogenesis. (A) Enhanced osteoclastogenesis and bone resorption in Stat1–/– mice. Bone morphometric analysis of tibia of 12-week-old mice shows the elevated values of markers of osteoclastic bone resorption. (N.Oc/B.Pm) Number of osteoclasts per 100 mm of bone perimeter; (ES/BS) erosive surface per bone surface; (Oc.S/BS) osteoclast surface per bone surface. (B) TRAP staining of the proximal epiphysis of the tibia. Excessive osteoclast formation is observed in Stat1–/– mice. (C) Increased bone volume and normal epiphyseal plate in the tibial bone of Stat1–/– mice. Toluidine blue staining of the sagittal section of proximal tibia of 12-week-old mice. Microcomputed tomography (μCT) of the middle diaphysis of tibia. The bone diameter of Stat1–/– mice is smaller than that of wild-type mice even if they are analyzed equally in the middle of long bones. (D, right) The radiographic analysis of lower extremities. (Left) The increased radio-opacity in Stat1–/– mice is prominent in long bones as shown in a high-magnification view of femurs. (E) Magnified view of the epiphyseal plates. There is no significant difference in the morphology of chondrocytes and the width of the growth plate.
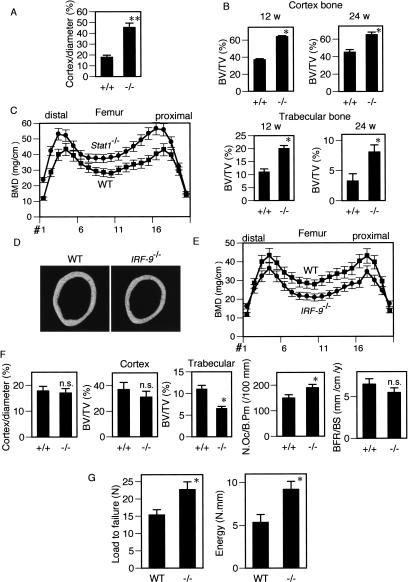
Interestingly, however, longitudinal sections and soft X-ray analysis of tibial bones of these mice suggested that the Stat1–/– mice have an increased bone mass (Fig. 1C,D). There was no abnormality in the microscopic analysis of the epiphyseal plate (Fig. 1E). Microcomputed tomography (μCT) analysis of the diaphysis of the long bone clearly shows that the cortex bone is markedly thickened (Fig. 1C). The ratio of cortex width to the bone diameter at the middle of tibia is greatly increased in the Stat1–/– mice (Fig. 2A). Furthermore, bone morphometric analysis showed that the increase in bone volume was observed in the trabecular bone as well as in the cortex bone (Fig. 2B), and the bone mineral density (BMD) of the femur was also up-regulated (Fig. 2C). We also examined the bone phenotype of mice lacking IRF-9 (IRF-9–/– mice), another essential molecule for ISGF3. In contrast to the Stat1–/– mice, the IRF-9–/– mice exhibited osteopenia, similar to the IFNAR1–/– mice, along with lower bone mineral density compared with the wild-type mice (Fig. 2D–F). These observations are consistent with our previous report, indicating the essential role of IRF-9 in the RANKL-dependent autoinhibition of osteoclast differentiation via induction of IFN-β (Takayanagi et al. 2002b). Unlike the Stat1–/– mice, there was no abnormality in osteoblast parameters in the bone morphometric analysis of the IRF-9–/– mice (see Supplementary Fig. S1). Taken together, these results suggest that Stat1 functions not only in the inhibition of osteoclast differentiation by IFN-β signaling, but also in the other limb of bone remodeling, osteoblast differentiation, in a context different from the IFN-activated ISGF3. The increased bone mass in the Stat1–/– mice suggests that this new regulatory function of Stat1 in the bone-forming osteoblast apparently dominates over its “classical” function in the osteoclasts in the overall bone-remodeling process.
Figure 2.
Enhanced bone formation and accelerated osteoblast differentiation in Stat1–/– mice. (A) Marked thickening of the cortex bone in Stat1–/– mice. Cortex/diameter denotes the ratio of the sum of the cortex width to the bone diameter at the middle of the tibia. (B) Cortex and trabecular bone volume was increased in Stat1–/– mice at the age of 12 wk and 24 wk. (BV/TV) Bone volume per tissue volume. (C) Bone mineral density (BMD) of 24-week-old Stat1–/– mice measured in 20 longitudinal divisions of femur. BMD was up-regulated in Stat1–/– mice from the proximal through the distal division (p < 0.05, #1–#18). (D) Bone phenotype of IRF-9–/– mice. μCT analysis in the middle of tibial diaphysis. (E) BMD is decreased in IRF-9–/– mice (p < 0.05, #3–#18). See panel C for details. (F) Cortex width and bone morphometric analysis of IRF-9–/– mice. Cortex width is not significantly altered in IRF-9–/– mice. Trabecular bone volumes decreased by 40%. (BFR/BS) Bone formation rate per bone surface. (G) Mechanical strength of the femur determined by the three-point bending test. Maximum load to failure and the energy resorption were greater in the femur of Stat1–/– mice than in those of wild-type mice, but the stiffness was not changed (data not shown).
The bone strength test indicates that the increased bone mass in the mutant mice leads to an increase in strength, distinct from pathological bone sclerosis or osteopetrosis, which induces fragility of the bone (Fig. 2G). It is notable that there was no severe abnormality in the gross development of skeletons, long bone length, or growth plate anatomy in Stat1–/– mice (Fig. 1C–E), suggesting that the Stat1–/– mice have no defect in the process of endochondral bone formation. Although Stat1 regulates chondrocyte proliferation in pathological conditions such as achondroplasia (Sahni et al. 1999), the above results indicate that Stat1 is dispensable for the physiological regulation of chondrocyte proliferation and differentiation. Consistent with this theory, we detected little, if any, expression of Stat1 mRNA in the normal skeletal tissue of developing embryos [embryonic days 12.5–16.5 (E12.5–E16.5); P.E. Bialek and G. Karsenty, unpubl.]. Furthermore, there was no difference in osteocalcin expression at E16.5 between wild-type and Stat1–/– mice (see Supplementary Fig. S2) and no acceleration of ossification in the newborn Stat1–/– mice (see Supplementary Fig. S3), suggesting that the regulatory function of Stat1 is physiologically critical for the bone remodeling at the postnatal stage.
Acceleration of osteoblast differentiation in the absence of Stat1
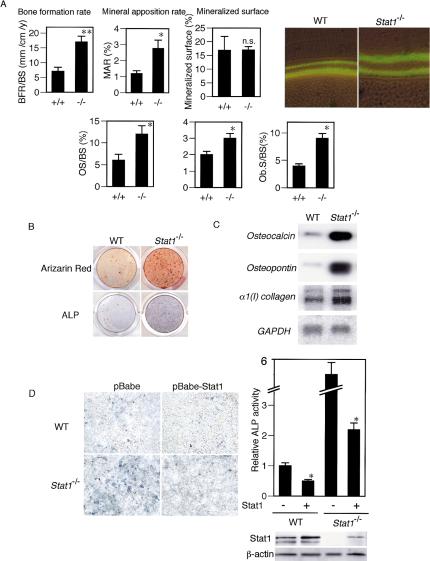
The unexpected bone phenotype of the Stat1–/– mice prompted us to examine the status of bone-forming osteoblasts in these mice. As shown in Figure 3A, bone morphometric analysis after calcein injection revealed a notable increase in bone formation rate (BFR). Osteoblast parameters such as osteoid surface/thickness and osteoblast surface were also up-regulated in the mutant mice, suggesting excessive osteoblast differentiation and matrix synthesis, compared with the wild-type mice (Fig. 3A).
Figure 3.
Increased bone formation and osteoblast differentiation in Stat1–/– mice. (A, left) Bone morphometric analysis performed after bone labeling with calcein injections (16 mg/kg) with a 7-d interval. BFR/BS is elevated because of an up-regulated mineral apposition rate (MAR). (Right) Calcein labeling indicates the increased bone formation in the cortex bone of Stat1–/– mice. (OS/BS) Osteoid surface per bone surface; (O.Th) osteoid thickness; (Ob.S/BS) osteoblast surface per bone surface. (B) Enhanced osteoblast differentiation derived from Stat1–/– mice shown by alizarin red and ALP staining. (C) mRNA expressions of osteoblast differentiation marker genes such as osteocalcin, osteopontin, and type I collagen are up-regulated in Stat1–/– osteoblasts (RNA blotting analysis). (D) Effect of retroviral expression of Stat1 gene in wild-type and Stat1–/– osteoblasts. Osteoblast differentiation assayed by ALP activity was suppressed by expression of Stat1 in both wild-type and Stat1–/– cells (* p < 0.05 vs. Stat1–). The expression level of Stat1 is analyzed by immunoblotting (lower right).
We next studied the in vitro osteoblast differentiation of precursor cells obtained from the calvarial bone of the wild-type and Stat1–/– mice. As shown in Figure 3B, osteoblast differentiation was significantly enhanced in the absence of Stat1, as revealed by notable increases in both alkaline phosphate (ALP) activity and bone nodule formation as determined by alizarin red staining. Furthermore, mRNAs for osteoblast differentiation marker genes such as osteocalcin, osteopontin, and type I collagen were all up-regulated in cultured osteoblasts from the Stat1–/– mice (Fig. 3C).
We also examined the rates of cell proliferation and apoptosis in wild-type and Stat1–/– osteoblasts; however, essentially no difference in cellular responses was observed between wild-type and Stat1–/– osteoblasts, supporting the theory that the loss of Stat1 selectively affects the osteoblast differentiation program (see Supplementary Fig. S4). To further address this issue, we ectopically expressed Stat1 by retroviral gene transfer in wild-type and Stat1–/– precursor cells, and examined their differentiation. As shown in Figure 3D, the expression of Stat1 suppressed osteoblast differentiation, without affecting the cell numbers of wild-type and Stat1–/– precursor cells. Collectively, these results suggest that Stat1 interferes with the osteoblast differentiation program and that the increased bone volume in the Stat1–/– mice is, indeed, caused by excessive bone formation by osteoblasts.
In view of the well-known fact that Stat1 is activated in response to IFNs and other cytokines, such as leukemia inhibitory factor (LIF), interleukin (IL)-6, and oncostatin M (Meraz et al. 1996; Bellido et al. 1997), we examined if these cytokines inhibit the differentiation of osteoblasts in vitro, but no such evidence was obtained (data not shown).
Enhanced response of osteoblasts to BMP-2 in the absence of Stat1
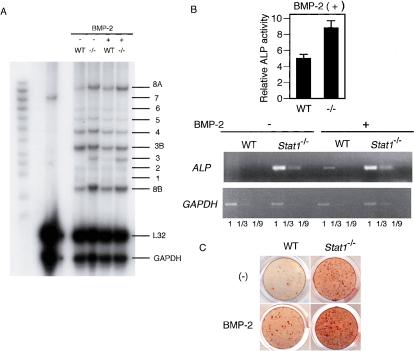
To gain insights into the mechanism by which Stat1 interferes with the osteoblast differentiation program, we screened the mRNA expression of BMP family genes in wild-type and Stat1–/– mice. However, there was no difference in the expression level of mRNA for BMP-2, BMP-4, BMP-6, and BMP-7, which are known to be potent stimulators of osteoblast differentiation (Fig. 4A). Although BMP-3 expression is up-regulated in Stat1–/– cells, BMP-3 is shown to negatively affect the bone mass (Daluiski et al. 2001). These results make it unlikely that Stat1 exerts its inhibitory effect on osteoblast differentiation by affecting the expression of BMPs that promote their differentiation.
Figure 4.
Effect of Stat1 deficiency on the expression of the BMP family and the response to BMP-2. (A) mRNA expression of BMP family genes in wild-type and Stat1–/– osteoblasts in the presence or absence of BMP-2 (RNase protection assay). There was no difference in the expression of osteogenic members such as BMP-2, BMP-4, and BMP-7. (B) The differentiation of wild-type and Stat1–/– osteoblasts in the presence of exogenous BMP-2. Stat1–/– osteoblasts show enhanced ALP activity and mRNA expression of the ALP gene even in the presence of excessive BMP-2. (C) Bone nodule formation is more significant in Stat1–/– osteoblasts than in wild-type osteoblasts even with stimulation of BMP-2 (alizarin red staining).
We next examined the differentiation of wild-type and Stat1–/– osteoblasts in the presence of exogenous BMP-2, one of the best-known BMPs that stimulate osteoblast differentiation. Interestingly, enhanced osteoblast differentiation was observed in precursor cells from the Stat1–/– mice relative to those from the wild-type mice when stimulated with exogenous BMP-2 (Fig. 4B,C), suggesting that Stat1 is involved in the negative regulation of the signaling pathway and/or transcriptional mechanism operating downstream of BMP-2.
Enhanced activation of Runx2 in Stat1–/– osteoblasts
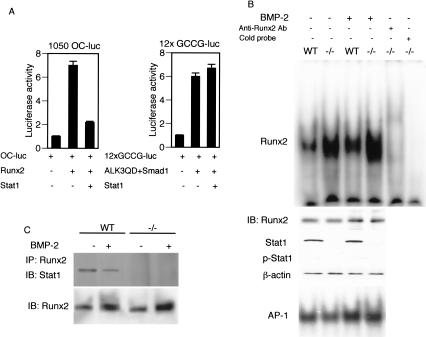
The above results provided us a cue to further analyze the molecular basis of excessive osteoblast differentiation in the absence of Stat1, that is, the role of Stat1 in BMP-2 signaling events. Briefly, BMP-2 binds to its receptor, which induces the phosphorylation of Smad family proteins such as Smad1, Smad5, and Smad8, resulting in the formation of a trimetric complex with Smad4 (Heldin et al. 1997; Massague 2000). The Smad complex translocates to the nucleus and cooperates with another class of transcription factor, Runx2, to activate the transcription of osteoblast-specific genes (Zhang et al. 2000; Ito and Miyazono 2003). Moreover, Runx2 expression is induced by BMP-2 (Lee et al. 2000). Then, we examined the effect of Stat1 on the activities of Runx2 and Smad using two luciferase reporter genes linked to a Runx2-dependent osteocalcin promoter (1050 OC-luc) or an artificial promoter that responds to Smads (12XGCCG-luc).
As shown in Figure 5A, the coexpression of Stat1 resulted in a strong inhibition of the Runx2-dependent activation of the osteocalcin promoter, without affecting the Smad1-dependent promoter activation, suggesting that Stat1 selectively interferes with the transcriptional activity of Runx2. In this regard, it is worth noting that no discernible Stat1-binding sites are present in the osteocalcin promoter (Javed et al. 1999), indicating that Stat1 interferes with Runx2 activity without binding to the promoter. In fact, an electrophoretic mobility shift assay (EMSA) using the Runx2-binding probe, revealed that the DNA-binding activity of Runx2 is up-regulated (by approximately fivefold) in Stat1–/– osteoblasts, although the Runx2 protein level remained the same as that in wild-type osteoblasts (Fig. 5B, upper panel). BMP-2 stimulation caused an increase in Runx2 protein level in both wild-type and Stat1–/– osteoblasts, and the DNA-binding activity of Runx2 was also markedly increased (by approximately fourfold) in Stat1–/– osteoblasts (Fig. 5B, upper panel). On the other hand, wild-type and Stat1–/– osteoblasts showed no difference in the DNA-binding activity of AP-1, which is also involved in osteoblast differentiation (Fig. 5B, lower panel). These results in toto indicate that Stat1 selectively interferes with the DNA-binding activity of Runx2.
Figure 5.
Runx2 transcriptional activity is inhibited by Stat1. (A) The effect of Stat1 on the promoter activation by Runx2 or Smad1. Stat1 inhibits Runx2 activation of the osteocalcin promoter, 1050Oc-luc, but not Smad1 activation of 12xGCCG-luc. Smad1 is cotransfected with ALK3QD, a plasmid that encodes a constitutively active form of BMP-2 receptor. (B) DNA-binding activity of Runx2 is up-regulated in Stat1–/– osteoblasts. Cell extracts of cultured osteoblasts derived from wild-type and Stat1–/– mice were analyzed by EMSA using an oligonucleotide probe containing the binding sequence with Runx2. Wild-type and Stat1–/– osteoblasts expressed the same level of Runx2, but the DNA-binding activity of Runx2 was enhanced in Stat1–/– osteoblasts. It is notable that Tyr 701 phosphorylation of Stat1 (p-Stat1) is not detectable in this condition. BMP-2 increases the Runx2 protein level in both wild-type and Stat1–/– osteoblasts, but Runx2 activity was still enhanced in Stat1–/– osteoblasts in the presence of BMP-2. AP-1 activity is not changed between these osteoblasts. (C) Association of endogenous Runx2 with Stat1 in primary osteoblasts. Cell extract was prepared from wild-type and Stat1–/– osteoblasts and analyzed by immunoprecipitation with anti-Runx2 antibody followed by blotting with anti-Stat1 antibody. The complex formation was reduced in the presence of BMP-2.
Is the inhibitory effect of Stat1 on Runx2 activity dependent on its activation by phosphorylation? In the context of the signaling of IFNs and other cytokines, Stat1 is activated by phosphorylation at Tyr 701. In addition, Ser 727 is also phosphorylated to enhance the transcriptional activity of Stat1 (Stark et al. 1998; O'Shea et al. 2002). Interestingly, the phosphorylation of Tyr 701 of Stat1 was barely detectable even in wild-type osteoblasts irrespective of BMP-2 stimulation, suggesting that the inhibition of Runx2 activity by Stat1 is independent of this phosphorylation (Fig. 5B; see also below). We also examined the phosphorylation of Ser 727, which is critical for its transcriptional activity (O'Shea et al. 2002); however, the serine phosphorylation of Stat1 was not detected with or without BMP-2 stimulation (data not shown), further suggesting that the effect of Stat1 on Runx2 activity is exerted without the phosphorylation-dependent activation of Stat1.
Association of Stat1 with Runx2 in regulation of Runx2-mediated transcription
To gain insights into the mechanism of how Stat1 interferes with Runx2 activity, we investigated the association between Stat1 and Runx2. As shown in Figure 5C, immunoprecipitation and immunoblotting analyses revealed that endogenous Stat1 interacts with endogenous Runx2 in primary osteoblasts. Interestingly, the interaction becomes weaker upon BMP-2 stimulation of the osteoblasts, in the face of an increase in total Runx2 protein level (Fig. 5C).
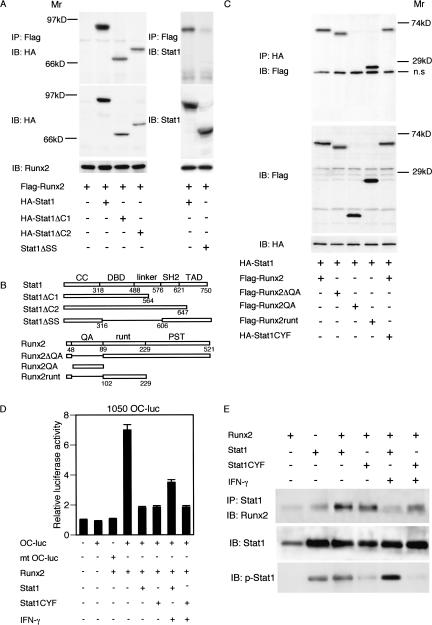
To determine the region of the Stat1 protein responsible for its association with Runx2, we constructed two deletion mutants (Stat1ΔC1 and Stat1ΔC2) lacking the C-terminal region containing TAD and SH2 (Fig. 6B; O'Shea et al. 2002). These proteins as well as Runx2 were epitope-tagged (see Materials and Methods for the details), and the intermolecular association was examined by transiently expressing them in 293T cells. We found that both C terminus mutants still associate with Runx2 (Fig. 6A, left). We then examined the Stat1 mutant, Stat1ΔSS, lacking the DNA-binding domain (DBD) and the linker domain (S316–S606; Wu et al. 2002), and found that this mutant does not associate with Runx2 (Fig. 6A, right). Thus, Stat1 requires the region containing DBD and the linker domain to associate with Runx2.
Figure 6.
Physical interaction of Stat1 with Runx2 interferes with Runx2 activity. (A) Association of Stat1 mutants with Runx2. C-terminal domains are dispensable, but the DBD/linker domain is essential for interaction with Runx2. (IP) Immunoprecipitation; (IB) immunoblotting. (B) A schematic of deletion mutants of Stat1 and Runx2. (C) Association of Runx2 mutants with Stat1. QA and PST domains are dispensable, but the remainder containing the runt domain is essential for interaction with Stat1. (n.s.) Nonspecific. (D) The inhibitory effect of Stat1 on Runx2 activity is phosphorylation-independent. The effect of Stat1 and Stat1CYF, the phosphorylation-deficient form of Stat1, on the Runx2-dependent activation of the osteocalcin promoter was examined in a transient assay in 293T cells. Stat1 and Stat1CYF equally have an inhibitory action on Runx2 activity. The inhibitory effect of Stat1 is reduced when it is tyrosine-phosphorylated by IFN-γ (see Fig. 5E). (E) Association of Stat1 and Stat1CYF with Runx2 and the effect of IFN-γ on their phosphorylation and interaction. Stat1 and Stat1CYF equally associate with Runx2 in 293T cells. Forty-eight hours after transfection, the cell lysate was prepared from cells cultured in the same condition for the luciferase assay (see Fig. 5D). IFN-γ-induced phosphorylation of Tyr 701 is observed in wild-type Stat1, but not in Stat1CYF. The interaction with Runx2 was reduced when Stat1 was phosphorylated by IFN-γ.
We further examined the region of Runx2 that is required for its association with Stat1, by using a similar coexpression assay. The Runx2 mutant lacking the QA domain, Runx2ΔQA (Thirunavukkarasu et al. 1998), can associate with Stat1, but the QA domain alone (Runx2QA) cannot associate with it (Fig. 6C). On the other hand, the Runx2 mutant lacking the QA and PST domains (Runx2runt) can bind with Stat1 (Fig. 6C). These observations suggest that the PST domain is dispensable for the association and that the runt domain may be responsible for its association with Stat1.
Phosphorylation of Stat1 and its inhibitory effect on Runx2 activity
Stat1 undergoes dimerization upon phosphorylation of Tyr 701 and translocates to the nucleus. To investigate the effect of this phosphorylation on Stat1 inhibition of Runx2, we used a mutant form of Stat1 in which Tyr 701 is converted to phenylalanine (Stat1CYF; Chatterjee-Kishore et al. 2000). As shown in Figure 6D, the overexpression of Stat1CYF inhibited the Runx2-dependent activation of the osteocalcin promoter, as efficiently as wild-type Stat1. Indeed, Stat1CYF can still interact with Runx2 (Fig. 6C,E), lending further support to the notion that the inhibitory function of Stat1 on Runx2 activity is dependent on its interaction with Runx2, but not on its phosphorylation.
Does activation of Stat1 affect its interaction with Runx2? Interestingly, IFN-γ stimulation partially relieved the suppressive effect of Stat1 on the Rnux2-dependent activation of the osteocalcin promoter (Fig. 6D) and also weakened the Stat1 interaction with Runx2 (Fig. 6E). On the other hand, this IFN-γ effect on Stat1CYF was not observed (Fig. 6D,E). These results suggest that Stat1 activation, which leads to its dimerization and nuclear translocation, results in the loss of its ability to interact with Runx2. Consistently, we observed that IFN-γ enhances in vitro osteoblast differentiation (H. Takayanagi, unpubl.). Although the physiological significance of these observations in bone remodeling is not clear at present, they lend further support to the idea that Stat1 inhibits Runx2 activity in its transcriptionally latent form.
Inhibition of nuclear translocation of Runx2 by Stat1
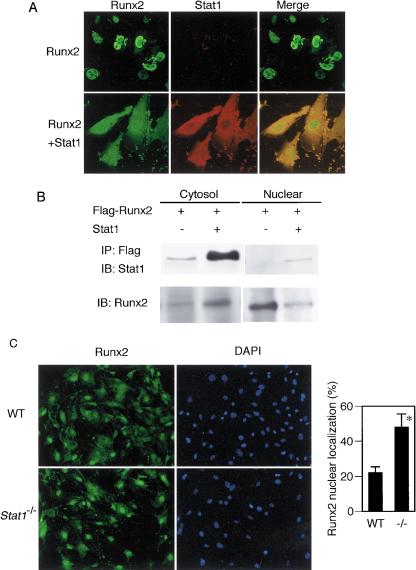
The above observations led us to hypothesize that unphosphorylated Stat1 residing in the cytoplasm associates with Runx2 and interferes with its nuclear translocation. As shown in Figure 7A, immunostaining analysis revealed that Runx2 efficiently undergoes nuclear translocation when it is expressed in 293T cells, but that the majority of Runx2 remains in the cytoplasm when Stat1 is coexpressed, an observation consistent with the results in Figure 6. This concept is further supported by the results of immunoblot analysis (Fig. 7B), which indicate that Runx2 is mainly detectable in the nuclear fraction in the absence of coexpressed Stat1, but this equilibrium is shifted to the cytoplasm in the presence of Stat1. In view of these results indicating an enhanced interaction between Stat1 and Runx2 upon cotransfection of the cDNAs encoding these two factors, it is likely that this intermolecular interaction is direct, although this issue needs to be clarified further.
Figure 7.
Cytoplasmic sequestration of Runx2 by Stat1. (A) Localization of Runx2 with or without coexpression of Stat1 in 293T cells. Immunostaining shows that Runx2 efficiently undergoes nuclear translocation when overexpressed in 293T cells, but the nuclear localization was inhibited by coexpression of Stat1, which is located mainly in the cytoplasm. (B) Detection of Runx2 protein in cytosolic and nuclear fractions by immunoblotting. If Runx2 is solely overexpressed in 293T cells, most of the Runx2 protein is detected in the nuclear fraction. Under this condition, the association of Runx2 with endogenous Stat1 is detected in the cytoplasm at a low level. When Stat1 is coexpressed, a great amount of Stat1 interacts with Runx2 in the cytoplasm, resulting in the sequestration of Runx2 in the cytoplasm. (C) Nuclear localization of Runx2 in wild-type and Stat1–/– osteoblasts. The percentage of cells that exhibit nuclear translocation of Runx2 is significantly higher in Stat1–/– osteoblasts than in wild-type osteoblasts.
Finally, we examined Runx2 nuclear localization in primary osteoblasts derived from the wild-type and Stat1–/– mice. Consistent with the EMSA data showing the enhanced DNA-binding activity of Runx2 in Stat1–/– cells (Fig. 5B), Runx2 nuclear translocation is more prominent in Stat1–/– osteoblasts than in wild-type osteoblasts (Fig. 7C), suggesting that Runx2 nuclear localization is regulated by the transcriptionally latent form of Stat1 in the cytoplasm.
Discussion
Stat1 as a negative regulator of bone formation
Skeletal homeostasis is controlled by the intricate coordination of constituent cells including chondrocytes, osteoblasts, and osteoclasts (Karsenty and Wagner 2002). We previously showed that Stat1 mediates the suppressive effect of IFNs on the differentiation of osteoclasts (Takayanagi et al. 2002b). In fact, excessive osteoclastogenesis, similar to that observed in IFNAR1–/– mice, was also found in Stat1–/– mice. Interestingly, however, bone mass is increased in the Stat1–/– mice, revealing that Stat1 also plays a critical role in the inhibition of Runx2-mediated osteoblast differentiation. The net increase in bone mass suggests that the function of Stat1 in osteoblasts dominates over that in osteoclasts in vivo. It has been reported that Stat1 plays a critical role in the FGF-mediated suppression of chondrocyte proliferation in disease conditions such as achondroplasia (Sahni et al. 1999, 2001). Our results showing the normal epiphyseal growth plate and longitudinal bone length in the adult Stat1–/– mice suggest that physiological chondrocyte proliferation is not markedly enhanced in the absence of Stat1.
Regulation of Runx2 by Stat1 at a postnatal stage
Runx2 has been identified as the central transcription factor for bone formation, but how its function is regulated remains largely unknown. We have shown that the loss of inhibition of Runx2 activity by Stat1 results in increased bone mass in the adult Stat1–/– mice without affecting bone formation during the developmental period, suggesting that Stat1 is selectively involved in the Runx2 regulation in bone remodeling at the postnatal stage. Since it is currently unknown how Runx2 is regulated during the development of the skeletal system, further studies are required to clarify the detailed mechanism of the temporal and spatial regulation of Runx2 for the normal development and homeostatic regulation of the skeletal system.
It has been reported that Runx2 overexpression in osteoblasts by the type I collagen promoter inhibits osteoblast differentiation at a late stage (Liu et al. 2001). Therefore, sustained Runx2 expression in the late stage of osteoblast differentiation may inhibit bone formation, and Runx2 expression should be strictly controlled in a stage-dependent manner during osteoblast differentiation (Komori 2002). In this regard, Stat1 deficiency does not modify Runx2 expression, but increases the activity of Runx2, whose expression is physiologically controlled. Thus, Stat1–/– mice may provide an interesting model in that the temporal Runx2 expression is normal but its activity is enhanced.
Involvement of BMP-2 in the regulation of Runx2 by Stat1
BMP-2 is a potent inducer of ectopic bone formation, but it remains to be elucidated whether BMP-2 is essential for osteoblast differentiation in vivo (Karsenty and Wagner 2002). In this study, we show that osteoblast differentiation is enhanced in Stat1–/– cells even in the presence of exogenous BMP-2. Enhanced osteoblast differentiation is also observed in the absence of exogenous BMP-2, but it remains to be clarified how BMP-2 contributes to the in vivo phenotype of Stat1–/– mice. Based on the fact that endogenous BMP-2 expression is detectable in cultured osteoblasts and BMP-2 induces and activates Runx2 (Zhang et al. 2000; Ito and Miyazono 2003), we propose that the enhanced response of osteoblasts to BMP-2 contributes, at least in part, to the excessive osteoblast differentiation in Stat1–/– mice. It is also interesting that BMP-2 induces Runx2 dissociation from Stat1 (Fig. 5C). Although the detailed mechanism underlying this phenomenon remains to be clarified, we infer that Runx2 dissociation from Stat1 upon BMP-2 stimulation is a consequence of Runx2 interaction with BMP-2-induced transcription factors, most likely the Smad proteins. In fact, Smad1, Smad5, and Smad8 are known to be activated by BMP-2, and these factors, together with Smad4, interact with Runx2 to induce the transcriptional program for osteoblast differentiation (Zhang et al. 2000). In other words, the Runx2 dissociation is a consequence of the swapping of the partner of Runx2 before and after BMP-2 stimulation. This issue obviously needs to be addressed further in a future study.
Stat1 as a cytoplasmic attenuator of Runx2
The mode of action of Stat1 is unique in that it interferes with Runx2 in the cytoplasm in its transcriptionally latent form. It has been shown previously that Stat1 functions in the nucleus in the absence of phosphorylation at Tyr 701 to mediate the constitutive expression of genes that respond to activated Stat1 (in the context of IFN signaling; Chatterjee-Kishore et al. 2000). On the other hand, our results indicate that Stat1 exerts its inhibitory effect on Runx2 mainly, if not exclusively, in the cytoplasm, although the possibility that the latent Stat1 additionally functions in the nucleus to interfere with Runx2 cannot be rigorously ruled out. Further work will be required to elucidate the precise inhibitory mechanism of Runx2 nuclear localization by Stat1. It is possible that Runx2 binding with Stat1 leads to the conformational change of Runx2, thus concealing the nuclear localization signal (NLS) domain. Supporting this view are the previous report that the Runx2 NLS is adjacent to the runt domain (Thirunavukkarasu et al. 1998) and our result suggesting that the runt domain may interact with Stat1. Because the DNA-binding activity but not the protein level of Runx2 is increased in osteoblasts from Stat1–/– mice, we infer that Stat1-associated Runx2 is also inactive in terms of its DNA-binding activity.
The inhibitory effect of Stat1 on Runx2 activity is unique in that a given transcription factor, in its latent form, regulates the function of another transcription factor in the cytoplasm. Collectively, we propose that Stat1 functions as a cytoplasmic attenuator of Runx2 in osteoblast differentiation. To date, most drug therapies for osteopenic diseases aim at the suppression of osteoclastic bone resorption. This study may provide an alternative approach to the modulation of bone formation through intervention of Stat1.
Materials and methods
Mice and bone analysis
The Stat1–/– and IRF-9–/– mice have been described (Kimura et al. 1996; Meraz et al. 1996). Mutant mice were backcrossed with C57BL/6 mice more than 10 times, and subjected to histomorphometric and microradiographic examinations as described (Takayanagi et al. 2002b). All mice were born and kept under pathogen-free conditions, and showed no abnormality in growth rate or body weight. Bone mineral density was determined by the dual energy X-ray absorptiometry (DXA) method using DCS-600R (Aloka). Bone mechanical strength testing was performed using a three-point bending method with Autograph AG2000-E (Shimazu). Biochemical parameters were determined from the load-deformation curve as described (Tanizawa et al. 2000). The maximum load to failure (N) indicated the magnitude of the load that caused fracture, and the energy resorption (N. mm) was determined by the area under the load-deformation curve up to the maximum load point. All data are shown as mean ± S.E. (n = 6). Statistical analysis was performed using one-way ANOVA or Student's t-test throughout the paper (n.s. denotes not significant; * and ** indicate p < 0.05 and p < 0.01, respectively).
In vitro osteoblast differentiation assay
Osteoblasts were isolated from the calvarial bone of newborn (3–5 d) mice by enzymatic digestion in α-MEM with 0.1% collagenase and 0.2% dispase as described (Ogata et al. 2000), and were cultured in α-MEM with 10% FBS. After 2 d, cells were reseeded (5 × 104 cells per well in a 24-well dish) and cultured in α-MEM with 10% FBS containing 50 μg/mL ascorbic acid, 10 mM β-glycerophosphate, and 10 nM dexamethasone for the osteoblast differentiation assay. After 7 d, ALP activity was assayed, and after 21 d, bone nodule formation was assayed by alizarin red staining as described (Ogata et al. 2000). After 14 d, we extracted mRNA from cultured osteoblasts with Sepasol (Nacalai), and performed semiquantitative reverse transcriptase PCR (RT–PCR) or RNA blotting analysis of osteoblast-specific genes as previously described (Ducy et al. 1997). PCR primers are as follows: ALP (sense, 5′-GACTGGTACTCGGATAAC GAGATGC-3′; antisense, 5′-TGCGGTTCCAGACATAGTG G-3′) and GAPDH (sense, 5′-AGGAGCGAGACCCCACTAAC-3′; antisense, 5′-TGCCAGTGAGCTTCCCGTTC-3′).
Retroviral gene transfer
Packaging and inoculation of retrovirus vectors, pBabe or pBabe-Stat1 [a gift from D. Stoiber (Institute of Pharmacology, University of Vienna, Vienna, Austria) and V. Sexl (Institute of Pharmacology, University of Vienna, Vienna, Austria)] were performed as described previously (Takayanagi et al. 2002b). Osteoblasts (1 × 105 cells in a well of a 6-well plate) were inoculated with retroviruses and cultured in α-MEM with 10% FBS. After 2 d of culture, the medium was changed to α-MEM with 10% FBS containing 50 μg/mL ascorbic acid, 10 mM β-glycerophosphate, and 10 nM dexamethasone. After 14 d of culture, cells were fixed for ALP assay or analyzed by immunoblotting.
RNase protection assay
For the RNase protection assay for the mRNAs of the BMP family members, we extracted total RNA from osteoblasts cultured for 7 d as described above, and analyzed the mRNA using an RNase protection assay kit (Pharmingen) according to the manufacturer's protocol. When we analyzed the osteoblast differentiation in the presence of recombinant BMP-2, we added BMP-2 to the culture of osteoblasts at a concentration of 100 ng/mL throughout the culture period.
Luciferase assay
We transfected the reporter plasmid containing the osteocalcin promoter, 1050Oc-luc, or mutated (mt) 1050Oc-luc (Javed et al. 1999), and the Runx2 expression vector, pEF-BOS-αA1, to 293T cells with or without the Stat1 expression vector, pSG91, using FuGENE6 (Roche). We similarly transfected 12xGCCG-luc, ALK3QD, and pcDNA3-Smad1 as described (Yoshida et al. 2000) with or without Stat1. When necessary, IFN-γ (100 U/mL) was added 36 h after transfection. The luciferase assay was performed 48 h after transfection as described previously (Takayanagi et al. 2002a). All data are expressed as mean ± S.E. (n = 5).
EMSA
After 7 d of culture of osteoblasts derived from WT or Stat1–/– mice, proteins were extracted with sonication method using lysis buffer [20 mM HEPES at pH 7.9, 50 mM NaCl, 10 mM EDTA, 10% (v/v) glycerol, 10 mM sodium molybdate, 10 mM ortho vanadate, 100 mM sodium fluoride, 0.1% (v/v) NP-40, 1 mM DTT, 100 μg/mL leupeptin, and 0.5 mM APMSF]. Then 35 μg of proteins was used to bind with the [γ-32P]ATP-labeled oligonucleotide probe containing the binding sequence with Runx2 as described (Kagoshima et al. 1996) or with AP-1 (Jochum et al. 2000). Supershift was performed with a mouse anti-Runx2 monoclonal antibody (a gift from Y. Ito, Institute of Molecular and Cell Biology, National University of Singapore, Singapore).
Immunoprecipitation and immunoblot analysis
Cell extracts were harvested from primary osteoblasts or 293T cells transfected with indicated expression vectors for 48 h in TNE buffer (10 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 2 mM Na3VO4, 10 mM NaF, and 10 μg/mL aprotinin). Cell extracts were incubated with 1 μg of anti-Runx2, anti-Stat1 (p84/91; Santa Cruz) or anti-Flag (Sigma) antibodies at 4°C for 1 h. Immune complexes were recovered with protein G Sepharose (Amersham), subjected to SDS-PAGE, and blotted with indicated antibodies. Nuclear and cytosolic extracts were prepared as described (Thirunavukkarasu et al. 1998). For immunoblotting, we used anti-HA antibody (Santa Cruz), anti-phospho-Stat1 (Tyr 701; New England Biolab), and Ser 727 antibodies (Upstate). HA-Stat1ΔC mutants were constructed by deleting the C terminus region of HA-Stat1 (a gift of X.Y. Fu) as described in Figure 6B. Stat1CYF (a gift of X.Y. Fu, Department of Pathology, Yale University School of Medicine, New Haven, CT) and Stat1ΔSS (a gift of Y. Eugene Chin, Department of Pathology and Laboratory Medicine, Brown University School of Medicine, Providence, RI) have been described (Wu et al. 2002). The construction strategy of Runx2 mutants has been previously described (Thirunavukkarasu et al. 1998).
Immunofluorescence staining
293T cells were fixed 24 h after transfection in 4% paraformaldehyde for 20 min and treated with 0.2% Triton X for 5 min. Primary osteoblasts were fixed 7 d after incubation in α-MEM containing 50 μg/mL ascorbic acid, 10 mM β-glycerophosphate, and 10 nM dexamethasone. Cells were sequentially incubated in 5% BSA/PBS for 30 min, 2 μg/mL anti-Runx2 monoclonal antibody or anti-Stat1 polyclonal antibody in PBST for 60 min, and then 4 μg/mL Alexa 488 or 546-labeled anti-mouse or anti-rabbit IgG antibody (Molecular Probe) for 60 min.
Acknowledgments
We thank R.D. Schreiber for the mutant mice, H. Murayama and Kureha Chemical Industries for technical assistance, andYamanouchi Pharmaceuticals for providing recombinant BMP-2. We also appreciate T. Komori, T. Furuichi, Y. Ito, K. Ito, J.B. Lian, X.Y. Fu, M. Kitagawa, G. Watanabe, K. Miyazono, K. Imamura, D. Stoiber, and V. Sexl for mice, materials, and discussion; and P.E. Bialek, A. Suematsu, M. Urushibara, A. Takaoka, K. Honda, S. Kano, and I. Kawai for technical assistance and discussion. This work was supported in part by a grant for Advanced Research on Cancer from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan; a grant from PRESTO, Japan Science and Technology Corporation (JST); Grants-in-Aid for Scientific Research from MEXT and JSPS; Health Sciences Research Grants from the Ministry of Health, Labour and Welfare of Japan; grants of the Virtual Research Institute of Aging of Nippon Boehringer Ingelheim; and a grant from Japan Orthopaedics and Traumatology Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1119303.
References
- Bellido, T., Borba, V.Z., Roberson, P., and Manolagas, S.C. 1997. Activation of the Janus kinase/STAT (signal transducer and activator of transcription) signal transduction pathway by interleukin-6-type cytokines promotes osteoblast differentiation. Endocrinology 138: 3666–3676. [DOI] [PubMed] [Google Scholar]
- Chatterjee-Kishore, M., Wright, K.L., Ting, J.P., and Stark, G.R. 2000. How Stat1 mediates constitutive gene expression: A complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 19: 4111–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daluiski, A., Engstrand, T., Bahamonde, M.E., Gamer, L.W., Agius, E., Stevenson, S.L., Cox, K., Rosen, V., and Lyons, K.M. 2001. Bone morphogenetic protein-3 is a negative regulator of bone density. Nat. Genet. 27: 84–88. [DOI] [PubMed] [Google Scholar]
- Deng, C., Wynshaw-Boris, A., Zhou, F., Kuo, A., and Leder, P. 1996. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 84: 911–921. [DOI] [PubMed] [Google Scholar]
- Ducy, P., Zhang, R., Geoffroy, V., Ridall, A.L., and Karsenty, G. 1997. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 89: 747–754. [DOI] [PubMed] [Google Scholar]
- Ducy, P., Schinke, T., and Karsenty, G. 2000. The osteoblast: A sophisticated fibroblast under central surveillance. Science 289: 1501–1504. [DOI] [PubMed] [Google Scholar]
- Heldin, C.H., Miyazono, K., and ten Dijke, P. 1997. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 390: 465–471. [DOI] [PubMed] [Google Scholar]
- Ito, Y. and Miyazono, K. 2003. RUNX transcription factors as key targets of TGF-β superfamily signaling. Curr. Opin. Genet. Dev. 13: 43–47. [DOI] [PubMed] [Google Scholar]
- Javed, A., Gutierrez, S., Montecino, M., van Wijnen, A.J., Stein, J.L., Stein, G.S., and Lian, J.B. 1999. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D-responsive transcription and contribute to chromatin organization. Mol. Cell. Biol. 19: 7491–7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochum, W., David, J.P., Elliott, C., Wutz, A., Plenk Jr., H., Matsuo, K., and Wagner, E.F. 2000. Increased bone formation and osteosclerosis in mice overexpressing the transcription factor Fra-1. Nat. Med. 6: 980–984. [DOI] [PubMed] [Google Scholar]
- Kagoshima, H., Akamatsu, Y., Ito, Y., and Shigesada, K. 1996. Functional dissection of the α and β subunits of transcription factor PEBP2 and the redox susceptibility of its DNA binding activity. J. Biol. Chem. 271: 33074–33082. [DOI] [PubMed] [Google Scholar]
- Karsenty, G. and Wagner, E.F. 2002. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell 2: 389–406. [DOI] [PubMed] [Google Scholar]
- Kimura, T., Kadokawa, Y., Harada, H., Matsumoto, M., Sato, M., Kashiwazaki, Y., Tarutani, M., Tan, R.S., Takasugi, T., Matsuyama, T., et al. 1996. Essential and non-redundant roles of p48 (ISGF3 γ) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells 1: 115–124. [DOI] [PubMed] [Google Scholar]
- Komori, T. 2002. Runx2, a multifunctional transcription factor in skeletal development. J. Cell Biochem. 87: 1–8. [DOI] [PubMed] [Google Scholar]
- Komori, T., Yagi, H., Nomura, S., Yamaguchi, A., Sasaki, K., Deguchi, K., Shimizu, Y., Bronson, R.T., Gao, Y.H., Inada, M., et al. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89: 755–764. [DOI] [PubMed] [Google Scholar]
- Kong, Y.Y., Feige, U., Sarosi, I., Bolon, B., Tafuri, A., Morony, S., Capparelli, C., Li, J., Elliott, R., McCabe, S., et al. 1999. Activated T cells regulate bone loss and joint destruction in adjuvant arthritis through osteoprotegerin ligand. Nature 402: 304–309. [DOI] [PubMed] [Google Scholar]
- Lee, K.S., Kim, H.J., Li, Q.L., Chi, X.Z., Ueta, C., Komori, T., Wozney, J.M., Kim, E.G., Choi, J.Y., Ryoo, H.M., et al. 2000. Runx2 is a common target of transforming growth factor β1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol. Cell. Biol. 20: 8783–8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W., Toyosawa, S., Furuichi, T., Kanatani, N., Yoshida, C., Liu, Y., Himeno, M., Narai, S., Yamaguchi, A., and Komori, T. 2001. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J. Cell Biol. 155: 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolagas, S.C. 2000. Birth and death of bone cells: Basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr. Rev. 21: 115–137. [DOI] [PubMed] [Google Scholar]
- Massague, J. 2000. How cells read TGF-β signals. Nat. Rev. Mol. Cell. Biol. 1: 169–178. [DOI] [PubMed] [Google Scholar]
- Meraz, M.A., White, J.M., Sheehan, K.C., Bach, E.A., Rodig, S.J., Dighe, A.S., Kaplan, D.H., Riley, J.K., Greenlund, A.C., Campbell, D., et al. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84: 431–442. [DOI] [PubMed] [Google Scholar]
- Ogata, N., Chikazu, D., Kubota, N., Terauchi, Y., Tobe, K., Azuma, Y., Ohta, T., Kadowaki, T., Nakamura, K., and Kawaguchi, H. 2000. Insulin receptor substrate-1 in osteoblast is indispensable for maintaining bone turnover. J. Clin. Invest. 105: 935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, B.R., Reginato, A.M., and Wang, W. 2000. Bone development. Annu. Rev. Cell. Dev. Biol. 16: 191–220. [DOI] [PubMed] [Google Scholar]
- O'Shea, J.J., Gadina, M., and Schreiber, R.D. 2002. Cytokine signaling in 2002: New surprises in the Jak/Stat pathway. Cell 109 Suppl: S121–S131. [DOI] [PubMed] [Google Scholar]
- Otto, F., Thornell, A.P., Crompton, T., Denzel, A., Gilmour, K.C., Rosewell, I.R., Stamp, G.W., Beddington, R.S., Mundlos, S., Olsen, B.R., et al. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89: 765–771. [DOI] [PubMed] [Google Scholar]
- Rodan, G.A. and Martin, T.J. 2000. Therapeutic approaches to bone diseases. Science 289: 1508–1514. [DOI] [PubMed] [Google Scholar]
- Rousseau, F., Bonaventure, J., Legeai-Mallet, L., Pelet, A., Rozet, J.M., Maroteaux, P., Le Merrer, M., and Munnich, A. 1994. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature 371: 252–254. [DOI] [PubMed] [Google Scholar]
- Sabatakos, G., Sims, N.A., Chen, J., Aoki, K., Kelz, M.B., Amling, M., Bouali, Y., Mukhopadhyay, K., Ford, K., Nestler, E.J., et al. 2000. Overexpression of ΔFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat. Med. 6: 985–990. [DOI] [PubMed] [Google Scholar]
- Sahni, M., Ambrosetti, D.C., Mansukhani, A., Gertner, R., Levy, D., and Basilico, C. 1999. FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT-1 pathway. Genes & Dev. 13: 1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni, M., Raz, R., Coffin, J.D., Levy, D., and Basilico, C. 2001. STAT1 mediates the increased apoptosis and reduced chondrocyte proliferation in mice overexpressing FGF2. Development 128: 2119–2129. [DOI] [PubMed] [Google Scholar]
- Stark, G.R., Kerr, I.M., Williams, B.R., Silverman, R.H., and Schreiber, R.D. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67: 227–264. [DOI] [PubMed] [Google Scholar]
- Su, W.C., Kitagawa, M., Xue, N., Xie, B., Garofalo, S., Cho, J., Deng, C., Horton, W.A., and Fu, X.Y. 1997. Activation of Stat1 by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature 386: 288–292. [DOI] [PubMed] [Google Scholar]
- Takayanagi, H., Iizuka, H., Juji, T., Nakagawa, T., Yamamoto, A., Miyazaki, T., Koshihara, Y., Oda, H., Nakamura, K., and Tanaka, S. 2000a. Involvement of receptor activator of nuclear factor κB ligand/osteoclast differentiation factor in osteoclastogenesis from synoviocytes in rheumatoid arthritis. Arthritis Rheum. 43: 259–269. [DOI] [PubMed] [Google Scholar]
- Takayanagi, H., Ogasawara, K., Hida, S., Chiba, T., Murata, S., Sato, K., Takaoka, A., Yokochi, T., Oda, H., Tanaka, K., et al. 2000b. T-cell-mediated regulation of osteoclastogenesis by signalling cross-talk between RANKL and IFN-γ. Nature 408: 600–605. [DOI] [PubMed] [Google Scholar]
- Takayanagi, H., Kim, S., Koga, T., Nishina, H., Isshiki, M., Yoshida, H., Saiura, A., Isobe, M., Yokochi, T., Inoue, J., et al. 2002a. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 3: 889–901. [DOI] [PubMed] [Google Scholar]
- Takayanagi, H., Kim, S., Matsuo, K., Suzuki, H., Suzuki, T., Sato, K., Yokochi, T., Oda, H., Nakamura, K., Ida, N., et al. 2002b. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-β. Nature 416: 744–749. [DOI] [PubMed] [Google Scholar]
- Takayanagi, H., Kim, S., and Taniguchi, T. 2002c. Signaling crosstalk between RANKL and interferons in osteoclast differentiation. Arthritis Res. 4 Suppl 3: S227–S232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, T., Ogasawara, K., Takaoka, A., and Tanaka, N. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19: 623–655. [DOI] [PubMed] [Google Scholar]
- Tanizawa, T., Yamaguchi, A., Uchiyama, Y., Miyaura, C., Ikeda, T., Ejiri, S., Nagal, Y., Yamato, H., Murayama, H., Sato, M., et al. 2000. Reduction in bone formation and elevated bone resorption in ovariectomized rats with special reference to acute inflammation. Bone 26: 43–53. [DOI] [PubMed] [Google Scholar]
- Teitelbaum, S.L. 2000. Bone resorption by osteoclasts. Science 289: 1504–1508. [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu, K., Mahajan, M., McLarren, K.W., Stifani, S., and Karsenty, G. 1998. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbfβ. Mol. Cell. Biol. 18: 4197–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, E.A., Rosen, V., D'Alessandro, J.S., Bauduy, M., Cordes, P., Harada, T., Israel, D.I., Hewick, R.M., Kerns, K.M., LaPan, P., et al. 1990. Recombinant human bone morphogenetic protein induces bone formation. Proc. Natl. Acad. Sci. 87: 2220–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T.R., Hong, Y.K., Wang, X.D., Ling, M.Y., Dragoi, A.M., Chung, A.S., Campbell, A.G., Han, Z.Y., Feng, G.S., and Chin, Y.E. 2002. SHP-2 is a dual-specificity phosphatase involved in Stat1 dephosphorylation at both tyrosine and serine residues in nuclei. J. Biol. Chem. 277: 47572–47580. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, A., Katagiri, T., Ikeda, T., Wozney, J.M., Rosen, V., Wang, E.A., Kahn, A.J., Suda, T., and Yoshiki, S. 1991. Recombinant human bone morphogenetic protein-2 stimulates osteoblastic maturation and inhibits myogenic differentiation in vitro. J. Cell Biol. 113: 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, Y., Tanaka, S., Umemori, H., Minowa, O., Usui, M., Ikematsu, N., Hosoda, E., Imamura, T., Kuno, J., Yamashita, T., et al. 2000. Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell 103: 1085–1097. [DOI] [PubMed] [Google Scholar]
- Zhang, Y.W., Yasui, N., Ito, K., Huang, G., Fujii, M., Hanai, J., Nogami, H., Ochi, T., Miyazono, K., and Ito, Y. 2000. A RUNX2/PEBP2αA/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc. Natl. Acad. Sci. 97: 10549–10554. [DOI] [PMC free article] [PubMed] [Google Scholar]