Abstract
The Drosophila insulin receptor (dInR) regulates cell growth and proliferation through the dPI3K/dAkt pathway, which is conserved in metazoan organisms. Here we report the identification and functional characterization of the Drosophila forkhead-related transcription factor dFOXO, a key component of the insulin signaling cascade. dFOXO is phosphorylated by dAkt upon insulin treatment, leading to cytoplasmic retention and inhibition of its transcriptional activity. Mutant dFOXO lacking dAkt phosphorylation sites no longer responds to insulin inhibition, remains in the nucleus, and is constitutively active. dFOXO activation in S2 cells induces growth arrest and activates two key players of the dInR/dPI3K/dAkt pathway: the translational regulator d4EBP and the dInR itself. Induction of d4EBP likely leads to growth inhibition by dFOXO, whereas activation of dInR provides a novel transcriptionally induced feedback control mechanism. Targeted expression of dFOXO in fly tissues regulates organ size by specifying cell number with no effect on cell size. Our results establish dFOXO as a key transcriptional regulator of the insulin pathway that modulates growth and proliferation.
Keywords: Insulin signaling, transcription factor, FOXO, Drosophila, growth, feedback
During the development of multicellular organisms, growth is tightly regulated by controlling cell number and cell size so that each organ reaches its appropriate dimensions in relation to the size of the organism. Many studies indicate that growth and proliferation are coordinated but distinct processes and that cells progress through the cell cycle only when sufficient mass, size, and macromolecular biosynthesis have been reached. (Hartwell et al. 1974; Johnston et al. 1977; Weigmann et al. 1997). Organism growth is controlled by coordinating both cell cycle progression and survival, which is modulated by nutrient availability, growth factors, and temperature. Growth factors can stimulate cell division and survival by activating the insulin receptor, which in turn acts through two main signal transduction cascades: the Ras/MAP kinase (Lee and McCubrey 2002) and the PI3K/Akt kinase pathways (Cantley 2002). Insulin-mediated activation of PI3K increases production of 3′-phosphorylated phosphoinositide lipids (PIP3) that serve as second messengers to recruit Akt to the plasma membrane (Datta et al. 1999). Once properly localized in the membrane, Akt becomes activated by phosphorylation and in turn phosphorylates a number of downstream targets that ultimately regulate cell growth. For example, Akt stimulates protein synthesis through activation of the target of rapamycin (TOR) kinase, which subsequently phosphorylates and inactivates the translational repressor eukaryotic initiation factor 4E-binding protein (4EBP; Gingras et al. 2001).
In addition to modulating translation, Akt regulates transcription through the forkhead-related FOXO family of transcription factors FOXO1, FOXO3a, and FOXO4 (Burgering and Kops 2002) by phosphorylating these proteins at three conserved serine/threonine residues. This leads to retention of FOXO transcription factors in the cytoplasm, thereby down-regulating RNA synthesis of specific target genes (Burgering and Kops 2002) that affect cell cycle progression (Kops et al. 1999; Alvarez et al. 2001) and apoptosis (Brunet et al. 1999; Dijkers et al. 2000) and modulate metabolic genes (Ayala et al. 1999; Durham et al. 1999; Guo et al. 1999; Hall et al. 2000; Nasrin et al. 2000; Schmoll et al. 2000; Nakae et al. 2001; Nadal et al. 2002). Thus, FOXO transcription factors play a critical role in regulating cell growth and survival.
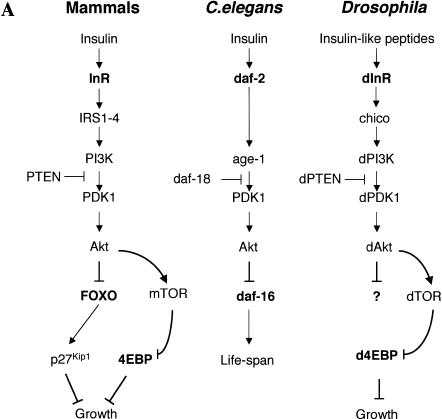
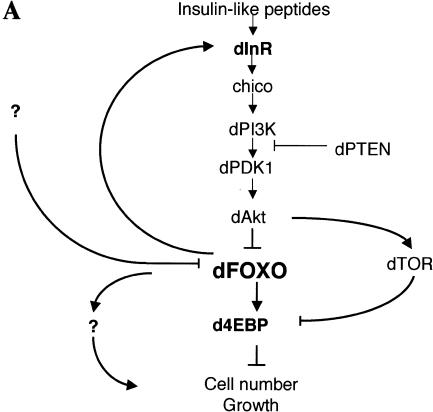
Recent genetic studies in Drosophila and Caenorhabditis elegans show that the InR/PI3K/Akt signaling pathway is largely conserved in metazoans (Fig. 1A). In invertebrates, this pathway apparently plays an essential role in regulating life span as well as body, organ, and cell size (Finch and Ruvkun 2001). C. elegans can enter the dauer state when food is limited; when conditions improve, growth is stimulated by activating the InR/PI3K/Akt signaling pathway. Worms with mutations in this pathway are small (Hekimi et al. 1998), their organs have fewer cells, and they live longer. Interestingly, mutations in the transcription factor DAF-16 suppress this phenotype (Lin et al. 1997; Ogg et al. 1997), suggesting that this key transcription factor is negatively regulated by the InR/PI3K/Akt pathway (Burgering and Kops 2002). In Drosophila, the dInR/dPI3K/dAkt pathway is also thought to regulate body size and life span (Fig. 1A). Flies with heteroallelic combinations of dInR mutations are reduced in size because of fewer and smaller cells (Fernandez et al. 1995; Chen et al. 1996; Brogiolo et al. 2001). Mutations in other components of the dInR/dPI3K/dAkt pathway produce similar phenotypes (Stocker and Hafen 2000; Johnston and Gallant 2002; Kozma and Thomas 2002). Although the identity and number of specific gene targets of the insulin signaling pathway in Drosophila remain unclear, one important downstream effector of insulin signaling appears to be the translational inhibitor d4EBP (Miron et al. 2001).
Figure 1.
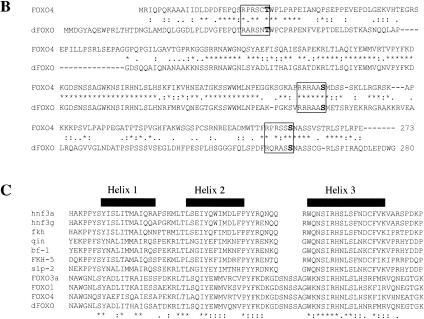
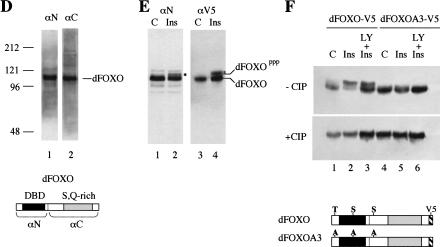
(A) The insulin receptor signaling pathway is conserved in mammals, C. elegans, and Drosophila. (B) Identification of a Drosophila homolog of FOXO/DAF-16. Sequence alignment showing the high degree of conservation displayed in the DNA-binding domains of FOXO4 and dFOXO. The three conserved Akt phosphorylation motifs are boxed, and the amino acids that can be phosphorylated are in boldface. Asterisks mark identical amino acids; colons mark conserved amino acid changes; dots indicate weakly conserved changes. (C) FOXO family members have a 5-amino-acid insertion between helices 2 and 3 in the DNA-binding domain, which is lacking in the rest of forkhead-related proteins. (D) Antibodies raised against recombinant C- and N-terminal regions of dFOXO recognize a band with similar mobility in Drosophila S2 cell extracts. A representation of dFOXO indicating the fragments used for antibody production is shown below. (E) Insulin treatment produces a slower-mobility form of dFOXO (marked with an asterisk) that is detected with endogenous (lanes 1,2) or overexpressed (lanes 3,4) dFOXO. (F) Overexpressed dFOXO is phosphorylated upon insulin treatment of S2 cells (lane 2). Pretreatment of samples with LY294002 before insulin treatment reduces the amount of dFOXO that is phosphorylated (lane 3). dFOXOA3 is not phosphorylated upon insulin treatment (lanes 4–6). The lower panel shows the same samples after phosphatase treatment. A scheme representing wild-type and mutant dFOXO is shown below.
Despite the importance of the dInR/dPI3K/dAkt pathway in regulating cell growth and proliferation in Drosophila, little is known about how signaling is controlled downstream of dAkt (Fig. 1A). In C. elegans and mammals, a critical member of this pathway downstream of Akt is the transcription factor DAF-16/FOXO, which counteracts insulin signaling. However, the Drosophila equivalent of DAF-16/FOXO has thus far not been described. In addition, the mechanisms, if any, that are used to provide feedback regulation of the InR pathway are unknown. Here, we describe the cloning and characterization of the Drosophila homolog of DAF-16/FOXO. We have investigated whether dFOXO is regulated by the dInR/dPI3K/dAkt pathway, searched for downstream target genes, and analyzed the biological role of dFOXO in Drosophila. Our results suggest that dFOXO is a key transcriptional regulator that controls both downstream target genes responsible for growth as well as upstream feedback targets in the insulin signaling pathway.
Results
Identification and cloning of dfoxo
Homology searches using FOXO4 or its DNA-binding domain revealed the presence of only one Drosophila gene with high similarity to FOXO transcription factors. We sequenced several cDNA clones, and the longest open reading frame encodes a protein of 613 amino acids with a calculated molecular mass of 67,412.12 D. We have designated this protein as dFOXO. The dfoxo gene contains 11 exons spanning 40 kb. The DNA-binding domain is located in the N-terminal region and has 45% identity with human FOXO4 (84% identity in the region that spans the three α-helices constituting the forkhead core domain; Fig. 1B). The homology with two other members of the FOXO family, FOXO1 and FOXO3a, is also significant in the DNA-binding domain. All four proteins share a characteristic five-amino-acid insertion (SNSSA) between α-helices 2 and 3 of the forkhead domain, which differentiates this subset from other forkhead-related family members (Fig. 1C). Importantly, the three residues phosphorylated by Akt in FOXO4 are conserved in dFOXO: T28, S193, and S258 in FOXO4 correspond to T44, S190, and S259 in dFOXO (Fig. 1B, boldface residues). The amino acids that define the motif recognized by Akt (RXRXXS/T) are also conserved (Fig. 1B, boxed residues). dFOXO has a C-terminal region with a high Ser and Gln content (11.7% and 13.4%, respectively), commonly found in transcription activation domains.
dFOXO is phosphorylated upon insulin treatment in S2 cells
Rabbit polyclonal antibodies raised against either the N- or C-terminal region of dFOXO recognize a single major polypeptide in Drosophila embryo extracts with an apparent molecular mass of 113 kD as determined by SDS-PAGE (Fig. 1D). To confirm that this protein corresponds to dFOXO, we expressed a V5-epitope-tagged version of dFOXO under control of the metallothionein promoter. When this Cu2+-inducible construct was transfected into Schneider line 2 (S2) cells, both N- and C-terminal antibodies as well as the V5 antibody specifically recognized a major polypeptide with the same mobility (113 kD) that was found in embryo extracts (data not shown). We have therefore designated this 113-kD protein as dFOXO. Western blot analysis confirmed that untransfected S2 cells express endogenous dFOXO at a level similar to that found in embryo extracts (data not shown).
If the dFOXO protein we have identified is a bona fide ortholog of mammalian FOXO, insulin should regulate its activity by phosphorylation via a cascade involving Akt (Brunet et al. 1999; Kops et al. 1999). Also, phosphorylation of the three specific serine/threonine residues should sequester this transcription factor in the cytoplasm. To test these properties, S2 cells were grown with or without insulin, and endogenous dFOXO was detected by Western blot analysis. A single band migrating with molecular mass 113 kD was recognized by both N- and C-terminal dFOXO antibodies in the untreated samples (Fig. 1E, lane 1; data not shown). In contrast, the insulin-treated sample revealed the presence of two bands, one migrating with the mobility of 113 kD and a second band with slower mobility (Fig. 1E, lane 2, marked with an asterisk). A similar insulin-induced shift was obtained with transfected dFOXO-V5 expressed in S2 cells and detected with a V5 antibody (Fig. 1E, cf. lanes 4 and 3). These results suggested that the slower-migrating band may correspond to a phosphorylated form of dFOXO.
To establish that the slower-migrating form of dFOXO induced by insulin treatment is indeed caused by dAkt-catalyzed phosphorylation, we constructed a mutant form of dFOXO in which all three putative dAkt phosphorylation sites (T44, S190, and S259) were mutated to alanine (dFOXOA3). Both wild-type (dFOXO-V5) and mutant (dFOXOA3-V5) proteins were expressed in S2 cells. After transient expression, the cells were subjected to three different treatments in parallel: insulin (Fig. 1F, lanes 2,5); pretreatment with LY294002 (a specific inhibitor of PI3K that counteracts the effects of insulin) followed by insulin treatment (Fig. 1F, lanes 3,6); or no treatment control (Fig. 1F, lanes 1,4). Extracts derived from cells treated with insulin contained the slower-migrating form of wild-type dFOXO when compared with control cells (Fig. 1F, upper panel, cf. lanes 2 and 1). Pretreatment with the PI3K inhibitor LY294002 reduced the amount of the slower-migrating form of dFOXO (Fig. 1F, lane 3). In contrast, no slower-migrating species was observed for the triple alanine mutant (dFOXOA3) when comparing control, insulin-treated, and LY294002 + insulin-treated samples (Fig. 1F, lanes 4–6). To further confirm that the slower-migrating form of dFOXO was caused by phosphorylation, cell extracts were incubated with calf intestinal phosphatase (CIP). Western blot analysis (Fig. 1F, lower panel) showed that the slower-migrating form of dFOXO was quantitatively converted to the 113-kD form after CIP treatment. Together, these results indicate that dFOXO is phosphorylated by insulin treatment and that this phosphorylation depends on the presence of the dAkt consensus residues T44, S190, and S259.
Insulin induces cytoplasmic localization of dFOXO
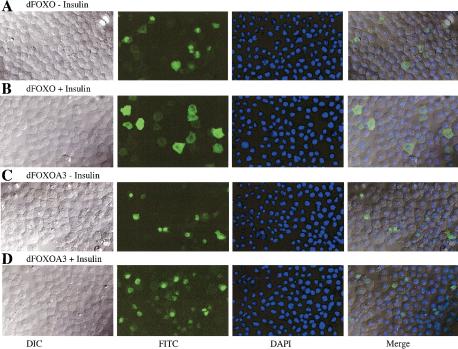
To test how dFOXO subcellular localization is affected by insulin-mediated phosphorylation, S2 cells expressing either wild-type dFOXO or mutant dFOXOA3 were incubated for 48 h in the absence of serum. Then insulin was added, and localization of transfected dFOXO was determined by confocal microscopy after staining with the V5 antibody. When S2 cells are incubated in the absence of serum and insulin, both dFOXO and dFOXOA3 are found predominantly in the nucleus (Fig. 2A,C). After insulin treatment, dFOXO is localized in the cytoplasm (Fig. 2B). In contrast, mutant dFOXOA3 remains nuclear even after insulin treatment (Fig. 2D). This result is consistent with the idea that subcellular localization of dFOXO is regulated by insulin.
Figure 2.
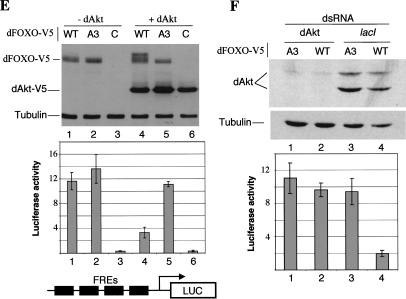
(A–D) Insulin regulates subcellular localization of dFOXO. S2 cells overexpressing dFOXO or dFOXOA3 were grown in the absence (A,C) or presence (B,D) of insulin. (E) Akt phosphorylates and inhibits dFOXO activity. (Top) S2 cells grown in the absence of serum/insulin were transfected with dFOXO [wild-type (WT) lanes 1,4], dFOXOA3 (A3, lanes 2,5), or with empty vector (C, lanes 3,6). Myr-dAkt-V5 was cotransfected in lanes 4–6. (Bottom) Luciferase assays performed with the samples from above and measured with a reporter containing four FOXO4 recognition elements. (F) Insulin inhibits dFOXO through dAkt. Cells were transfected with dFOXO (lanes 2,4) or dFOXOA3 (lanes 1,3) and dsRNA against dAkt (lanes 1,2) or lacI (lanes 3,4) was added. (Bottom) Luciferase activity was measured with the same reporter as in E.
dAkt phosphorylates dFOXO and inhibits its activity
We next asked whether dFOXO phosphorylation is regulated through the dPI3K/dAkt pathway. We used a constitutively active form of dAkt in which a myristoylation signal has been fused to the N terminus of dAkt (Verdu et al. 1999). Myr-dAkt tagged with V5 epitope was cotransfected in S2 cells grown in the absence of serum and insulin with either dFOXO or dFOXOA3, and the phosphorylation state of both proteins was analyzed by Western blot analysis. In the absence of dAkt, both dFOXO and dFOXOA3 remain unphosphorylated (Fig. 2E, upper panel, lanes 1,2). When Myr-dAkt was present in the cells, dFOXO but not dFOXOA3 becomes phosphorylated even in the absence of insulin (Fig. 2E, upper panel, lanes 4,5). This result indicates that Myr-dAkt can phosphorylate dFOXO in S2 cells. To assess the effect of dFOXO phosphorylation by Myr-dAkt, we made use of a reporter construct containing four tandem FOXO4-binding sites upstream of the alcohol dehydrogenase distal core promoter driving the luciferase gene (pGL4xFRE). In the absence of Myr-dAkt, cells cotransfected with wild-type or mutant dFOXO constructs incubated without serum displayed comparable luciferase activity (Fig. 2E, lower panel, lanes 1,2). In contrast, when Myr-dAkt is present, cells cotransfected with wild-type dFOXO displayed luciferase activity that was reduced by more than 65% (Fig. 2E, lower panel, lane 4), whereas activity of the mutant dFOXOA3 remained essentially unchanged (Fig. 2E, lower panel, cf. lanes 2 and 5).
The results presented above suggest that insulin induces dFOXO phosphorylation through dAkt, which leads to cytoplasmic localization and transcriptional inactivation of dFOXO. To further confirm that insulin inhibits dFOXO activity through dAkt, we performed RNAi experiments. S2 cells transfected with either dFOXO or dFOXOA3 and cotransfected with the luciferase reporter pGL4xFRE were grown in the presence of insulin and treated with dsRNA directed against dAkt. As a control, dsRNA against lactose repressor (lacI) was used. As expected, dFOXO activity is not inhibited by insulin when cells are depleted of dAkt by dsRNA treatment, but it is inhibited in the lacI control (Fig. 2F, cf. lanes 2 and 4). These results confirm that dAkt mediates insulin inhibition of dFOXO.
Expression of constitutively active dFOXO arrests cell growth
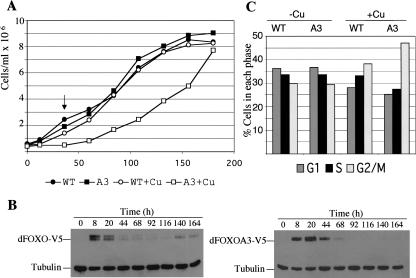
Studies with several components of the dInR/dPI3K/dAkt pathway have shown that insulin promotes growth and proliferation by activating dPI3K and dAkt (Kozma and Thomas 2002). We have shown that dFOXO activity is inhibited by dAkt upon insulin treatment (Fig. 2E). We therefore wanted to know whether induction of dFOXO would inhibit growth in S2 cells. To do so, we cultured S2 cells stably transfected with either dFOXO or mutant dFOXOA3 in the presence of serum and insulin for 48 h. Then exogenous dFOXO and dFOXOA3 expression was induced with CuSO4 for a period of 8 h. Subsequently, Cu2+ was removed, and cells were then allowed to divide for 1 wk with samples taken every 12 h to measure cell numbers. Cells stably transfected with the wild-type protein proliferated at the same rate irrespective of dFOXO induction (Fig. 3A, •, ○). In contrast, cells stably transfected with the mutant protein, dFOXOA3, displayed a significantly slower growth rate when compared with the same cells grown without dFOXOA3 induction (Fig. 3A, ▪, □). Because insulin was present throughout these experiments (inactivating dFOXO but not dFOXOA3), our findings suggest that the constitutively active dFOXOA3 can induce cell arrest. Importantly, cells expressing dFOXOA3 arrest growth during the first 44 h after induction when dFOXOA3 is present. After 44 h, when dFOXOA3 has apparently been turned over (Fig. 3B, lower panel, 68–164 h), cells recover and start dividing normally again (Fig. 3A, □, 68–164 h). FACS analysis of samples taken during the different time points indicates that S2 cells arrest their growth at G2/M (Fig. 3C; data not shown). These results indicate that activation of dFOXO can induce cell cycle arrest and this effect is mitigated by insulin.
Figure 3.
dFOXO arrests cell growth. (A) S2 cells stably transfected with either wild-type dFOXO (•, ○) or A3 mutant (▪, □) were grown in the presence of insulin. To induce dFOXO expression, an 8-h pulse of Cu2+ was performed (○, □). Only cells expressing dFOXOA3 stop growing during the first 44 h. (B) Western blot of cell extracts obtained from cells induced with Cu2+ in A. (C) Frequencies for G1, S, or G2/M as assayed by FACS for cells after 36 h of Cu2+ addition (arrow in A).
dFOXO activates dInR and d4EBP transcription
The results presented above suggest that dFOXO regulates cell cycle arrest possibly by transcriptionally activating genes implicated in cell division or in cell growth. As an initial attempt to identify target genes of dFOXO, DNA microarrays were used to assess gene expression profiles in S2 cells stably transfected with mutant dFOXO and grown in the presence of insulin. Cells expressing wild-type dFOXO or untransfected S2 cells subjected to the same treatment were assayed as controls.
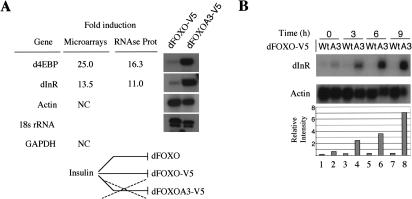
We found that 277 genes were up-regulated in dFOXOA3-expressing cells when compared with dFOXO-expressing cells or untransfected S2 cells. Interestingly, two genes that were consistently and specifically up-regulated in these conditions were the dInR gene (13.5-fold) and the d4EBP gene (25-fold; Fig. 4A). Both genes have been implicated in the regulation of cell growth by insulin (Fernandez et al. 1995; Miron et al. 2001). As expected, experimental control genes such as actin or GAPDH remained unchanged under these conditions (Fig. 4A). To confirm that dInR and d4EBP are bona fide transcriptional targets of dFOXO, we performed the same experiment described above but in the presence of cycloheximide to inhibit translation. As expected, both dInR and d4EBP continued to be transcriptionally activated (2.5- and 3.1-fold, respectively) by dFOXOA3 but not dFOXO in the insulin-repressed state. This result suggests that dFOXO, when released from control by the insulin/dAkt cascade, is involved in transcription from the dInR and d4EBP promoters.
Figure 4.
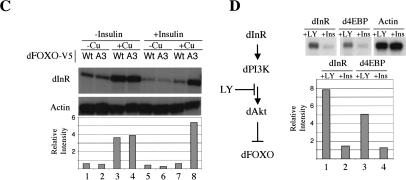
dInR and d4EBP are up-regulated by dFOXO. (A) DNA microarrays detected dInR and d4EBP as putative targets for dFOXO. Actin and GAPDH controls remained unchanged. RNase protection confirmed microarray data. (B) RNase protection shows a rapid induction of dInR messenger by dFOXOA3. (C) Cu2+ or insulin treatments per se do not induce transcription of dInR. In the absence of insulin, both wild-type and A3 mutant induce transcription of dInR. In the presence of insulin, only A3 mutant up-regulates this mRNA. (D) The PI3K inhibitor LY294002 induces transcription of dInR and d4EBP, suggesting that endogenous dFOXO is responsible for this effect. For all the panels, the graphic below represents the quantification of data performed on the RNase protection experiment shown above.
To confirm these microarray results and to independently quantitate the increase in mRNA transcription, we performed RNase protection assays with mRNAs extracted from cells stably transfected with either dFOXO or dFOXOA3. Indeed, dFOXOA3 stimulated transcription of d4EBP and dInR by 16.3- and 11-fold, respectively. A time-course experiment (Fig. 4B) confirmed that dInR mRNA increased rapidly upon dFOXOA3 expression: 3 h after CuSO4 addition, there is already an 8-fold increase, reaching 20-fold after 9 h of CuSO4 induction. Similar results were obtained for d4EBP (data not shown).
To exclude the possibility that treatment with insulin and/or CuSO4 per se causes the effects seen on transcription of dInR and d4EBP, cells stably transfected with dFOXO or dFOXOA3 were grown in the presence of either insulin or CuSO4, or with both insulin and CuSO4 (Fig. 4C). In the absence of insulin and presence of CuSO4, both dFOXO and dFOXOA3 activate dInR and d4EBP transcription (Fig. 4C, lanes 3,4; data not shown). However, in the presence of insulin and CuSO4, only the constitutively active dFOXOA3 induced transcription of these two target genes (Fig. 4C, lanes 7,8; data not shown). In the absence of CuSO4, no activation is seen (Fig. 4C, lanes 1,2,5,6). These results rule out the possibility that insulin/Cu2+ treatment by itself activated transcription of these genes. Instead, our experiments suggest that dFOXO expression specifically activates both dInR and d4EBP transcription, thus unmasking an important feedback control mechanism in this pathway involving dFOXO and dInR.
Having obtained evidence that exogenously transfected dFOXO responds to insulin and regulates both the downstream target gene d4EBP and the feedback control target dInR, we next wanted to know if endogenous dFOXO would also activate transcription of these genes. We used the PI3K inhibitor LY294002 to activate endogenous dFOXO or insulin to deactivate it. S2 cells grown in the absence of serum for 48 h were treated either with LY294002 or insulin. Total RNA was extracted and RNase protection was performed to detect dInR and d4EBP mRNAs. Both mRNA levels are significantly increased after LY294002 treatment (5.3-fold for dInR and 4-fold for d4EBP) when compared with insulin treatment (Fig. 4D). This result provides further evidence indicating that the dPI3K–dAkt pathway regulates dInR and d4EBP transcription via dFOXO.
dFOXO binds to both the d4EBP and the dInR promoters and activates their transcription
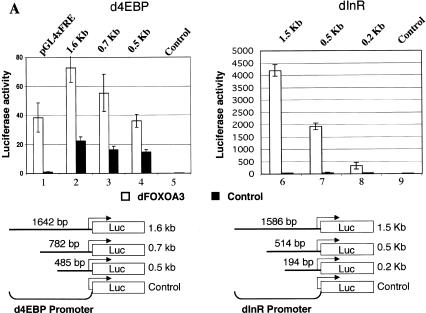
We next wanted to determine whether dFOXO directly binds to the promoters of d4EBP and dInR. To identify the DNA region recognized by dFOXO in these two promoters, we inserted a 1708-bp fragment of the d4EBP promoter and a 1562-bp fragment of the dInR promoter into a luciferase reporter vector. When transfected into S2 cells, these fragments responded to dFOXO activation (3-fold for d4EBP, >200-fold for dInR; Fig. 5A, lanes 2,6). A series of deletions lacking upstream sequences still responded to dFOXO activation, albeit more weakly (Fig. 5A, lanes 3,4,7,8), suggesting that dFOXO can bind the DNA in a region close to the start of transcription (485 bp for the d4EBP promoter and 194 bp for the dInR promoter). In contrast, dFOXO completely fails to activate a reporter construct in which upstream activating sequences (UAS) for the transcription factor GAL4 are fused to the luciferase gene (data not shown), confirming that transcription activation is specific for both d4EBP and dInR promoters.
Figure 5.
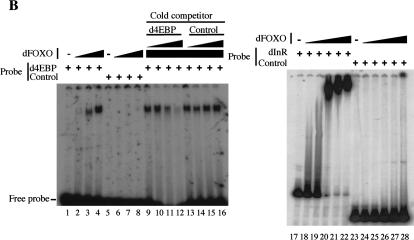
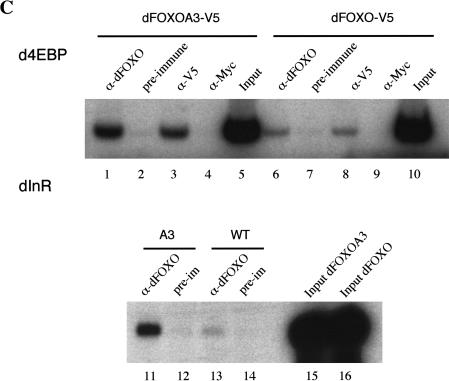
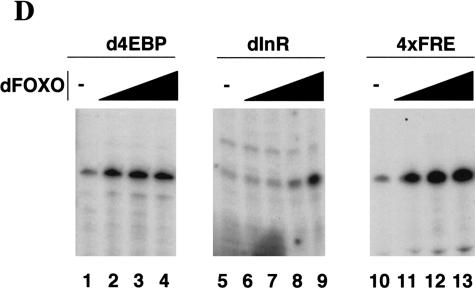
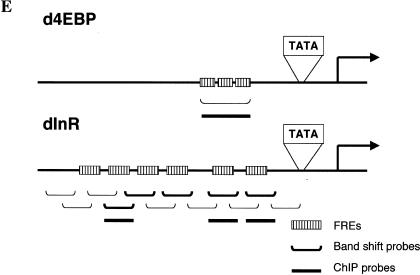
dFOXO directly activates transcription of dInR and d4EBP. (A) Luciferase assays showed activation of d4EBP and dInR promoters in S2 cells after cotransfection with dFOXOA3 (white bars). (Black bars) Empty vector. (B) Band shift performed with d4EBP and dInR promoters showed that recombinant dFOXO binds specifically to these promoters. (C) dFOXO binds specifically to d4EBP and dInR promoters in vivo. ChIP of cross-linked extracts of S2 cells expressing dFOXO or dFOXOA3 grown in prescence of insulin. (D) Recombinant dFOXO activates transcription of d4EBP (lanes 1–4) and dInR (lanes 5–9) promoters and of a synthetic promoter bearing 4 FREs (lanes 10–13) in vitro. (E) Schematic representation of the d4EBP and dInR promoters showing the putative FREs (striped boxes) located upstream of the transcription start sites (indicated by arrows). The probes used in band shifts are indicated below as horizontal brackets, and those probes bound by dFOXO are shown as thick lines. Thick horizontal bars indicate the DNA regions bound in vivo by dFOXO as analyzed by ChIP.
Interestingly, 125 bp upstream of the transcription start site of the d4EBP promoter there are three tandem copies of a putative FOXO4 recognition element (FRE; Fig. 5E). These elements are reminiscent of the ones present in the human glucose-6-phosphatase promoter, previously shown to bind FOXO4 (Yang et al. 2002). This was reassuring because dFOXO and FOXO4 share 85% identity in the core of the forkhead DNA-binding domain (Fig. 1B). Similarly, several putative FRE sequences appear in the dInR promoter in the region comprising nucleotides –1434 to –70 (Fig. 5E).
To determine whether dFOXO binds these putative FREs, we performed band shift experiments with a 113-bp DNA probe encompassing the d4EBP FRE motifs and with 12 separate DNA probes (ranging from 100 to 150 bp) spanning a region of 1.4 kb from the dInR promoter (Fig. 5E). Purified recombinant dFOXO expressed in Escherichia coli efficiently binds the 113-bp FRE-containing fragment from the d4EBP promoter (Fig. 5B, lanes 1–4) compared with control DNA fragments (Fig. 5B, lanes 5–8). Furthermore, dFOXO binding to the d4EBP promoter fragment can be efficiently competed with an unlabeled 113-bp d4EBP promoter fragment (Fig. 5B, lanes 9–12) but not with nonspecific DNA (Fig. 5B, lanes 13–16). Similarly, purified recombinant dFOXO binds efficiently to 5 out of 12 of the DNA fragments located within the dInR promoter (Fig. 5B, lanes 17–22; data not shown). As expected, each of the five DNA fragments bound by dFOXO contains putative FREs. Thus, dFOXO can specifically bind to both promoters in vitro. To determine whether dFOXO also binds these same DNA regions in vivo, we performed chromatin immunoprecipitation (ChIP) experiments with S2 cells expressing either dFOXO or dFOXOA3. Cells were incubated with serum, and dFOXO expression was induced with the addition of CuSO4. After 6 h, cells were cross-linked with formaldehyde, and extracts were prepared and immunoprecipitated. After reversal of cross-links, DNA was recovered, and PCR was performed with primers encompassing regions containing putative FREs in both promoters (Fig. 5E). Our results indicate that dFOXO can directly bind to both the d4EBP and dInR promoters in vivo (Fig. 5C, lanes 1,3,11; data not shown). In contrast, no specific d4EBP or dInR promoter fragments were precipitated using preimmune serum or an unrelated antibody (Fig. 5C, lanes 2,4,12). Control experiments performed with probes for the U6 snRNA promoter showed that dFOXO binding to dInR and d4EBP promoters was specific (data not shown). These results establish that dFOXO can specifically bind the d4EBP and dInR promoters both in vitro and in vivo.
To demonstrate that dFOXO can directly activate transcription of these promoters in vitro, we used the constructs that contain 485 bp of the d4EBP promoter region and 514 bp of the dInR promoter region, respectively. Addition of purified recombinant dFOXO to in vitro reactions activates transcription of these promoters by at least 3-fold (d4EBP) and 5.5-fold (dInR; Fig. 5D, lanes 1,2,5,9), which is comparable to the activation observed in vivo (Fig. 5A). Under our in vitro transcription conditions, activation of the d4EBP promoter by dFOXO becomes rapidly saturated with increasing amounts of dFOXO (Fig. 5D, lanes 2–4). As expected, dFOXO also activates (up to sixfold; Fig. 5D, lanes 10–13) a synthetic promoter bearing four FOXO4-binding sites placed upstream of the alcohol dehydrogenase distal promoter. Together these results show that transcription of d4EBP and dInR can be directly activated by dFOXO in vitro.
dFOXO regulates organ size in Drosophila by regulating cell number
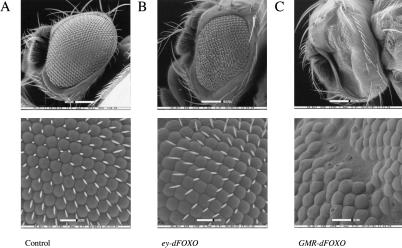
Taken together, our data strongly suggest that dFOXO is a key regulator of the insulin signaling pathway. As expected, dFOXO activity is inhibited via the dInR/dPI3K/dAkt pathway. We have also found that dFOXO activates transcription of a major downstream target (d4EBP) of this pathway. In Drosophila, this pathway has been linked to cell size and cell number regulation. We therefore wondered whether dFOXO expression in vivo would affect these same parameters. To address this point, we generated transgenic flies that overexpressed dFOXO by using the UAS/GAL4 system (Brand and Perrimon 1993). When dFOXO expression was directed to the eye by using eyeless-GAL4 (Hazelett et al. 1998), eyes became significantly smaller than wild type (Fig. 6A,B). Interestingly, the reduction in eye size (35%) was caused by a reduction in cell number (673 ± 24 ommatidia in control eyes vs. 438 ± 50 ommatidia in ey-GAL4/UAS-dFOXO), but no significant change in cell size was observed (85.8 ± 5.9 area units/ommatidia in control eyes vs. 91.2 ± 4.8 area units/ommatidia in ey-GAL4/UAS-dFOXO). When we used GMR-GAL4, which directs expression in cells posterior to the morphogenetic furrow (Hay et al. 1994), to drive dFOXO expression, we observed a more severe phenotype. Many ommatidia were lost, and the remaining ommatidia lacked bristles and appeared disorganized, altering the general structure of the eye (Fig. 6C). These results suggest that dFOXO overexpression can severely affect normal development of the eye.
Figure 6.
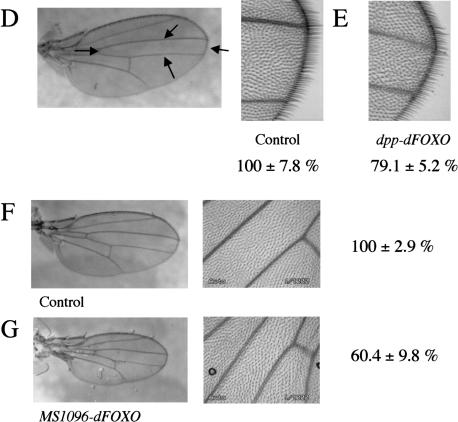
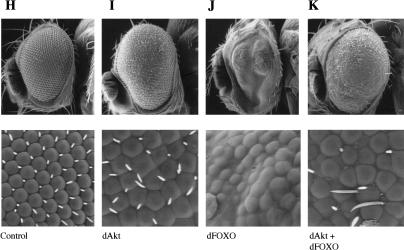
dFOXO controls growth by affecting cell number. (A,B) Expression of dFOXO in fly eyes reduces eye size without affecting ommatidia size. (A) ey-Gal4/+. (B) ey-Gal4/UAS-dFOXO. (C) Overexpression of dFOXO in eye discs before the morphogenetic furrow (GMR-Gal4/UAS-dFOXO) causes a striking reduction of the eye size with severe ommatidia loss. (D–G) Overexpression of dFOXO in fly wings causes reduction in wing size by only affecting cell number. Values are the total average area in percent. (D) dpp-Gal4/+ (100% ± 7.8%). Arrows indicate the area affected by the driver. (E) dpp-Gal4/UAS-dFOXO (79.1% ± 5.2%). (F) MS1096-Gal4/+ (100% ± 2.9%). (G) MS1096-Gal4/UAS-dFOXO (60.4% ± 9.8%). (H–K) dAkt partially rescues the phenotype produced by dFOXO expression. (H) GMR-Gal4/+ +/+. (I) GMR-Gal4/+ UAS-dAkt/+. (J) GMR-Gal4/UAS-dFOXO +/+. (K) GMR-Gal4/UAS-dFOXO UAS-dAkt/+.
We next tested the effect of dFOXO overexpression in an organ other than the eye. We used dpp-GAL4 (Staehling-Hampton and Hoffmann 1994) to direct expression of dFOXO in the wing region encompassed by the third and fourth longitudinal veins (Fig. 6D). As expected, dFOXO overexpression resulted in a significant reduction of compartment size (20%; Fig. 6D,E). Again, we confirmed that this reduction in size was caused by a reduction in cell number but not in cell size (71 ± 7.5 cells/area unit in the control vs. 71 ± 5.5 cells/area unit in dpp-GAL4/UAS-dFOXO). Ectopic expression of dFOXO in the wing using MS1096-GAL4 (Capdevila and Guerrero 1994) produced a more striking reduction in wing size (40%; Fig. 6F,G), again because of loss of cell number with no significant variation in cell size (70.6 ± 6.5 cells/area unit in the control vs. 74.8 ± 11.8 cells/area unit in MS1096-GAL4/UAS-dFOXO).
We have shown that dAkt can phosphorylate and inactivate dFOXO in S2 cells (Fig. 2E). We wanted to know whether dAkt also inhibits the phenotypic effects of dFOXO in flies. We obtained flies expressing dFOXO, dAkt, or both (Fig. 6H–K). Indeed, dAkt expression partially rescues the eye phenotype observed with dFOXO (Fig. 6, cf. J and K), showing that they interact genetically. These results provide further evidence of both proteins acting in the same pathway.
Discussion
Feedback regulation of the insulin signaling pathway by dFOXO
Although a good deal has been learned about insulin-regulated signal transduction pathways in mammals and other metazoans, many of the regulatory steps and mechanisms remain unclear. By studying the InR signaling pathway in Drosophila and focusing our attention on a key downstream component, the transcriptional activator dFOXO, we have uncovered novel aspects of this important signaling cascade. First, we describe the cloning and functional characterization of dFOXO, the Drosophila homolog of DAF-16/FOXO, a transcriptional regulator of the dInR/dPI3K/dAkt pathway. Surprisingly, dFOXO transcriptionally activates downstream as well as upstream targets of this signaling cascade, providing the first evidence for a transcriptional feedback mechanism in the InR pathway that regulates cell growth and proliferation. Furthermore, we have found that dFOXO modulates the dInR signaling pathway by transcriptionally activating two key elements of this signaling cascade: the downstream effector d4EBP and dInR itself (Fig. 7A). Activation of dInR provides an interesting way to modulate the dInR/dPI3K/dAkt pathway via a feedback regulatory loop that may have important implications during development.
Figure 7.
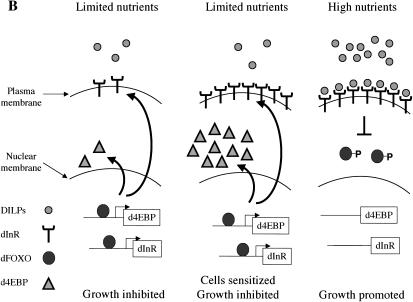
(A) dFOXO is a key element of insulin signaling in Drosophila. The insulin receptor inactivates dFOXO through dPI3K/dAkt. Activation of d4EBP may explain growth inhibition by dFOXO, whereas activation of dInR may provide a novel transcriptionally induced feedback control mechanism for the pathway. (B) Model to explain regulation of growth by a feedback mechanism involving dInR and dFOXO.
It has been shown that regulation of growth during development depends on the availability of nutrients (Britton et al. 2002) and that food limitation decreases the Drosophila insulin-like peptide (DILP) levels (Ikeya et al. 2002; Rulifson et al. 2002). An activated dInR pathway promotes growth, whereas mutations in this pathway impair normal development. For instance, flies mutant for chico, which encodes the Drosophila homolog of IRS1-4, are developmentally delayed, have severely reduced body size and increased fat accumulation (Bohni et al. 1999). Likewise, mutations in several other components of the dInR pathway produce related phenotypes. Our finding that dFOXO is involved with feedback activation of dInR provides a novel mechanism for the cells to regulate growth by responding rapidly to changes in nutrient conditions. When nutrients are abundant, elevated levels of DILPs are secreted to activate the dInR pathway, and the resulting downstream signaling promotes growth, in part by inhibiting dFOXO. These favorable nutrient conditions would allow growth and development (Fig. 7B). However, in a situation in which nutrients are limiting, DILPs would be secreted at a reduced rate, the dInR pathway would not be activated, and dFOXO would remain dephosphorylated, nuclear, and active. As a result, growth would be inhibited, in part by dFOXO activation of d4EBP. On the other hand, because dFOXO would be active when nutrients are limited, dInR becomes up-regulated and primed to signal when triggered by changes in DILPs levels. In this way, when nutrient conditions change, the cells would be highly sensitized and able to respond rapidly by turning on the mechanisms that stimulate growth, including shutting down dFOXO via dAkt phosphorylation. In addition, dFOXO-mediated dInR transcriptional activation presents a highly sensitive way to regulate the dInR/dPI3K/dAkt signaling pathway in response to subtle developmental cues that modulate DILP levels. High levels of DILPs would activate dInR, which would lead to the inhibition of its own transcription, turning off the pathway. Conversely, reduced levels of DILPs would activate dInR transcription. This would sensitize the pathway and provide a mechanism to amplify growth factor signals by allowing detection of lower levels of DILPs. Once this pathway is activated, feedback regulation via dFOXO would dampen dInR expression and signaling. Growth and development through the dInR pathway could thus be exquisitely balanced and regulated. Interestingly, overexpression of dFOXO under the control of strong promoters (GMR, tubulin) in flies results in severe morphological defects (Fig. 6C; data not shown). These results suggest that abnormally high levels of dFOXO may produce growth arrest in developing organs. Surprisingly, flies with loss-of-function mutations in dFOXO appear to develop normally (M. Jünger and E. Hafen, pers. comm.), indicating that dFOXO is not essential during fly development. These results suggest the existence of additional mechanisms that modulate insulin signaling and underscore the complexity of such developmental pathways.
dAkt inhibits d4EBP activity and down-regulates its transcription via dFOXO
In mammals, it has been reported that Akt promotes protein synthesis through TOR-mediated phosphorylation and subsequent inactivation of the translational inhibitor 4EBP. Hypophosphorylated 4EBP interacts strongly with eIF4E, providing a mechanism for Akt to regulate 4EBP via TOR (Gingras et al. 1998, 1999). In flies, a similar mechanism has been reported (Miron et al. 2001). In contrast, the role of dFOXO in transcriptionally modulating d4EBP has not been previously described. Our finding that dFOXO directly regulates d4EBP transcription provides an alternative and parallel mechanism for dAkt to inhibit d4EBP function. Under conditions in which the insulin pathway is active, dAkt sequesters dFOXO in the cytoplasm, and d4EBP transcription is turned off. When the dInR/dPI3K/dAkt pathway is inactive, dFOXO is free to stimulate transcription of d4EBP and inhibit protein synthesis. It has been previously reported that overexpression of 4EBP slows growth in mammalian cells (Rousseau et al. 1996), and now we have shown that dFOXO overexpression leads to growth arrest in Drosophila S2 cells. It is therefore likely that up-regulation of d4EBP by dFOXO contributes to the observed growth arrest. However, we cannot rule out additional mechanisms (i.e., induction of apoptosis by dFOXO), which could also contribute to the observed phenotype.
dFOXO regulates cell number without affecting cell size
It has been well documented that the dInR/dPI3K/dAkt pathway regulates cell number and cell size in Drosophila. However, the precise mechanisms by which the insulin pathway controls these parameters remain unknown. Mutations in some members of this pathway affect cell size as well as cell number, whereas mutations in other members appear to affect only cell size. For example, mutations in both dInR and dPI3K produce smaller flies with reduced numbers of cells and smaller cell size (Leevers et al. 1996; Bohni et al. 1999). Mutations in the negative regulator dPTEN produce bigger cells and increased proliferation (Gao et al. 2000). In contrast, mutations in dAkt produce smaller organs without affecting cell number, only cell size (Verdu et al. 1999). Overexpression of mutant d4EBP with increased binding affinity for eIF4E produces flies with smaller and fewer cells (Miron et al. 2001). Thus, until now, none of the components of the dInR pathway has been found to regulate only cell number without influencing cell size. It was therefore intriguing to find that overexpression of dFOXO produces a reduction in cell number without any measurable effect on cell size. This is reminiscent of the transcription factor c-myc, which in mammals regulates cell number without altering cell size (Trumpp et al. 2001) but in Drosophila affects both cell size and number (Johnston and Gallant 2002). Taken together, these results reveal the species specific complexity of the mechanisms that regulate cell growth and proliferation. Indeed, our results suggest that the InR/PI3K/Akt pathway is far from being a simple linear cascade. Instead, dAkt appears to regulate numerous targets, each one with its own set of downstream effectors. In addition, it is conceivable that dFOXO may be regulated by kinases other than dAkt. In mammals, FOXO4 has been shown to be regulated by the Ras/MAP kinase pathway (Kops et al. 1999), and a similar mechanism may exist in flies. Interestingly, our microarray experiments identified >200 genes in addition to d4EBP that are up-regulated by dFOXO (data not shown), which increases the complexity of transcriptional regulation affected by dFOXO. Additional studies will be necessary to determine the multiple mechanisms by which the insulin signaling cascade dictates cell number and size during development of the metazoan body plan.
Materials and methods
Constructs, Drosophila strains, and antibodies
Clones LD05569 and LD19191 contain the complete cDNA of dFOXO (accession no. AF426831). dFOXO was cloned in pMTV5-HisA (Invitrogen), giving pMTdFOXO. QuikChange (Stratagene) was used to mutate the three Akt phosphorylation sites, producing pMTdFOXOA3. Clone SD10374 containing full-length dAkt was subcloned in pAc5V5-HisA (Invitrogen). An src myristoylation signal (MGSSKSKPK) was added at the N terminus by PCR, giving the pAcMyrdAkt vector. The luciferase reporter pGL4xFRE was constructed by inserting the ADH distal promoter (–42 to +40) in pGL3Basic (Promega). Four FOXO4-binding sites (AGTTTGTTTGTCGATTAAATAAA CATGTAAACACTTTGTTTTGTTGATACAAACAAAA) were added upstream by PCR. A DNA fragment of the d4EBP promoter (–1460 to +197) and a 1586-bp DNA fragment of the dInR promoter were amplified by PCR and cloned in pGL3Basic, producing pGLd4EBP1.6 and pGLdInR1.5, respectively. Shorter versions of these DNA fragments were produced in a similar way. As negative control, a construct containing Upstream Activating Sequences (UAS) for the transcription factor GAL4 and an E1B TATA sequence upstream of a luciferase reporter gene was used. This construct was activated by GAL4 but not by dFOXO. All the constructs were verified by sequencing.
dFOXO-V5 was subcloned in a pUAST vector, and transgenic flies were obtained by injecting the construct into yw recipients. Four independent lines were established. UAS-dFOXOA3 was injected in the same way; however, no transformant line could be obtained for this construct. w;UAS-dAkt flies were a kind gift from Morris Birnbaum (Howard Hughes Medical Institute, University of Pennsylvania, Philadelphia, PA; Verdu et al. 1999). y1 w*; wgSp-1/CyO; P{w+mW.hs=GAL4-dpp.blk1}40C.6/MKRS (Bloomington 1553b) was obtained from the Bloomington Stock Center. MS1096 (Capdevila and Guerrero 1994) was a gift from Y. Nibu (Division of Genetics and Development, Department of Molecular and Cell Biology, University of California, Berkeley, CA). ey-GAL4 (Hazelett et al. 1998) and GMR-GAL4 (Hay et al. 1994) were obtained from L. Michaut (Biozentrum, University of Basel, Basel, Switzerland).
Antibodies were raised in rabbits against amino acids 1–213 and 214–613 of dFOXO.
Cell culture and extract preparation
Drosophila Schneider line S2 cells (ATCC CRL-1963) were grown in M3 medium (Sigma) with 10% heat-inactivated fetal bovine serum, penicillin, and streptomycin at 25°C in suspension. Stable transfected S2 cells were grown in the presence of 300 μg/mL hygromycin (Clontech). To remove serum from the medium, cells were pelleted and washed once with phosphate buffer saline (PBS). Then cells were incubated in M3 medium without serum for at least 48 h. Next, 1 × 106 cells were plated per well in a 24-well plate. Treatments with insulin (1 μg/mL; Roche) dissolved in H2O or LY294002 (20 μM; Calbiochem) dissolved in ethanol were performed by incubating the cells for 6 h prior to manipulation. In Figure 1F, insulin was incubated for 40 min and LY294002 for 10 min prior to insulin treatment. Extracts were obtained by washing S2 cells with PBS and resuspending them in passive lysis buffer (Promega).
Transfection
For transfection, 1 × 106 S2 cells were plated per well in a 24-well plate. Cells were transfected using effectene reagent (QIA-GEN). For this, 200 ng of expression vector (pMTdFOXO, pMTdFOXOA3, or pMTV5-HisA), 50 ng of reporter vector (pGL4xFRE, pGLdInR1.5, or pGLd4EBP1.6), and 50 ng of pAcMyrdAkt (when necessary) were used. Stable transfectants for pMTdFOXO or pMTdFOXOA3 were obtained as recommended (Invitrogen), and 600 μM CuSO4 was used to induce dFOXO or dFOXOA3 expression.
Phosphatase treatment and Western blot
Western blot detection and phosphatase treatment were performed as described (Miron et al. 2001). Membranes were probed with anti-dFOXO (1:1000), anti-V5 (1:5000; Invitrogen), or anti-tubulin (1:10.000; Sigma).
Immunolocalization
S2 cells grown in M3 medium without serum were grown on coverslips and incubated for 6 h (±insulin). Cells were washed with PBS, fixed with 2% formaldehyde in PBS for 10 min, and permeabilized with 0.1% saponin in PBS for 20 min. Cells were stained with anti-V5 antibody (1:5000 dilution) and anti-mouse Alexa 488 (1:200; Molecular Probes). Cells were mounted with vectashield containing DAPI (5 μg/mL) for observation under a confocal microscope (LSM510, Zeiss).
Cell growth analysis
Cultures of cells stably transfected with constructs pMTdFOXO or pMTdFOXOA3 were seeded at 0.5 × 106 cells/mL in the presence of serum and insulin. CuSO4 was added 48 h later and then removed after another 8 h by pelleting the cells and washing them in PBS. Cells were pelleted again and resuspended in M3 medium with serum and insulin. At this point and every 12 h after, cells were counted. The data in Figure 3 are representative of three independent experiments. Western blotting was performed with V5 antibody for each 24-h time point to detect dFOXO or dFOXOA3 levels. FACS analysis was performed for every 24-h time point. Briefly, cells were washed with PBS, fixed in 90% ethanol, and kept at –20°C until all samples were collected. An equal number of cells for each point was resuspended in 800 μL of PBS, and 100 μL of RNAse A (10 mg/mL) and 50 μL of propidium iodide (1 mg/mL) were added. Cells were incubated at 37°C for 1 h. Flow cytometry analysis was performed with a Beckman-Coulter XL apparatus.
RNA interference
Nucleotides 850–1590 of dAkt or the full-length E. coli lacI gene were cloned in pCR4 Topo (Invitrogen) and transcribed independently with T7and T3 RNApol. Equal molar amounts of each RNA strand were annealed by heating at 65°C for 30 min and cooling slowly to room temperature. Then 5 μg of dsRNA was used per well in a 24-well plate. dsRNAs were added and cells were incubated at 25°C for 2 d. Subsequently, cells were transfected, and luciferase activity was measured as described before.
DNA microarrays
Cells stably transfected with pMTdFOXO or pMTdFOXOA3, or untransfected S2 cells, were grown in suspension in M3 medium in the presence of serum and insulin. After 6 h of CuSO4 induction, cells were pelleted, and total RNA was extracted with Trizol (Invitrogen). cRNA synthesized from total mRNA was used to probe Affymetrix microarrays containing 13,500 Drosophila transcripts. When necessary, cycloheximide (35 μM) was added 2 h after induction and incubated for 5 h. All steps for microarray analysis were done following protocols by Affymetrix. Genes activated by dFOXO were defined as genes upregulated in dFOXOA3 cells but not in dFOXO or S2 cells.
RNase protection
RNase protection assays were carried out with the RPAIII kit (Ambion). DNA fragments including exonic and intronic sequences of d4EBP, dInR, actin, and 18s rRNA were amplified by PCR and cloned in pBluescript II SK+ (Stratagene) for in vitro transcription; 10 μg of total RNA was used per sample. The experiment of Figure 4C was performed by incubating dFOXO or dFOXOA3 transfected cells with or without CuSO4 and with or without insulin for 6 h. To analyze endogenous dFOXO activity, S2 cells were grown in the absence of serum for 48 h. Then insulin or LY294002 was added, and 6 h later, total RNA was extracted and analyzed by RNase protection as described above.
Band shift analysis
Band shift experiments were performed with a DNA fragment of 113 bp spanning nucleotides –190 to –77 of the d4EBP promoter. A 107-bp fragment from the multiple cloning site of pBluescript SK II (between XbaI and XhoI sites) was blunt-ended and used as negative control. For analysis of the dInR, DNA fragments –21–119, –94–194, –172–273, –249–369, –346–514, –488–627, –597–854, –828–980, –955–1128, –1103–1216, –1193–1435, and –1410–1586 were used. Binding reactions were performed as described (Coleman and Pugh 1997) with labeled probe (20 nM), increasing amounts of recombinant purified dFOXO (40–600 nM), and cold competitor (200 nM to 2 μM). Complexes were resolved on 4.35% polyacrylamide (37:1), 2.5% glycerol, 1× TBE, and 4 mM MgCl2 at 4°C.
Chromatin immunoprecipitation
Samples were treated as described (Andrulis et al. 2000) except no CsCl gradient was performed. Instead, samples were diluted 1:10 with PBS prior to immunoprecipitation.
In vitro transcription
Drosophila nuclear extract was prepared essentially as described (Biggin and Tjian 1988). The H.4 fraction was immunodepleted of endogenous dFOXO by incubation with Protein-A Sepharose beads containing antibodies raised against the N terminus of dFOXO. Transcription analysis was performed by primer extension as described (Hansen et al. 1997).
Fly analysis, cell count, and area measurements
All measurements were done with females. Electron micrographs were taken following standard protocols (Bozzola and Russell 1999). Ommatidia were counted in eight (mutant) or seven (control) flies. Seven ommatidia were taken from each eye to calculate their area by using the histogram function of Adobe Photoshop. Wings were analyzed as described (Miron et al. 2001). A total of 30 (dpp) and 15 (MS1096) wings were counted for either phenotype.
Acknowledgments
We thank M. Birnbaum, L. Michaut, Y. Nibu, and the Bloomington Drosophila Stock center for fly strains; Y. Isogai for help with confocal microscopy; G. Vrdoljak from the Electron Microscopy Laboratory at UC Berkeley for his help with electron microscopy; D. Schichnes from the Biological Imaging Facility for helping with the confocal and optical imaging; and H. Nolla from the Flow Cytometry Facility for helping with FACS. We also thank R. Freiman, K. Geles, B. Glover, M. Jünger, Y. Isogai, W-L. Liu, S. Martin, D. Rio, D. Taatjes, B. Weinert, and J. Ziegelbauer for comments on the manuscript. Members of the Tjian and Levine labs are acknowledged for helpful ideas and technical assistance. O.P. is supported by EMBO and HHMI postdoctoral fellowships. M.T.M. is supported by a fellowship from the Damon Runyon Cancer Research Foundation (DRG-#1684). This work was funded by grants from the NIH and the Howard Hughes Medical Institute to R.T.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1098703.
Note added in proof
Recent work by Jünger et al. (2003) reports similar findings that dFOXO is a major regulator on the insulin signaling pathway in Drosophila.
References
- Alvarez, B., Martinez, A.C., Burgering, B.M., and Carrera, A.C. 2001. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature 413: 744–747. [DOI] [PubMed] [Google Scholar]
- Andrulis, E.D., Guzman, E., Doring, P., Werner, J., and Lis, J.T. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes & Dev. 14: 2635–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala, J.E., Streeper, R.S., Desgrosellier, J.S., Durham, S.K., Suwanichkul, A., Svitek, C.A., Goldman, J.K., Barr, F.G., Powell, D.R., and O'Brien, R.M. 1999. Conservation of an insulin response unit between mouse and human glucose-6-phosphatase catalytic subunit gene promoters: Transcription factor FKHR binds the insulin response sequence. Diabetes 48: 1885–1889. [DOI] [PubMed] [Google Scholar]
- Biggin, M.D. and Tjian, R. 1988. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell 53: 699–711. [DOI] [PubMed] [Google Scholar]
- Bohni, R., Riesgo-Escovar, J., Oldham, S., Brogiolo, W., Stocker, H., Andruss, B.F., Beckingham, K., and Hafen, E. 1999. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell 97: 865–875. [DOI] [PubMed] [Google Scholar]
- Bozzola, J.J. and Russell, L.D. 1999. Electron microscopy, principles and techniques for biologists, 2nd ed. Jones and Bartlett, Sudbury, MA.
- Brand, A.H. and Perrimon, N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Britton, J.S., Lockwood, W.K., Li, L., Cohen, S.M., and Edgar, B.A. 2002. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2: 239–249. [DOI] [PubMed] [Google Scholar]
- Brogiolo, W., Stocker, H., Ikeya, T., Rintelen, F., Fernandez, R., and Hafen, E. 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11: 213–221. [DOI] [PubMed] [Google Scholar]
- Brunet, A., Bonni, A., Zigmond, M.J., Lin, M.Z., Juo, P., Hu, L.S., Anderson, M.J., Arden, K.C., Blenis, J., and Greenberg, M.E. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857–868. [DOI] [PubMed] [Google Scholar]
- Burgering, B.M. and Kops, G.J. 2002. Cell cycle and death control: Long live Forkheads. Trends Biochem. Sci. 27: 352–360. [DOI] [PubMed] [Google Scholar]
- Cantley, L.C. 2002. The phosphoinositide 3-kinase pathway. Science 296: 1655–1657. [DOI] [PubMed] [Google Scholar]
- Capdevila, J. and Guerrero, I. 1994. Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J. 13: 4459–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Jack, J., and Garofalo, R.S. 1996. The Drosophila insulin receptor is required for normal growth. Endocrinology 137: 846–856. [DOI] [PubMed] [Google Scholar]
- Coleman, R.A. and Pugh, B.F. 1997. Slow dimer dissociation of the TATA binding protein dictates the kinetics of DNA binding. Proc. Natl. Acad. Sci. 94: 7221–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, S.R., Brunet, A., and Greenberg, M.E. 1999. Cellular survival: A play in three Akts. Genes & Dev. 13: 2905–2927. [DOI] [PubMed] [Google Scholar]
- Dijkers, P.F., Medema, R.H., Lammers, J.W., Koenderman, L., and Coffer, P.J. 2000. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 10: 1201–1204. [DOI] [PubMed] [Google Scholar]
- Durham, S.K., Suwanichkul, A., Scheimann, A.O., Yee, D., Jackson, J.G., Barr, F.G., and Powell, D.R. 1999. FKHR binds the insulin response element in the insulin-like growth factor binding protein-1 promoter. Endocrinology 140: 3140–3146. [DOI] [PubMed] [Google Scholar]
- Fernandez, R., Tabarini, D., Azpiazu, N., Frasch, M., and Schlessinger, J. 1995. The Drosophila insulin receptor homolog: A gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. 14: 3373–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch, C.E. and Ruvkun, G. 2001. The genetics of aging. Annu. Rev. Genomics Hum. Genet. 2: 435–462. [DOI] [PubMed] [Google Scholar]
- Gao, X., Neufeld, T.P., and Pan, D. 2000. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev. Biol. 221: 404–418. [DOI] [PubMed] [Google Scholar]
- Gingras, A.C., Kennedy, S.G., O'Leary, M.A., Sonenberg, N., and Hay, N. 1998. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes & Dev. 12: 502–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.C., Gygi, S.P., Raught, B., Polakiewicz, R.D., Abraham, R.T., Hoekstra, M.F., Aebersold, R., and Sonenberg, N. 1999. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes & Dev. 13: 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., and Sonenberg, N. 2001. Regulation of translation initiation by FRAP/mTOR. Genes & Dev. 15: 807–826. [DOI] [PubMed] [Google Scholar]
- Guo, S., Rena, G., Cichy, S., He, X., Cohen, P., and Unterman, T. 1999. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J. Biol. Chem. 274: 17184–17192. [DOI] [PubMed] [Google Scholar]
- Hall, R.K., Yamasaki, T., Kucera, T., Waltner-Law, M., O'Brien, R., and Granner, D.K. 2000. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J. Biol. Chem. 275: 30169–30175. [DOI] [PubMed] [Google Scholar]
- Hansen, S.K., Takada, S., Jacobson, R.H., Lis, J.T., and Tjian, R. 1997. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell 91: 71–83. [DOI] [PubMed] [Google Scholar]
- Hartwell, L.H., Culotti, J., Pringle, J.R., and Reid, B.J. 1974. Genetic control of the cell division cycle in yeast. Science 183: 46–51. [DOI] [PubMed] [Google Scholar]
- Hay, B.A., Wolff, T., and Rubin, G.M. 1994. Expression of baculovirus P35 prevents cell death in Drosophila. Development 120: 2121–2129. [DOI] [PubMed] [Google Scholar]
- Hazelett, D.J., Bourouis, M., Walldorf, U., and Treisman, J.E. 1998. decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development 125: 3741–3751. [DOI] [PubMed] [Google Scholar]
- Hekimi, S., Lakowski, B., Barnes, T.M., and Ewbank, J.J. 1998. Molecular genetics of life span in C. elegans: How much does it teach us? Trends Genet. 14: 14–20. [DOI] [PubMed] [Google Scholar]
- Ikeya, T., Galic, M., Belawat, P., Nairz, K., and Hafen, E. 2002. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 12: 1293. [DOI] [PubMed] [Google Scholar]
- Johnston, L.A. and Gallant, P. 2002. Control of growth and organ size in Drosophila. Bioessays 24: 54–64. [DOI] [PubMed] [Google Scholar]
- Johnston, G.C., Pringle, J.R., and Hartwell, L.H. 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105: 79–98. [DOI] [PubMed] [Google Scholar]
- Jünger, M.A., Rintelen, F., Stocker, H., Radimerski, T., Wasserman, J.D., Greenberg, M.E., and Hafen, E. 2003. Drosophila FOXO is not essential for size control but mediates growth inhibition associated with reduced insulin signaling. J. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Kops, G.J., de Ruiter, N.D., De Vries-Smits, A.M., Powell, D.R., Bos, J.L., and Burgering, B.M. 1999. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398: 630–634. [DOI] [PubMed] [Google Scholar]
- Kozma, S.C. and Thomas, G. 2002. Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. Bioessays 24: 65–71. [DOI] [PubMed] [Google Scholar]
- Lee Jr., J.T. and McCubrey, J.A. 2002. The Raf/MEK/ERK signal transduction cascade as a target for chemotherapeutic intervention in leukemia. Leukemia 16: 486–507. [DOI] [PubMed] [Google Scholar]
- Leevers, S.J., Weinkove, D., MacDougall, L.K., Hafen, E., and Waterfield, M.D. 1996. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 15: 6584–6594. [PMC free article] [PubMed] [Google Scholar]
- Lin, K., Dorman, J.B., Rodan, A., and Kenyon, C. 1997. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278: 1319–1322. [DOI] [PubMed] [Google Scholar]
- Miron, M., Verdu, J., Lachance, P.E., Birnbaum, M.J., Lasko, P.F., and Sonenberg, N. 2001. The translational inhibitor 4E-BP is an effector of PI3K/Akt signalling and cell growth in Drosophila. Nat. Cell Biol. 3: 596–601. [DOI] [PubMed] [Google Scholar]
- Nadal, A., Marrero, P.F., and Haro, D. 2002. Down-regulation of the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene by insulin: The role of the forkhead transcription factor FKHRL1. Biochem. J. 366: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae, J., Kitamura, T., Silver, D.L., and Accili, D. 2001. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J. Clin. Invest. 108: 1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin, N., Ogg, S., Cahill, C.M., Biggs, W., Nui, S., Dore, J., Calvo, D., Shi, Y., Ruvkun, G., and Alexander-Bridges, M.C. 2000. DAF-16 recruits the CREB-binding protein coactivator complex to the insulin-like growth factor binding protein 1 promoter in HepG2 cells. Proc. Natl. Acad. Sci. 97: 10412–10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg, S., Paradis, S., Gottlieb, S., Patterson, G.I., Lee, L., Tissenbaum, H.A., and Ruvkun, G. 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994–999. [DOI] [PubMed] [Google Scholar]
- Rousseau, D., Gingras, A.C., Pause, A., and Sonenberg, N. 1996. The eIF4E-binding proteins 1 and 2 are negative regulators of cell growth. Oncogene 13: 2415–2420. [PubMed] [Google Scholar]
- Rulifson, E.J., Kim, S.K., and Nusse, R. 2002. Ablation of insulin-producing neurons in flies: Growth and diabetic phenotypes. Science 296: 1118–1120. [DOI] [PubMed] [Google Scholar]
- Schmoll, D., Walker, K.S., Alessi, D.R., Grempler, R., Burchell, A., Guo, S., Walther, R., and Unterman, T.G. 2000. Regulation of glucose-6-phosphatase gene expression by protein kinase Bα and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J. Biol. Chem. 275: 36324–36333. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton, K. and Hoffmann, F.M. 1994. Ectopic decapentaplegic in the Drosophila midgut alters the expression of five homeotic genes, dpp, and wingless, causing specific morphological defects. Dev. Biol. 164: 502–512. [DOI] [PubMed] [Google Scholar]
- Stocker, H. and Hafen, E. 2000. Genetic control of cell size. Curr. Opin. Genet. Dev. 10: 529–535. [DOI] [PubMed] [Google Scholar]
- Trumpp, A., Refaeli, Y., Oskarsson, T., Gasser, S., Murphy, M., Martin, G.R., and Bishop, J.M. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414: 768–773. [DOI] [PubMed] [Google Scholar]
- Verdu, J., Buratovich, M.A., Wilder, E.L., and Birnbaum, M.J. 1999. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat. Cell Biol. 1: 500–506. [DOI] [PubMed] [Google Scholar]
- Weigmann, K., Cohen, S.M., and Lehner, C.F. 1997. Cell cycle progression, growth and patterning in imaginal discs despite inhibition of cell division after inactivation of Drosophila Cdc2 kinase. Development 124: 3555–3563. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Whelan, J., Babb, R., and Bowen, B.R. 2002. An mRNA splice variant of the AFX gene with altered transcriptional activity. J. Biol. Chem. 277: 8068–8075. [DOI] [PubMed] [Google Scholar]