Abstract
Cytokine-dependent gene transcription greatly depends on the tyrosine phosphorylation (“activation”) of Stat proteins at the cell membrane. This rapidly leads to their accumulation in the nucleus by an unknown mechanism. We performed microinjections of recombinant Stat1 protein to show that nuclear accumulation of phosphorylated Stat1 can occur without cytokine stimulation of cells. Microinjection of Stat1 antibody and treatment of cells with kinase or phosphatase inhibitors revealed that nuclear accumulation is a highly dynamic process sustained by Stat1 nucleocytoplasmic cycling and continuous kinase activity. By characterizing nuclear accumulation mutants, it is demonstrated that nuclear import and nuclear retention are two separate steps leading up to nuclear accumulation, with nonspecific DNA binding of activated Stat1 being sufficient for nuclear retention. Critical for nuclear buildup of Stat1 and the subsequent nuclear export is the point of time of tyrosine dephosphorylation, because our data indicate that activated Stat1 is incapable of leaving the nucleus and requires dephosphorylation to do so. It is demonstrated that the inactivation of Stat1 is controlled by its exchange reaction with DNA, whereby DNA binding protects Stat1 from dephosphorylation in a sequence-specific manner. Thus, during nuclear accumulation, a surprisingly simple mechanism integrates central aspects of cytokine-dependent gene regulation, for example, receptor monitoring, promoter occupancy, and transcription factor inactivation.
Keywords: Stat1, nuclear accumulation, DNA binding, dephosphorylation, transcription, cytokine signaling
Responses of cells to extracellular polypeptide signals, for example, cytokines like interferons or interleukins, entail the transient activation and nuclear accumulation of members of the signal transducer and activator of transcription (Stat) family of transcription factors (Darnell 1997). Canonical signaling through the Janus tyrosine kinase (Jak)/Stat pathway begins with the engagement of cytokines with their cognate receptor at the cell membrane (Ihle et al. 1998). This triggers the phosphorylation of signature tyrosine residues in the intracellular receptor tails by noncovalently attached Jak kinases, which also autophosphorylate on tyrosines (Briscoe et al. 1996), and the phosphorylated receptors are thus turned into binding partners for the Stat SH2 domain. Stat1 monomers form high-avidity reciprocal homo- or heterodimers and detach from their receptor docking site after phosphorylation of tyrosine residue 701 at their C terminus (Shuai et al. 1993, 1994; Greenlund et al. 1995). This sequence of events results within minutes in the nuclear translocation of activated Stats, where they can bind to palindromic DNA recognition sites (GAS) and directly induce transcription (Darnell et al. 1994). Stat1 consists of well-defined structural domains. The N-terminal domain engages in protein binding such as cooperative dimer–dimer interactions (Vinkemeier et al. 1996; Xu et al. 1996). It is connected to a large core domain containing the DNA-binding module (Horvath et al. 1995), an SH2 domain responsible for dimer formation (Shuai et al. 1994), and a linker domain implicated in DNA binding (Yang et al. 2002). A transferable transactivation domain is located at the C terminus (Müller et al. 1993).
The regulated nucleocytoplasmic translocation of Stat proteins is only beginning to be understood, with Stat1 in interferon γ (IFNγ) signaling as the best characterized example. Recently, we and others described an unusual dimer-specific nuclear localization signal (dsNLS) comprising residues 407–413 in the DNA-binding domain that mediates binding of Stat1 dimers to importin α/β and subsequent nuclear import (Fagerlund et al. 2002; McBride et al. 2002; Meyer et al. 2002a). Prior to stimulation with IFNγ, Stat1 is found in both the cytoplasm and the nucleus because of signal-independent nucleocytoplasmic shuttling (Meyer et al. 2002a,2002b). The mechanism by which this occurs is not resolved, but differs from the signal-induced nuclear import of activated Stat1 (Sekimoto et al. 1997; Meyer et al. 2002a). Thus, nuclear import does not necessarily result in nuclear accumulation, which is only observed with tyrosine-phosphorylated Stat1 dimers containing a functional SH2 domain (Mowen and David 1998). Loss of DNA binding was also described to cause defects in nuclear accumulation, although conflicting data with various DNA-binding mutants were reported (Herrington et al. 1999; McBride et al. 2000; Melén et al. 2001; Lillemeier et al. 2001). A leucine-rich export signal (NES) in the N-terminal region contributes to nuclear export of Stat1, but pharmacological inactivation of the NES-receptor CRM1 will not induce nuclear accumulation or suppress nucleocytoplasmic shuttling of the unphosphorylated molecule (Begitt et al. 2000; Meyer et al. 2002a). Nuclear accumulation of Stat1 during IFNγ signaling is transient, yet appears to remain stable for several hours. Multiple mechanisms are involved in limiting receptor signaling temporally. Receptor-associated protein tyrosine phosphatases and the targeted degradation of the IFNγ receptor chains play a prominent role (Kisseleva et al. 2002). Pulse-chase experiments demonstrated that Stat1 was not phosphorylated on tyrosine when it reappeared in the cytoplasm after nuclear accumulation (Haspel et al. 1996), and a phosphatase was identified recently that participates in the dephosphorylation of activated Stat1 in the nucleus (Haspel and Darnell 1999; ten Hoeve et al. 2002).
Here, a coherent model is provided that describes Stat transcription factors as shuttling proteins that accumulate in the nucleus because of their nonspecific affinity for DNA. We identify and characterize nuclear retention mutants and show that tyrosine-phosphorylated Stat molecules are barred from nuclear exit. DNA-bound Stat1 is protected from dephosphorylation and is thus transiently retained in the nucleus. These data describe a mechanism that allows nuclear Stats to monitor receptor activity and that at the same time selectively promotes occupation of target regulatory elements.
Results
The role of receptor activation and tyrosine phosphorylation in nucleocytoplasmic shuttling and nuclear accumulation of Stat1
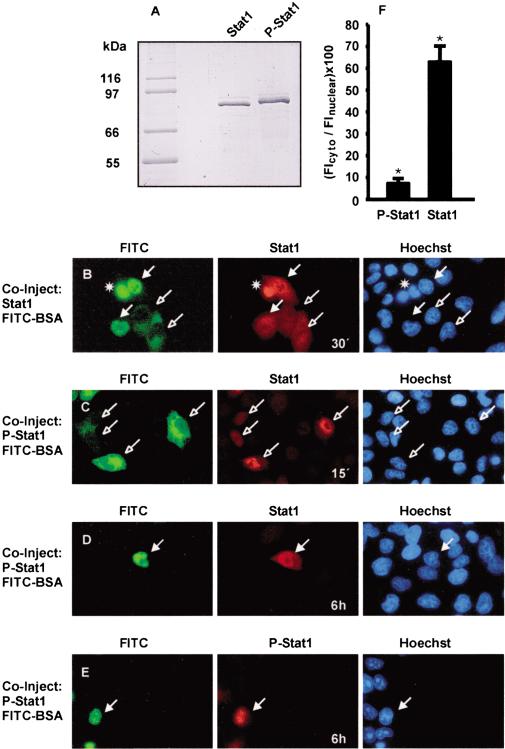
The analysis of nucleocytoplasmic transport of Stat1 has revealed its high, energy-independent mobility in the cytoplasm, which is indicative of random diffusion (Lillemeier et al. 2001). On the other hand, processes that require receptor activation with vesicle formation or complexation with ligand have been proposed to precede nuclear accumulation of Stat3 and Stat1, respectively (Subramaniam et al. 2000; Bild et al. 2002). To investigate nuclear translocation, we purified the human Stat1 protein from baculovirus-infected Sf9 cells (Fig. 1A). Additionally, the recombinant protein was phosphorylated in vitro, and a pure preparation of correctly tyrosine-phosphorylated Stat1 was isolated (Fig. 1A). These proteins were microinjected into HeLa cells and the cell line U3A (data not shown), which does not express Stat1 (Müller et al. 1993). We initially microinjected unphosphorylated Stat1 into the cytoplasm of HeLa cells and followed its intracellular redistribution. The Stat1 protein entered the nucleus rapidly and showed a pancellular distribution after ∼30 min (Fig. 1B). This situation was stable, and nuclear accumulation did not occur even after 3 h (data not shown). Nuclear microinjection of unphosphorylated Stat1 similarly led to a pancellular distribution ∼30 min after injection (Fig. 1B).
Figure 1.
Nucleocytoplasmic shuttling and nuclear accumulation of Stat1 in resting cells. HeLa cells were comicroinjected with unphosphorylated (Stat1) or tyrosine-phosphorylated (P-Stat1) Stat1 and FITC-BSA. After incubation for the indicated times and fixation of the cells, immunocytochemistry and staining of nuclei with Hoechst dye followed. Injected cells are indicated with filled (nuclear injection) or unfilled (cytoplasmic injection) arrows. (A) SDS-PAGE analysis of the Stat1 preparations (1 μg each) used in B–E. The migration of Coomassie-stained molecular size markers is indicated to the left. (B) Unphosphorylated Stat1 was injected into the nucleus or cytoplasm, and the cells were then incubated at 37°C for 30 min. Cells were stained with anti-Stat1 antibody. (*) Dead cell. (C) Tyrosine-phosphorylated Stat1 was injected into the cytoplasm, and the cells were subsequently incubated at 37°C for 15 min. Cells were stained with anti-Stat1 antibody. (D–F) Tyrosine-phosphorylated Stat1 was injected into the nucleus, and the cells were incubated at 37°C for 6 h before fixation. Immunocytochemistry was performed with an antibody recognizing Stat1 or P-Stat1. Representative cells are shown in D and E. For 12 cells of each, the ratio of cytoplasmic and nuclear Stat1 immunofluorescence density was determined and is depicted in the bar diagram (F). Statistically significant differences are indicated by *.
Next, tyrosine-phosphorylated Stat1 was injected into the cytosol of untreated HeLa cells and U3A cells. Figure 1C shows the nuclear accumulation that resulted 15 min after injection. Serum starvation for 4 h prior to microinjection resulted in identical accumulation (data not shown). After nuclear microinjection of tyrosine-phosphorylated Stat1, a slow release of Stat1 into the cytosol was observed. We traced the microinjected material with antibodies directed against unphosphorylated as well as phosphorylated Stat1. Even 2 h after nuclear microinjection, we were unable to detect Stat1 immunoreactivity in the cytoplasm (data not shown). However, after 6 h, considerable amounts of Stat1 were found outside of the nucleus, because an almost pancellular distribution was observed with a Stat1-specific antibody (Fig. 1D,F). In contrast, the remaining tyrosine-phosphorylated Stat1 was confined to the nuclear compartment (Fig. 1E,F). These data confirm that unphosphorylated Stat1 is capable of nucleocytoplasmic shuttling, yet accumulation only occurred with the tyrosine-phosphorylated species. Conversely, nuclear export required dephosphorylation. Notably, nuclear translocation and the subsequent accumulation of tyrosine-phosphorylated Stat1 were entirely independent of IFN stimulation or receptor activation.
Stat1 nuclear accumulation is a dynamic process sustained by continuous shuttling and kinase activity
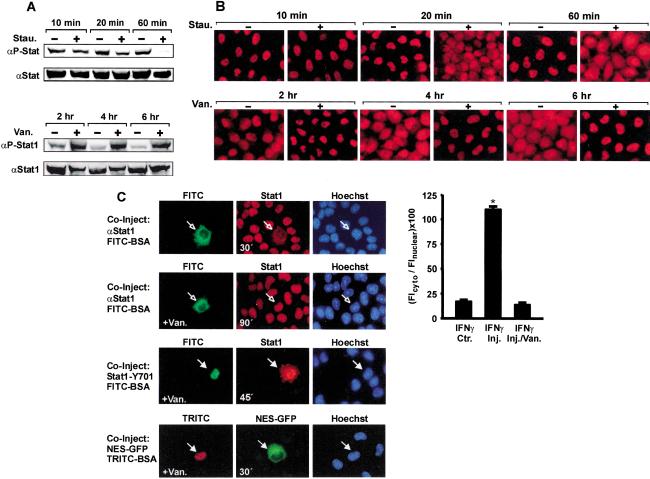
Nuclear accumulation of Stat1 in response to stimulation of cells with IFNγ can last for several hours. Given that Stat1 has a slow turnover time (t1/2 > 8 h; Lee et al. 1997) but dephosphorylation is rapid (t1/2 < 15 min; Haspel et al. 1996), we wanted to determine how long Stat1 molecules remained nuclear. We first observed the effects of aborted receptor signaling on the persistence of nuclear accumulation. Nuclear accumulation was induced by treating HeLa cells with IFNγ for 30 min. Subsequently, the medium was replaced with medium containing staurosporine to block further tyrosine kinase activity (Shuai et al. 1992). This resulted in the complete dephosphorylation of Stat1 after 60 min and a collapse of nuclear accumulation that closely mirrored the kinetics of tyrosine dephosphorylation (Fig. 2A,B, top panels). When we additionally included leptomycin B, an inhibitor of the export receptor CRM1 (Kudo et al. 1998), the rapid termination of nuclear accumulation was not prevented, confirming CRM1-independent nuclear export (Begitt et al. 2000; data not shown). On the contrary, inactivating tyrosine phosphatases by treatment of cells with pervanadate (Gordon 1991) not only suppressed Stat1 dephosphorylation but also extended the duration of nuclear accumulation (Fig. 2A,B, bottom panels).
Figure 2.
Stat1 nuclear accumulation is a dynamic process driven by nucleocytoplasmic shuttling and antagonistic kinase and phosphatase activities. (A) Time course of Stat1 tyrosine phosphorylation in HeLa cells. Cells were stimulated with IFNγ for 30 min to induce tyrosine phosphorylation, at which point IFNγ was withdrawn. The incubation was continued for the indicated times without or with staurosporine (upper panel) or vanadate/H2O2 (lower panel), respectively. Whole-cell extracts were analyzed by Western blotting with anti-P-Stat1 antibody and reprobed with anti-Stat1 antibody. (B) Time course of accumulation of Stat1 in the nuclei of HeLa cells after 30 min of IFNγ stimulation. Treatment of cells was identical to that in A. At the indicated times, Stat1 was detected in fixed cells by indirect immunofluorescence microscopy using an anti-Stat1 antibody. (C, first and second rows) HeLa cells were stimulated with IFNγ for 30 min before anti-Stat1 antibodies and FITC-labeled BSA were comicroinjected into the cytosol. Subsequently, the incubation was continued for another 30 min with IFNγ alone (first row), or for 90 min in the additional presence of vanadate/H2O2 (second row). Shown are representative cells and the corresponding bar diagram with a quantitative analysis of the ratio of cytoplasmic and nuclear Stat1 immunofluorescence density for noninjected cells (control) and for injected cells (n = 12 each). Statistically significant differences are indicated by *. (Third row) Purified Stat1 Tyr701F was coinjected with FITC-BSA in the nucleus of HeLa cells. At the time point of microinjection, the cells had been pretreated with IFNγ for 60 min with vanadate/H2O2 present after 30 min. The microinjected cells were incubated with IFNγ/vanadate for another 45 min before fixation and immunocytochemistry. (Fourth row) Stat1 export signal fused to GFP-GST (NES-GFP) was coinjected with TRITC-BSA in the nucleus of cells that were pretreated with IFNγ/vanadate as before. After microinjection, the incubation was continued for another 30 min in the presence of IFNγ/vanadate before the fusion protein was located in fixed cells by its GFPautofluorescence.
We then used an antibody-microinjection assay, which we introduced earlier (Meyer et al. 2002a), to reveal ongoing nucleocytoplasmic shuttling of Stat1 during stable nuclear accumulation. Cells treated with IFNγ for 30 min were injected with Stat1 antibodies in the cytoplasm and then incubated in the continuous presence of IFNγ for a further 30 min. As can be seen from Figure 2C (first row), this resulted in the significant loss of Stat1 from the nucleus because of antibody-induced precipitation in the cytoplasm. However, treatment of cells with the tyrosine phosphatase inhibitor vanadate precluded the antibody-induced nuclear clearance of Stat1, because even after 90 min of incubation there was no translocation of Stat1 to the cytoplasm (Fig. 2C, second row). The bar diagram in Figure 2C gives a quantitative summary of the above injection data. Additionally, the export specifically of unphosphorylated Stat1 from the nucleus of vanadate-treated cells was investigated. We made use of a mutated Stat1, the tyrosine phosphorylation site of which, Tyr 701, was defective (Shuai et al. 1993). We prepared purified protein of this mutant and injected it into the nucleus of cells that had been stimulated with IFNγ and treated with vanadate/H2O2 (Fig. 2C, third row). Contrary to tyrosine-phosphorylated wild-type Stat1, which was retained in the nucleus by vanadate treatment (second row), the unphosporylated Stat1Y701F was still capable of nuclear export during phosphatase inhibition. In another control experiment, we performed nuclear microinjections of a canonical Stat1-derived nuclear export signal linked to a fusion of green-fluorescent protein with glutathione-S-transferase (GFP-NES-GST). This reporter construct also was quickly exported into the cytoplasm irrespective of vanadate/H2O2 (Fig. 2C, fourth row). These observations demonstrated the highly dynamic nature of Stat1 nuclear accumulation, with kinase and phosphatase activities being the antagonistic determinants for its duration. Importantly, tyrosine-phosphorylated Stat1 was blocked from nuclear exit. In addition, these data showed that repeated cycles of nuclear import and export of Stat1 during the accumulation phase were required to maintain a stable accumulation in the nucleus.
Stat1 DNA-binding mutants discriminate nuclear retention from nuclear import
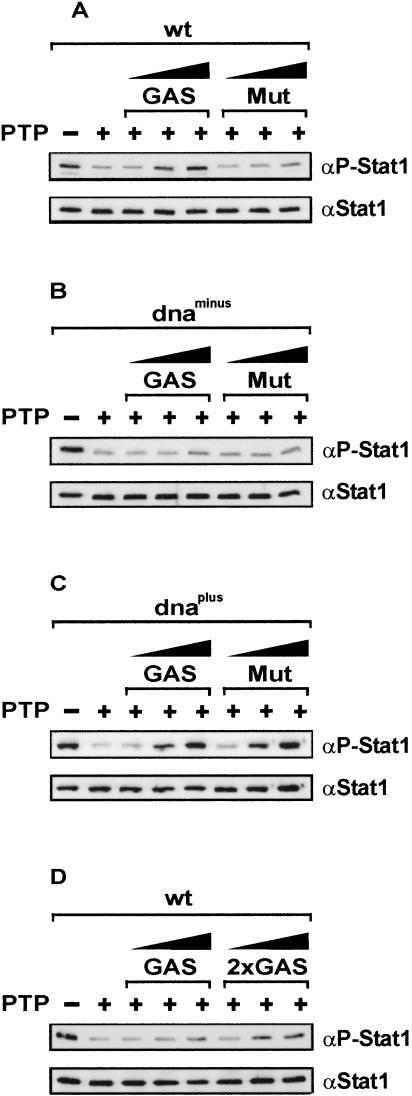
Nuclear accumulation was shown to depend on the integrity of the DNA-binding domain, which can be attributed to the need for a functional dsNLS located here (Fagerlund et al. 2002; Meyer et al. 2002a). In addition, vicinal residues unrelated to the dsNLS were also demonstrated to influence nuclear buildup of activated Stat1 (McBride et al. 2000). Therefore, we analyzed the effects that DNA binding has on Stat1 nuclear retention and also took nonspecific protein–DNA interactions into account. Guided by the Stat1–DNA cocrystal structure (Chen et al. 1998), we mutated residues in the DNA-binding domain that potentially make backbone contacts with phosphate groups in the DNA and are not part of the dsNLS (Fig. 3A). We reasoned that introducing positive charges in these positions (termed Stat1dnaplus; Thr327Arg; Val426His; Thr427His) might increase electrostatic attraction to the polyanionic DNA, whereas negative charges (termed Stat1dnaminus; Val426Asp; Thr427Asp) might potentially reduce electrostatic interactions with DNA. The Stat1 variants were expressed well and phosphorylated on tyrosine 701 in response to IFNγ (Fig. 3B). Additionally, we included another DNA binding mutant, named Stat1dnaoff here, that was previously characterized by Darnell and coworkers (Yang et al. 2002) and displays a markedly increased off-rate from GAS sites. First, we observed the nucleocytoplasmic distribution of GFP-fusion proteins before and after IFNγ stimulation and found accumulation of Stat1dnaplus and dnaoff, whereas the Stat1dnaminus mutant failed to accumulate in the nucleus in response to IFNγ (Fig. 3C). Addition of a negative charge in position 327 (Thr to Glu) of Stat1dnaminus also resulted in defective nuclear accumulation, albeit less severely, because a partial accumulation could still be induced by IFNγ (data not shown). We therefore used the double mutant dnaminus for further experimentation.
Figure 3.
Influence of DNA binding on nuclear accumulation of Stat1. (A) Domain structure of Stat1wt and the mutants used in this work. (N) N-domain; (CC) coiled-coil domain; (DNA) DNA-binding domain; (L) linker domain; (SH2) SH2 domain. The position of residue Tyr 701, and the C-terminal transactivation domain (TAD) are indicated, as well as the location of the dsNLS. (B) U3A cells transiently expressing the indicated Stat1–GFPvariants were treated with IFNγ for 30 min. Shown is a Western blot analysis with anti-P-Stat1 and anti-Stat1 antibodies of whole-cell extracts. (C) HeLa cells expressing the indicated Stat1–GFPvariant proteins before and after 60 min of treatment with IFNγ (+IFN). Where indicated, vanadate/H2O2 was added after 30 min (+IFN +Van.). Shown are the GFPfluorescence and the Hoechst-stained nuclei of fixed cells. (D) Mobility shift assay with a Stat1-specific M67 probe of wild-type Stat1 and DNA-binding mutants. The position of the Stat1–DNA complex is indicated by an arrow; * marks a nonspecific band. Cytoplasmic extracts from transiently transfected and IFNγ-treated (30 min) U3A cells were used. (E) Precipitation of Stat1–GFP variant proteins with agarose-bound oligonucleotides comprising tandem M67 binding sites (GAS) or an unrelated sequence (Mut). Unmodified agarose beads served as a control (beads). Beads were incubated with cell extracts from transfected and IFNγ-treated U3A cells as described. The precipitate (pull-down) was analyzed by Western blotting with anti-Stat1 antibody. Four percent (v/v) of the cell extract input was blotted and probed with anti-P-Stat1 antibody. (F) The dimerization of Stat1dnaminus was detected by coimmunoprecipitation of GFP- and Flag-tagged variants. HeLa cells coexpressing GFP- and Flag-tagged variants of Stat1wt, Stat1dnaminus, or Stat1Y701F (a dimerization-incompetent mutant) were stimulated with IFNγ. Shown are Western blots with anti-Stat1 antibody of whole-cell extracts before immunoprecipitation (Input), and of the immunoprecipitates obtained with anti-Flag antibody (IP). The position of GFP- and Flag-tagged Stat1 is indicated. (G) Western blot of cytoplasmic (C) and nuclear (N) extracts from IFNγ-treated (30 min) U3A cells transiently expressing wild-type Stat1 or dnaminus. Extracts were probed with anti-P-Stat1 antibody. Parallel blots with anti-Eps15 and anti-Sp1 antibody were used to probe for extract purity. (H) Anti-GFPantibodies and TRITC-BSA were comicroinjected into the nucleus of unstimulated HeLa cells transiently expressing Stat1dnaminus–GFP(top) or Stat1wt–GFP(bottom). Incubation with IFNγ immediately commenced, and after 30 min, Stat1 was located in fixed cells by GFPautofluorescence. Arrows indicate injected cells.
DNA-binding activity was investigated in mobility shift assays with an optimal Stat1-binding site (M67), and both Stat1dnaplus and dnaminus failed to bind (Fig. 3D). Because this experiment can only reveal sequence-specific DNA interactions, we used a different approach to also determine nonspecific binding. We used oligonucleotides harboring either tandem optimal Stat1-binding sites or a TTC-polymer with a single site weakly resembling a cognate GAS element (see Materials and Methods). These were coupled to agarose beads and then mixed with extracts from IFNγ-treated U3A cells expressing similar amounts of tyrosine-phosphorylated wild-type Stat1 (Stat1wt) or either of these mutants. As expected, Stat1wt showed a strong preference for the optimal binding site (Fig. 3E). However, the mutant Stat1dnaplus was capable of binding to both the specific and the nonspecific site with almost equal avidity (Fig. 3E). In contrast, Stat1dnaminus showed only a very weak interaction with either sequence (Fig. 3E). Thus, Stat1dnaplus bound to DNA, albeit nonspecifically, whereas dnaminus did not. To exclude the possibility that the observed defect in DNA binding was caused by an inability to dimerize, we cotransfected U3A cells with Stat1dnaminus tagged with either GFPor Flag and confirmed the formation of GFP/Flag-tagged heterodimers (Fig. 3F). We also tested nuclear extracts from IFNγ-stimulated U3A cells for the presence of tyrosine-phosphorylated Stat1dnaminus and detected ample amounts similar to wild type (Fig. 3G). Additionally, we were able to induce the nuclear accumulation of Stat1dnaminus– GFPafter nuclear microinjection of GFPantibodies into IFNγ-stimulated HeLa cells (Fig. 3H). Importantly, the antibody-induced nuclear accumulation of Stat1wt and Stat1dnaminus could not be discriminated, thus indicating unperturbed import of the mutant. Moreover, blocking of tyrosine dephosphorylation with vanadate/H2O2 during 30 min of IFNγ stimulation rescued the accumulation defect of Stat1dnaminus (Fig. 3C).
These results identified nonspecific DNA binding as a sufficient requirement for nuclear accumulation and distinguished STAT1dnaminus from a recently characterized import mutant (Meyer et al. 2002a). Thus, we determined defective retention as a cause preventing accumulation in the nucleus.
DNA-bound Stat1 is protected from tyrosine dephosphorylation
The above experiments demonstrated the need for tyrosine dephosphorylation prior to nuclear export of Stat1, and they identified diminished DNA-binding activity as the cause of defective nuclear retention. At first we therefore examined the influence of DNA binding on the dephosphorylation of Stat1 variants in vitro. Highly pure recombinant tyrosine-phosphorylated Stat1 was incubated in the absence and presence of DNA with purified TC-PTPa (also called TC45; Ibarra-Sanchez et al. 2000), a ubiquitously expressed Stat1-specific nuclear tyrosine phosphatase (ten Hoeve et al. 2002). We used recombinant full-length TC45 as well as a truncated version that lacks a noncatalytic segment of 35 amino acids at its C terminus. This variant enzyme was demonstrated to exhibit increased phosphatase activity (Hao et al. 1997). The two preparations differed in their Stat1 dephosphorylation activity, but the specificity of the reaction was unchanged by the truncation (data not shown). The Stat concentration was set equal to the Kd value of 1 nM (Vinkemeier et al. 1996). As can be seen in Figure 4A, Stat1 was efficiently dephosphorylated by TC45. In the presence of DNA, however, the tyrosine dephosphorylation of Stat1 was strongly reduced in a concentration-dependent manner (Fig. 4A). Notably, the dephosphorylation reaction was sensitive also to alterations of the DNA sequences used, because GAS sites protected Stat1 better from dephosphorylation than unrelated DNA sequences (Fig. 4A). Control reactions with an artificial substrate showed that TC45 was not generally inhibited in the presence of DNA (see Materials and Methods). We then examined the in vitro dephosphorylation of the two DNA-binding mutants Stat1dnaminus and dnaplus, which display oppositional accumulation behavior. Similar to Stat1wt, the mutants were efficiently dephosphorylated by TC45 in the absence of DNA, and the addition of DNA to the reaction afforded protection from dephosphorylation (Fig. 4B,C). However, Stat1dnaminus, the mutant that shows deficient accumulation, was only very poorly protected from dephosphorylation even at the highest DNA concentrations used (25-fold molar excess) and regardless of its sequence (Fig. 4B). An entirely different result was found with the accumulating mutant Stat1dnaplus, which was strongly protected from dephosphorylation by DNA. Moreover, opposite to Stat1wt, the protective influence of DNA had no sequence-specific component, as both the GAS-containing oligonucleotide and a nonspecific DNA were equally efficient in preventing Stat1 inactivation (Fig. 4C). Additionally, we explored the influence of tandem GAS sites on the dephosphorylation of wild-type Stat1. Expectedly, effective protection from dephosphorylation at subsaturating DNA concentrations was observed only with tandem GAS sites (Fig. 4D). In conclusion, Stat1 that was not bound to DNA underwent tyrosine dephosphorylation at maximum rate, whereas DNA binding protected this molecule from inactivation in a sequence-dependent manner.
Figure 4.
DNA binding protects Stat1 from dephosphorylation. (A) Purified tyrosine-phosphorylated Stat1wt at a concentration of 1 nM was incubated without (–) or with (+) the Stat1-specific tyrosine phosphatase TC45 (PTP) at 30°C for 1 h. Increasing concentrations of a duplex 37-mer DNA (0.5 nM, 5 nM, 25 nM) harboring either a single GAS site (GAS) or an unrelated sequence (Mut) were included in the reaction where indicated. Reaction products were analyzed by Western blotting using anti-P-Stat1. The blots were subsequently reprobed with anti-Stat1 antibody. (B,C) Identical experimental setup as in A, with the indicated Stat1 DNA-binding mutants replacing Stat1wt. (D) Comparison of the effects of single (GAS) or tandem GAS sites (2×GAS) on the dephosphorylation of purified wild-type Stat1. Experimental setup as in A, except that GAS sites were included at concentrations of 0.125 nM, 0.625 nM, and 1.25 nM, respectively.
DNA binding determines the duration of nuclear accumulation
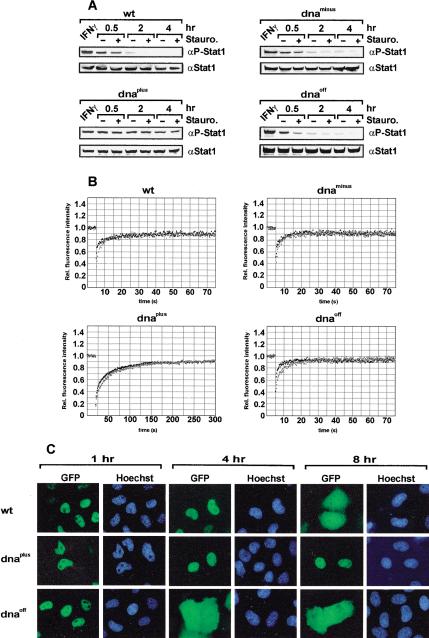
We then examined in vivo the rate of tyrosine dephosphorylation and its influence on the intranuclear mobility of Stat1 as well as on the duration of nuclear accumulation. Reconstituted U3A cells were treated with IFNγ for 30 min to induce Stat1 phosphorylation. Thereafter, the medium was replaced with growth medium alone or medium containing staurosporine to expose tyrosine dephosphorylation by blocking continuous kinase activity. As is shown in Figure 5A, the initial concentrations of activated Stats were similar for all variants after 30 min of IFNγ treatment, indicating their comparable facility to become tyrosine-phosphorylated. However, impeding tyrosine kinase activity with staurosporine revealed the rapid dephosphorylation not only of Stat1wt but also of dnaminus and dnaoff. In contrast, Stat1dnaplus was much less sensitive to the cessation of kinase activity, as the level of activated protein remained high, indicating strongly reduced tyrosine dephosphorylation (Fig. 5A).
Figure 5.
Influence of DNA binding on the dephosphorylation of Stat1, its intranuclear mobility, and the duration of nuclear accumulation. (A) Time course of Stat1 tyrosine dephosphorylation in U3A cells transiently expressing Stat1 variant proteins. Cells were prestimulated with IFNγ for 30 min to induce tyrosine phosphorylation (lane IFNγ). After removal of IFN, the cells were treated without or with staurosporine for 0.5, 2, or 4 h as indicated. Whole-cell extracts were analyzed by Western blotting with anti-P-Stat1 antibody and reprobed with anti-Stat1 antibody. (B) FRAPanalysis of the nuclear fluorescence in IFNγ-stimulated HeLa cells expressing Stat1–GFPvariant proteins. Shown is the fluorescence recovery in the bleach area in the absence (black squares) or presence (gray squares) of vanadate/H2O2. Note the extended time scale for Stat1dnaplus. (C) HeLa cells expressing the indicated Stat1–GFPvariant proteins were stimulated with IFNγ for 30 min to induce nuclear accumulation. Thereafter, the incubation continued in the absence of IFN for the indicated times. Shown are the GFPfluorescence and the Hoechst-stained nuclei of fixed cells.
Next, the mobility of the Stat1 DNA-binding mutants before and during nuclear accumulation was determined by FRAPanalysis (fluorescence recovery after photobleaching). As is shown in Table 1, in unstimulated cells, the Stat1 variants did not differ from one another because we determined almost identical recovery rates. Interestingly, mobility was consistently lower in the cytoplasm. Yet, after IFNγ stimulation, mobility differences between the Stat1 mutants became apparent. Wild-type Stat1 was highly mobile in the nucleus, as were the dnaoff and dnaminus mutants. These proteins had recovery times of ∼20 sec (Fig. 5B). Remarkably, the mutant Stat1dnaplus showed a significantly reduced intranuclear mobility, and the recovery time was extended to ∼200 sec. Thus, DNA binding differences were reflected in an altered mobility in the nuclear compartment, with enhanced binding correlating with reduced mobility. Importantly, fluorescence recovery was complete for all Stat1 variants, indicating that no immobile population was present within the nucleus during accumulation.
Table 1.
Intracellular mobility of Stat1 variant proteins
| Cytosol unstimulated (sec) | Nucleus unstimulated (sec) | Nucleus IFN (sec) | Nucleus IFN + vanadate (sec) | |
|---|---|---|---|---|
| wt | 45.5 ± 8.1 (n = 9) | 14.7 ± 3.3 (n = 9) | 18.6 ± 3.2 (n = 12) | 46.4 ± 11.2 (n = 9)* * |
| dnaplus | 41.8 ± 14.7 (n = 7) | 14.3 ± 3.7 (n = 4) | 206.7 ± 30.1 (n = 8)* | 233.7 ± 46.6 (n = 7)* |
| dnaminus | 37.8 ± 7.4 (n = 4) | 20.8 ± 6.5 (n = 4)* | 20.2 ± 4.1 (n = 4) | 36.6 ± 14.9 (n = 4)* |
| dnaoff | 39.6 ± 6.3 (n = 4) | 15.3 ± 7.8 (n = 5) | 22.8 ± 5.8 (n = 4) | 39.5 ± 5.4 (n = 4)* * |
Fluorescence recovery times of Stat1-GFP as determined by FRAP. They are expressed as mean ± SEM. Unpaired Student's t tests were performed to compare the intracellular mobility between Stat1 wt (row 1) and the mutants (rows 2-4). This was done for unstimulated cells (columns 1 and 2) and for cells treated with IFNγ (columns 3 and 4). Statistically significant differences (p < 0.05) with Stat1 wt are indicated by *. In addition, the recovery time recorded for each variant in the presence of IFNγ (column 3) was compared with the respective recovery time after the addition of vanadate/H2O2 (column 4). Here for each variant, significant differences (p < 0.05) are indicated by * *.
Figures 1 and 2 indicated that tyrosine-phosphorylated Stat1 was trapped in the nucleus. We therefore examined how the inhibition of tyrosine phosphatases influences the mobility of Stat1 inside the nucleus. This was achieved by treating IFNγ-stimulated cells with vanadate/H2O2. Strikingly, preventing tyrosine dephosphorylation had little impact on the intranuclear mobility of all Stat1 proteins investigated (Fig. 5B), because recovery times increased only by a factor of 2.5 or less (Table 1). Notably, fluorescence recovery of all Stat1 variant proteins was complete even in the absence of phosphatase activity, and the mobility continued to differ between the DNA binding mutants (Fig. 5B). Compared with other studies, the mobility of Stat1 was much faster than the core histones and similar to other inducible transcriptional regulators (McNally et al. 2000; Kimura and Cook 2001; Lillemeier et al. 2001). In conjunction with the in vitro dephosphorylation data, we conclude that the release of Stat1 from DNA did not require enzymatic activity. Because progression of the enzyme reaction was proportionate to the Stat–DNA exchange reaction, it follows that the dephosphorylation of Stat1 was kinetically controlled.
The above data for the DNA-binding mutants indicated that the inactivation of Stat1 is inversely correlated in vitro and in vivo with the avidity of the interactions of Stat1 with DNA. Hence, the resulting differences in terms of tyrosine dephosphorylation ought to be reflected in varied nuclear retention. Because a complete loss of DNA binding was associated with a radical abortion of nuclear accumulation (see Stat1dnaminus in Fig. 3C), more subtle modifications of the Stat1–DNA interactions might influence the duration of nuclear accumulation less severely. As Figure 5C shows, this is, in fact, the case. Although the IFNγ stimulus was identical for all mutants, the duration of Stat1 nuclear accumulation differed predictably according to the avidity of Stat1 DNA binding. The mutant Stat1dnaplus with improved binding to nonspecific DNA showed transient, yet prolonged nuclear accumulation, whereas the diminished DNA binding of Stat1dnaoff led to an abbreviated nuclear accumulation phase (Fig. 5C). We infer the following: Impaired DNA binding resulted in accelerated inactivation of Stat1 in the nucleus. Therefore, in the face of decreasing Stat1 rephosphorylation because of terminated receptor signaling, the steady-state concentration of activated Stat1 required for an observable nuclear accumulation was depleted sooner. The opposite of this was true for improved DNA binding interactions.
Interferon-induced nuclear accumulation is not indicative of transcriptional activity
To our knowledge, there are only examples in which IFNγ-induced nuclear accumulation of full-length Stat1 is simultaneously accompanied by the induction of responsive target genes. Therefore, we tested the gene-activation ability of the mutant Stat1dnaplus, which shows prolonged nuclear accumulation, and its direct opposite Stat1dnaminus, which is incapable of nuclear accumulation. As shown in Figure 6, the two Stat1 variants did not differ in their inability to induce a GAS-driven reporter gene in response to IFNγ. Therefore, nuclear accumulation does not restore target gene recognition if the Stat1 dimer has lost the capacity to discriminate GAS sites from unrelated sequences.
Figure 6.
Accumulation of Stat1 in the nucleus is not sufficient to induce transcription. Wild-type Stat1 or the indicated DNA-binding mutants were coexpressed with an IFNγ-inducible luciferase reporter plasmid in U3A cells. Luciferase activity was determined 36 h posttransfection in unstimulated cells (gray bars) or after stimulation with IFNγ for 6 h (black bars). Error bars represent standard deviations for six independent experiments.
Discussion
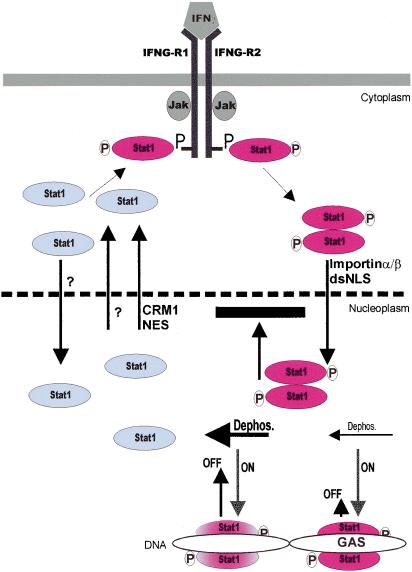
Signal transduction from the cell membrane to the nucleus is organized in regulatory circuits (Brivanlou and Darnell 2002). These circuits allow the nucleus to precisely detect and respond to changes of the primary signal input at the cell membrane. A model is shown in Figure 7 that explains the molecular basis of the signal-dependent nuclear accumulation of the transcription factor Stat1 and outlines the constitutive cycling of Stat1 both in resting cells and during stimulation with IFNγ.
Figure 7.
A model indicating that nucleocytoplasmic shuttling of Stat1 is controlled in a promoter-specific manner by the exchange reaction of Stat1 with DNA. Unphosphorylated Stat1 is depicted in blue, phosphorylated Stat1 dimers are in red. Arrow lengths symbolizing the on and off rates of the Stat1 exchange reaction with DNA are drawn proportionately (arbitrary units) to the respective rate constants at GAS-consensus (GAS) or unrelated sites. Progression of the Stat1 dephosphorylation reaction (Dephos.) at those binding sites is indicated by arrow size and Stat1 coloring. Absence of mechanistic insight is denoted by a question mark. See text for details.
Nuclear translocation and accumulation of activated Stat1 can occur without receptor activation, as demonstrated by microinjection of in vitro phosphorylated protein (Fig. 1C). This observation extends the previous finding that dimerization is sufficient for nuclear import of activated Stat1 (Milocco et al. 1999). These results do not rule out that there are events besides the routine import machinery that contribute to the nuclear translocation of activated Stat1. However, it seems unlikely that such events are necessary for anything other than tyrosine phosphorylation. These data are in agreement with a proposed “soft-wired” mechanism for cytoplasmic transport of Stat1 (Lillemeier et al. 2001).
Our results indicate that nucleocytoplasmic shuttling of Stat1 is crucial even during nuclear accumulation. However, shuttling was observed only with the unphosphorylated molecule. Blocking of tyrosine dephosphorylation by vanadate induced nuclear accumulation of the retention mutant Stat1dnaminus (Fig. 3C). The nuclear export of wild-type Stat1 was blocked after stimulation with IFNγ in the presence of vanadate, whereas nuclear export of both a Stat1 variant with a mutated tyrosine phosphorylation site and an export reporter construct were not affected by this treatment (Fig. 2C). In addition, we did not observe nuclear export of phosphorylated Stat1 after its nuclear microinjection (Fig. 1D,E). Interestingly, the dephosphorylation of recombinant Stat1 in the nucleus was slow in comparison to the endogenous protein. This could be caused by the high concentration of the injected material or by the absence of Arg 31 methylation, which has been shown to hamper Stat1 dephosphorylation (Zhu et al. 2002). We conclude that tyrosinephosphorylated dimers were incapable of nucleocytoplasmic shuttling and required inactivation before release into the cytoplasm. Nevertheless, the cycling rate was high, as cytoplasmic trapping depleted nuclear Stat1 in ∼30 min (Fig. 2C), and blocking of kinase activity led to a collapse of nuclear accumulation in <60 min (Fig. 2B). This is in good accord with the progression of tyrosine dephosphorylation (t1/2 < 15 min; Haspel et al. 1996). Therefore, Stat1 must continuously shuttle in and out of the nucleus with constant reactivation at the cell membrane to maintain a steady level of transcriptionally active dimers in the nucleus. These results explain why protracted nuclear export of unphosphorylated Stat1 resulted in reduced transcription (Begitt et al. 2000). Fast dephosphorylation with concomitant nuclear export and reactivation in the cytoplasm are both essential for dynamic signaling. In this way, the degree of receptor activity at the cell membrane is immediately reflected by the level of activated Stat1 in the nucleus. Such shuttling was also proposed for Stat6 in interleukin-4 signaling (Andrews et al. 2002).
An essential conceptual addition to the standard model of Stat function is the distinction between nuclear import and nuclear retention of activated Stat1. Judged by several criteria, such as the abundance of phosphorylated Stat1 in nuclear extracts, the antibody-induced nuclear precipitation after IFNγ stimulation, and the rapid nuclear accumulation induced by an export block through preventing tyrosine dephosphorylation (Fig. 3), the IFNγ-induced nuclear import of Stat1dnaminus proceeded unaltered, but nevertheless the mutant was incapable of nuclear accumulation. These results indicated that nuclear accumulation was not caused simply by accelerated nuclear import after cytokine stimulation. Rather, there must exist a retention mechanism affecting nuclear export. By modulating the nonspecific affinity of dimeric Stat1 for DNA, we were able to engineer mutants with increased or abolished nuclear retention. This was exemplified by the opposed nuclear accumulation abilities of Stat1dnaminus and dnaplus, neither of which could discriminate GAS elements from nonspecific sites, although they differed from one another in their overall DNA affinity. To date, nonspecific DNA binding of Stats has received little attention. We show that nonspecific interactions with DNA are observable for wild-type Stat1 (Fig. 3E), and binding to DNA outside of consensus binding elements is common for transcriptional regulators (Lin and Riggs 1975). We also point out the fact that GAS sites are generally poorly conserved, making it difficult to predict actual chromatin binding sites (Decker et al. 1997), indicating that the distinction between “specific” and “nonspecific” binding sites is a gradual one. Figure 5 demonstrates that nuclear accumulation of Stat1dnaoff is shortened, despite the fact that this mutant was activated to a similar extent as wild-type Stat1 and despite its normal ability to recognize GAS sites (Fig. 3D). Moreover, we found that microinjection of phosphorylated Stat1wt into the cytoplasm of unstimulated cells (Fig. 1C) or IFNγ-stimulated cells (data not shown) gave similar results. We therefore consider it unlikely that the amount of nuclear Stat1 is regulated by the availability of GAS binding sites. In addition, several DNA-binding mutants that cannot recognize GAS sites have been reported, but not all of them have lost the ability to accumulate in the nucleus (McBride et al. 2000; Lillemeier et al. 2001). There are no data available concerning the nonspecific DNA binding of these mutants, but it could well account for the observed differences.
Nonetheless, because tyrosine-phosphorylated Stat1 was not exported readily but was retained in the nucleus, DNA binding per se cannot account for nuclear retention. Rather, the nuclear export was controlled by tyrosine dephosphorylation. This together with the observation that loss of DNA binding resulted in loss of nuclear accumulation led to the conclusion that DNA binding and dephosphorylation were interrelated. And, indeed, DNA-bound Stat1 was protected from phosphatase attack both in vitro and in vivo (Figs. 4A, 5A). Thus, although the enzymatic reaction was required for nuclear export, it could not be the causative step for the release of Stat1 from its nuclear DNA tether. In support of this idea, we found that inactivation of tyrosine phosphatases reduced intranuclear mobility of Stat1 only marginally (Fig. 5B; Table 1). For these reasons, DNA binding is sufficient but not necessary for nuclear accumulation. Any process that interferes with the dephosphorylation of Stat1 in the nucleus, for example, destruction by mutation of the Stat1 interface with the phosphatase, should cause retention in the nucleus (see also vanadate treatment in Fig. 3C). A negative influence of DNA binding on Stat dephosphorylation was noted earlier also for Stat5. Binding to the glucocorticoid receptor enhanced DNA binding of Stat5, which correlated with its prolonged tyrosine phosphorylation in vivo (Wyszomierski et al. 1999).
Central to the understanding of nuclear accumulation was the observation of sequence-specific inhibitory effects of DNA on Stat1 dephosphorylation. It was found that activated Stat1 is more efficiently protected by optimal GAS sites than by nonconsensus oligonucleotides (Fig. 4A). Quantitative analysis of the Stat1–DNA exchange reaction had revealed identical affinities for various sites, but despite having similar apparent Kd values, the binding with DNA may differ significantly in rates of association with and dissociation from the Stat1 protein (Vinkemeier et al. 1996). The Stat1 protein achieves equilibrium in DNA binding very rapidly (≪30 sec); however, although the protein–DNA complex had a half-life of no more than 3 min for any of the GAS sites investigated, the off-times varied at least sixfold. Cooperative DNA binding further stabilized the Stat1–DNA complex (see also Fig. 4D). Importantly, the data we present here show that conservation of the active transcription factor was determined by the DNA site to which it was bound. Therefore, Stat1 dimers bound to favorable sites, for example, the promoter region of cytokine-regulated genes, are likely to be preserved comparatively better than dimers occupying nonfunctional sites. As a result, the intrinsic preference of Stat1 for its target genes would be transformed into longevity of the transcriptionally active species. This way, the process of nuclear accumulation might confer a selection advantage on Stat1 dimers bound to GAS consensus promoter sites. Thus, in contrast to other gene regulators (Tansey 2001), control of transcription by Stat1 appears to be governed primarily by chance, simply by falling off the DNA with concomitant inactivation at a frequency dictated by the particular binding site.
Materials and methods
cDNAs, cell culture, and reagents
Plasmids expressing Stat1 fused to GFP (pStat1–GFP) or GST-NES–GFP(pGFP-h4C-GST) have been described (Begitt et al. 2000). Site-directed mutagenesis of pStat1–GFP(QuikChange, Stratagene) generated (1) pStat1dnaplus, (2) pStat1dnaminus, (3) Stat1dnaoff, and (4) pStat1-Y701F, in which (1) the codon for T327 was replaced by R, codons V426 and T427 by HH, (2) V426 and T427 by DD, (3) K544 and E545 by AA, and (4) codon Y701 by F. Mutations were verified by DNA sequencing. The coding sequence of full-length human TC45 (residues 2–387) was amplified by PCR from IMAGE clone IMAGp998B124644Q3 (RZPD Deutsches Ressourcenzentrum) and cloned into the EcoRI and BamHI sites of expression vector pASK-IBA3 (IBA). Cell culture and transient transfections with lipofectamine were as described (Begitt et al. 2000). Cells were treated with IFNγ (5 ng/mL; Biomol), staurosporine (500 nM; Sigma), leptomycin B (10 ng/mL; Sigma), Na-vanadate (0.8 mM), and H2O2 (0.2 mM) as indicated in the figures.
Pull-down assay with biotinylated oligos
Duplex oligonucleotides (0.5 mL, 28 pmole/μL, with a 5′ biotin on one strand) containing either two optimal GAS sites (5′-GAGACTCAGTTTCCCGTAAATCGTCCAGTTTCCCGTAA AGACTATGC and its antisense), or a single GAS-like site [5′-(TTC)4TAC(TTC)15 and its antisense] were conjugated to streptavidin agarose (0.5 mL packed vol; Pierce) at 4°C for 1 h. Confluent U3A cells (one 10-cm dish) transiently coexpressing c-Eyk kinase (Besser et al. 1999) and a Stat1–GFPvariant were stimulated for 35 min with IFNγ and 15 min in the additional presence of vanadate/H2O2 before lysis in 400 μL of lysis buffer A (Meyer et al. 2002a). Stat1 tyrosine phosphorylation was assessed by Western blotting, and the extracts were diluted accordingly with buffer A. After preclearing with streptavidin agarose, 250 μL of the normalized extracts were rotated with 50 μL (packed vol) of DNA-conjugated beads at 4°C for 1.5 h. The beads were washed with lysis buffer A (three times with 500 μL), and bound proteins were eluted by boiling in SDS sample buffer and analyzed by Western blotting.
Electrophoretic mobility shift assay (EMSA) and analysis of transcription
EMSA was performed as described with 5 μL of cytoplasmic extracts. Luciferase reporter gene assays have been described (Begitt et al. 2000).
Antibody microinjections, Western blotting, and immunoprecipitation
Western blotting, immunoprecipitation, and antibody injections were done as described (Meyer et al. 2002a). HeLa-S3 cells and U3A cells were injected with anti-Stat1 antibody C-136 (Santa Cruz) or the monoclonal anti-GFPantibody 2A3 (2.5 mg/mL; a kind gift of M. Vigneron and C. Kedinger). After the indicated incubation period, Stat1 was detected immunocytochemically with antibody C-24 (Santa Cruz).
Purification and microinjection of recombinant Stat1
Baculoviruses expressing Stat1wt, Stat1dnaplus, Stat1dnaminus, or Stat1Y701F, respectively, were produced with the Bac-to-Bac system as specified by the manufacturer (GIBCO). Protein expression in baculovirus-infected Sf9 insect cells, protein purification, and in vitro tyrosine phosphorylation were done essentially as described (Vinkemeier et al. 1996) with the following modifications. Incubation of epidermal growth factor (EGF) receptor with EGF and N-ethylmaleimide treatment of unphosphorylated Stat1 were omitted. Homogeneous tyrosine phosphorylation of the final preparation was verified by MALDI analysis, as was the absence of Arg 31 methylation. Purified protein (1 mg/mL in injection buffer with 2 mM DTT) was microinjected with identical outcome into U3A cells and HeLa cells as described (Begitt et al. 2000). Afterward, the cells were left at 37°C in a humidified incubator for the indicated times. Fixation with 3.7% formaldehyde (10 min) and permeabilization with 0.2% Triton X-100 (5 min) were at room temperature, before immunocytochemistry with anti-Stat1 antibody C-136 (1:1000) or C-24 (1:10000). Tyrosine-phosphorylated Stat1 was detected with a specific antibody (1:1000; Upstate) after 10 min of fixation in methanol at –20°C. Endogenous Stat1 protein in HeLa cells was not detectable immunocytochemically at the high antibody dilutions used.
Dephosphorylation assays
In vitro dephosphorylation reactions were performed at 30°C (or at 37°C with the full-length TC45) in 20 μL of reaction buffer (25 mM Tris-HCl at pH 7.5, 0.5 mg/mL BSA, 10 mM DTT, 50 mM KCl, 5 mM EDTA, Complete protease inhibitors; Roche) with 0.5 nM Stat1 dimers, and 1.5 U of the truncated version of human TC45 (residues 1–352; Sigma) or 15 U of the full-length phosphatase. Full-length TC45 was expressed in bacteria (BL21pLysS) and purified by virtue of a C-terminal Strep-tag as recommended by the manufacturer (IBA). Duplex oligonucleotides (GAS: 5′-AAGTCGTTTCCCGGAAATAGAA GATTATTATCATTAT-3′ and its antisense; Mut: 5′-AAGTC GAGGTACAGGTAAAGAAGAACCTCGTTGTCAC-3′ and its antisense; 2×GAS: 5′-GTTTCCCCGAAATTGACGGATTTC CCCGAA AC-3′ and its antisense) were added where indicated. The DNA concentrations are given in the figures. Reactions were stopped after 60 min by boiling in SDS sample buffer and analyzed by Western blotting. Dephosphorylation of the control substrate p-nitrophenyl phosphate (pNPP) was not influenced by DNA (25 nM GAS; data not shown) For experimental details, see Sigma product T 1196 information. The method described there was used to determine the catalytic activity of full-length TC45. As expected, full-length and truncated TC45 dephosphorylated pNPP with identical rates (Hao et al. 1997). In vivo dephosphorylation assays were performed with U3A cells transiently expressing the indicated Stat1 variant proteins. Cells growing on 6-cm dishes were transferred to six-well plates 24 h posttransfection and cultivated for another 24 h, before stimulation with IFNγ for 30 min. Subsequently, the medium was replaced by medium without or with staurosporine, and the incubation was continued for the indicated times. Washed cells were lysed in lysis buffer A (without glycerol phosphate) and extracts representing similar cell numbers were analyzed by Western blotting.
Fluorescence microscopy and FRAP
Conventional fluorescence microscopy was as described (Begitt et al. 2000). FRAP(Axelrod et al. 1976) was done with a LSM510 inverted confocal laser scanning microscope (Carl Zeiss Jena) with a 100×/1.3 objective and an argon laser (λ = 488 nm) as excitation source. Fluorescence images were scanned as a time series with 250-msec intervals, and the relative fluorescence intensity was measured in a bleached region of interest (ROI, 10 μm2). Fluorescence intensity differences (Fu – Fb) between unbleached ROI (Fu) and adjoining bleached ROI (Fb) were calculated and used for an exponential curve fitting (prism program) to determine recovery times. Recovery was considered complete if Fu – Fb ≤ 0.01 Fu. Cells were treated with IFNγ for 30 min, at which point incubation was continued without or with vanadate/H2O2. FRAPwas performed before or 60 min after IFNγ addition.
Quantification of immunofluorescence intensities
Images were acquired by confocal microscopy as described (Meyer et al. 2002b). Stat1 immunofluorescence densities were determined both in the nucleus and in the cytoplasm of the median slice (x/y image). Correction for background fluorescence was done. For a number of cells (n), the ratio of (cytoplasmic fluorescence density)/(nuclear fluorescence density) was calculated and depicted in a bar diagram. The values are the mean and the standard deviation. The data were analyzed by Student's t-test (Fig. 1) or by ANOVA followed by the Tukey multiple comparison test (Fig. 2). Differences were considered statistically significant at p < 0.05.
Acknowledgments
We thank M. Kummerow, S. Meyer, M. van Rossum, and P. Striedelmeyer for expert technical assistance, and E. Krause for MALDI analysis. We are grateful to the following individuals for providing useful reagents: D. Besser and J. E. Darnell (Rockefeller University); M. Vigneron and C. Kedinger (Université de Strasbourg); G. Stark (Lerner Research Institute), and I. Kerr (ICRF). This work was supported by a Bio-Future grant from the Bundesministerium für Bildung und Forschung. U.V. is an EMBO-Young-Investigator.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.268003.
References
- Andrews, R.P., Ericksen, M.B., Cunningham, C.M., Daines, M.O., and Hershey, G.K. 2002. Analysis of the life cycle of stat6. Continuous cycling of Stat6 is required for IL-4 signaling. J. Biol. Chem. 277: 36563–36569. [DOI] [PubMed] [Google Scholar]
- Axelrod, D., Koppel, D.E., Schlessinger, J., Elson, E., and Webb, W.W. 1976. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16: 1055–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begitt, A., Meyer, T., van Rossum, M., and Vinkemeier, U. 2000. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc. Natl. Acad. Sci. 97: 10418–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser, D., Bromberg, J.F, Darnell Jr., J.E., and Hanafusa, H. 1999. A single amino acid substitution in the v-Eyk intracellular domain results in activation of Stat3 and enhances cellular transformation. Mol. Cell. Biol. 19: 1401–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild, A.H., Turkson, J., and Jove, R. 2002. Cytoplasmic transport of Stat3 by receptor-mediated endocytosis. EMBO J. 21: 3255–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe, J., Guschin, D., Rogers, N.C., Watling, D., Müller, M., Horn, F., Heinrich, P., Stark, G.R., and Kerr, I.M. 1996. JAKs, STATs and signal transduction in response to the interferons and other cytokines. Philos. Trans. R. Soc. Lond. B Biol. Sci. 351: 167–171. [DOI] [PubMed] [Google Scholar]
- Brivanlou, A.H. and Darnell Jr., J.E. 2002. Signal transduction and the control of gene expression. Science 295: 813–818. [DOI] [PubMed] [Google Scholar]
- Chen, X., Vinkemeier, U., Zhao, Y., Jeruzalmi, D., Darnell Jr., J.E., and Kuriyan, J. 1998. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93: 827–839. [DOI] [PubMed] [Google Scholar]
- Darnell Jr., J.E. 1997. STATs and gene regulation. Science 277: 1630–1635. [DOI] [PubMed] [Google Scholar]
- Darnell Jr., J.E., Kerr, I.M., and Stark, G.R. 1994. Jak–STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264: 1415–1421. [DOI] [PubMed] [Google Scholar]
- Decker, T., Kovarik, P., and Meinke, A. 1997. GAS elements: A few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res. 17: 121–134. [DOI] [PubMed] [Google Scholar]
- Fagerlund, R., Melén, K., Kinnunen, L., and Julkunen, I. 2002. Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and importin α 5. J. Biol. Chem. 277: 30072–30078. [DOI] [PubMed] [Google Scholar]
- Gordon, J.A. 1991. Use of vanadate as protein-phosphotyrosine phosphatase inhibitor. Methods Enzymol. 201: 477–482. [DOI] [PubMed] [Google Scholar]
- Greenlund, A.C., Morales, M.O., Viviano, B.L., Yan, H., Krolewski, J., and Schreiber, R.D. 1995. Stat recruitment by tyrosine-phosphorylated cytokine receptors: An ordered reversible affinity-driven process. Immunity 2: 677–687. [DOI] [PubMed] [Google Scholar]
- Hao, L., Tiganis, T., Tonks, N.K., and Charbonneau, H. 1997. The noncatalytic C-terminal segment of the T cell protein tyrosine phosphatase regulates activity via an intramolecular mechanism. J. Biol. Chem. 272: 29322–29329. [DOI] [PubMed] [Google Scholar]
- Haspel, R.L. and Darnell Jr., J.E. 1999. A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc. Natl. Acad. Sci. 96: 10188–10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haspel, R.L., Salditt-Georgieff, M., and Darnell Jr., J.E. 1996. The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends upon a protein tyrosine phosphatase. EMBO J. 15: 6262–6268. [PMC free article] [PubMed] [Google Scholar]
- Herrington, J., Rui, L., Luo, G., Yu-Lee, L.Y., and Carter-Su, C. 1999. A functional DNA binding domain is required for growth hormone-induced nuclear accumulation of Stat5B. J. Biol. Chem. 274: 5138–5145. [DOI] [PubMed] [Google Scholar]
- Horvath, C.M., Wen, Z., and Darnell Jr., J.E. 1995. A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes & Dev. 9: 984–994. [DOI] [PubMed] [Google Scholar]
- Ibarra-Sanchez, M.J., Simoncic, P.D., Nestel, F.R., Duplay, P., Lapp, W.S., and Tremblay, M.L. 2000. The T-cell protein tyrosine phosphatase. Semin. Immunol. 12: 379–386. [DOI] [PubMed] [Google Scholar]
- Ihle, J.N., Thierfelder, W., Teglund, S., Stravapodis, D., Wang, D., Feng, J., and Parganas, E. 1998. Signaling by the cytokine receptor superfamily. Ann. NY Acad. Sci. 865: 1–9. [DOI] [PubMed] [Google Scholar]
- Kimura, H. and Cook, P.R. 2001. Kinetics of core histones in living human cells: Little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153: 1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva, T., Bhattacharya, S., Braunstein, J., and Schindler, C.W. 2002. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 285: 1–24. [DOI] [PubMed] [Google Scholar]
- Kudo, N., Wolff, B., Sekimoto, T., Schreiner, E.P., Yoneda, Y., Yanagida, M., Horinouchi, S., and Yoshida, M. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242: 540–547. [DOI] [PubMed] [Google Scholar]
- Lee, C.K., Bluyssen, H.A., and Levy, D.E. 1997. Regulation of interferon-α responsiveness by the duration of Janus kinase activity. J. Biol. Chem. 272: 21872–21877. [DOI] [PubMed] [Google Scholar]
- Lillemeier, B.F., Köster, M., and Kerr, I.M. 2001. STAT1 from the cell membrane to the DNA. EMBO J. 20: 2508–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. and Riggs, A.D. 1975. The general affinity of lac repressor for E. coli DNA: Implications for gene regulation in procaryotes and eucaryotes. Cell 4: 107–111. [DOI] [PubMed] [Google Scholar]
- McBride, K.M., McDonald, C., and Reich, N.C. 2000. Nuclear export signal located within the DNA-binding domain of the STAT1 transcription factor. EMBO J. 19: 6196–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, K.M., Banninger, G., McDonald, C., and Reich, N.C. 2002. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-α. EMBO J. 21: 1754–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, J.G., Müller, W.G., Walker, D., Wolford, R., and Hager, G.L. 2000. The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science 287: 1262–1265. [DOI] [PubMed] [Google Scholar]
- Melén, K., Kinnunen, L., and Julkunen, I. 2001. Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J. Biol. Chem. 276: 16447–16455. [DOI] [PubMed] [Google Scholar]
- Meyer, T., Begitt, A., Lödige, I., van Rossum, M., and Vinkemeier, U. 2002a. Constitutive and IFN-γ-induced nuclear import of STAT1 proceed through independent pathways. EMBO J. 21: 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, T., Gavenis, K., and Vinkemeier, U. 2002b. Cell type-specific and tyrosine phosphorylation-independent nuclear presence of STAT1 and STAT3. Exp. Cell Res. 272: 45–55. [DOI] [PubMed] [Google Scholar]
- Milocco, L.H., Haslam, J.A., Rosen, J., and Seidel, H.M. 1999. Design of conditionally active STATs: Insights into STAT activation and gene regulatory function. Mol. Cell. Biol. 19: 2913–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowen, K. and David, M. 1998. Role of the STAT1-SH2 domain and STAT2 in the activation and nuclear translocation of STAT1. J. Biol. Chem. 273: 30073–30076. [DOI] [PubMed] [Google Scholar]
- Müller, M., Laxton, C., Briscoe, J., Schindler, C., Improta, T., Darnell Jr., J.E., Stark, G.R., and Kerr, I.M. 1993. Complementation of a mutant cell line: Central role of the 91 kDa polypeptide of ISGF3 in the interferon-α and -γ signal transduction pathways. EMBO J. 12: 4221–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto, T., Imamoto, N., Nakajima, K., Hirano, T., and Yoneda, Y. 1997. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 16: 7067–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai, K., Schindler, C., Prezioso, V.R., and Darnell Jr., J.E. 1992. Activation of transcription by IFN-γ: Tyrosine phosphorylation of a 91-kD DNA binding protein. Science 258: 1808–1812. [DOI] [PubMed] [Google Scholar]
- Shuai, K., Stark, G.R., Kerr, I.M., and Darnell Jr., J.E. 1993. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-γ. Science 261: 1744–1746. [DOI] [PubMed] [Google Scholar]
- Shuai, K., Horvath, C.M., Huang, L.H., Qureshi, S.A., Cowburn, D., and Darnell Jr., J.E. 1994. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell 76: 821–828. [DOI] [PubMed] [Google Scholar]
- Subramaniam, P.S., Larkin III, J., Mujtaba, M.G., Walter, M.R., and Johnson, H.M. 2000. The COOH-terminal nuclear localization sequence of interferon γ regulates STAT1 α nuclear translocation at an intracellular site. J. Cell Sci. 113: 2771–2781. [DOI] [PubMed] [Google Scholar]
- Tansey, W.P. 2001. Transcriptional activation: Risky business. Genes & Dev. 15: 1045–1050. [DOI] [PubMed] [Google Scholar]
- ten Hoeve, J., de Jesus Ibarra-Sanchez, M., Fu, Y., Zhu, W., Tremblay, M., David, M., and Shuai, K. 2002. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell. Biol. 22: 5662–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkemeier, U., Cohen, S.L., Moarefi, I., Chait, B.T., Kuriyan, J., and Darnell Jr., J.E. 1996. DNA binding of in vitro activated Stat1 α, Stat1 β and truncated Stat1: Interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 15: 5616–5626. [PMC free article] [PubMed] [Google Scholar]
- Wyszomierski, S.L., Yeh, J., and Rosen, J.M. 1999. Glucocorticoid receptor/signal transducer and activator of transcription 5 (STAT5) interactions enhance STAT5 activation by prolonging STAT5 DNA binding and tyrosine phosphorylation. Mol. Endocrinol. 13: 330–343. [DOI] [PubMed] [Google Scholar]
- Xu, X., Sun, Y.L., and Hoey, T. 1996. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science 273: 794–797. [DOI] [PubMed] [Google Scholar]
- Yang, E., Henriksen, M.A., Schaefer, O., Zakharova, N., and Darnell Jr., J.E. 2002. Dissociation time from DNA determines transcriptional function in a STAT1 linker mutant. J. Biol. Chem. 277: 13455–13462. [DOI] [PubMed] [Google Scholar]
- Zhu, W., Mustelin, T., and David M. 2002. Arginine methylation of STAT1 regulates its dephosphorylation by T cell protein tyrosine phosphatase. J. Biol. Chem. 277: 35787–35790. [DOI] [PubMed] [Google Scholar]