Abstract
Fanconi anemia (FA) is a genetic disorder characterized by hypersensitivity to DNA damage, bone marrow failure, congenital defects, and cancer. To further investigate the in vivo function of the FA pathway, mice with a targeted deletion in the distally acting FA gene Fancd2 were created. Similar to human FA patients and other FA mouse models, Fancd2 mutant mice exhibited cellular sensitivity to DNA interstrand cross-links and germ cell loss. In addition, chromosome mispairing was seen in male meiosis. However, Fancd2 mutant mice also displayed phenotypes not observed in other mice with disruptions of proximal FA genes. These include microphthalmia, perinatal lethality, and epithelial cancers, similar to mice with Brca2/Fancd1 hypomorphic mutations. These additional phenotypes were not caused by defects in the ATM-mediated S-phase checkpoint, which was intact in primary Fancd2 mutant fibroblasts. The phenotypic overlap between Fancd2-null and Brca2/Fancd1 hypomorphic mice is consistent with a common function for both proteins in the same pathway, regulating genomic stability.
Keywords: Fanconi anemia, cancer, Fancd2, Brca2, DNA repair, chromosome pairing
Fanconi anemia (FA) is an autosomal recessive disorder characterized by cellular sensitivity to DNA damage, bone marrow failure, and predisposition to cancer (Fanconi 1967; D'Andrea and Grompe 2003). Patients may also have congenital malformations including short stature, small gonads, microphthalmia, and skeletal defects (Alter 1993). At least eight complementation groups exist (FA-A through FA-G, including D1 and D2; Joenje et al. 1997; Timmers et al. 2001), and the genes for seven of these have been identified (FANCA, FANCC, FANCD2, FANCE, FANCF, FANCG, and BRCA2; Strathdee et al. 1992; Lo Ten Foe et al. 1996; de Winter et al. 1998, 2000a,b; Timmers et al. 2001; Howlett et al. 2002).
The precise functions of the FA, BRCA1, and BRCA2 proteins remain unclear, but there is evidence for overlapping roles. FANCA, FANCC, FANCE, FANCF, and FANCG interact in a multisubunit nuclear complex (Kupfer et al. 1997; Garcia-Higuera et al. 1999; de Winter et al. 2000c; Medhurst et al. 2001). Genes constituting this complex act epistatically as shown by the redundant phenotypes of Fanca, Fancc, Fancg, and Fanca/Fancc double knockout mice (Noll et al. 2002). FANCD2 acts downstream of the nuclear complex. After DNA damage (Garcia-Higuera et al. 2001) and during the S phase of the cell cycle (Taniguchi et al. 2002a), FANCD2 is monoubiquitinated at Lys 561 but not in cells lacking any member of the upstream FA complex (Garcia-Higuera et al. 2001). Monoubiquitination of FANCD2 is required for its colocalization with RAD51 and BRCA1 in nuclear foci. FANCD2 can also be phosphorylated at Ser 222 by ATM (ataxia telengiectasia mutated). This modification requires wild-type nibrin (Nakanishi et al. 2002) and has been reported to mediate the damage-induced arrest of DNA synthesis in SV40-transformed fibroblasts (Taniguchi et al. 2002b).
The identification of BRCA2 as an FA protein has suggested a potential function for the pathway. BRCA2 binds single-stranded DNA (Yang et al. 2002), interacts directly with RAD51 (Marmorstein et al. 1998), and modulates its activity (Davies et al. 2001). Cells lacking functional BRCA2 are deficient in homology-directed repair of DNA double-strand breaks (Moynahan et al. 2001b; Tutt et al. 2001), and therefore the FA/BRCA pathway may function to modulate DNA repair by homologous recombination.
Mouse genetics has been useful for investigating the function of the FA/BRCA pathway. Knockouts of Fancc (Chen et al. 1996; Whitney et al. 1996), Fanca (Cheng et al. 2000; Noll et al. 2002), and Fancg (Yang et al. 2001; Koomen et al. 2002) all share a defect in germ-cell development and demonstrate cellular sensitivity to agents that produce DNA interstrand cross-links (ICLs). In contrast to human FA patients, however, FA mice do not develop anemia and do not have skeletal defects; nor are they at an increased risk for developing cancer. Targeted deletions have also been generated for the murine homolog of BRCA2. Whereas homozygous null mutants lacking Brca2 (Sharan et al. 1997) die at midgestation, mice homozygous for a truncating mutation at the C terminus of Brca2 are viable and show some phenotypic overlap with Fanca, Fancc, and Fancg knockout mice (Connor et al. 1997; McAllister et al. 2002). Here, we report the phenotype of mice homozygous for a null allele of the Fancd2 locus.
Results
Generation of Fancd2 knockout mice
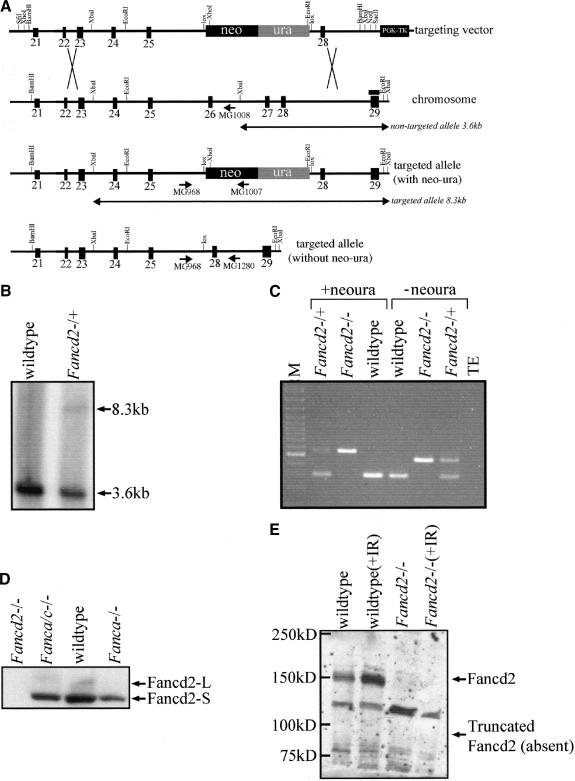
Murine Fancd2 is highly homologous to human FANCD2 and consists of 44 exons (GenBank accession no. BC042619). The targeting construct (Fig. 1A) was designed to delete exons 26 and 27(220 bp) of the 44-exon gene and is predicted to disrupt the reading frame of the mRNA, resulting in a premature nonsense codon at 54 bp downstream of the exon 25 and exon 28 junction. Chimeric males were bred to generate Fancd2 heterozygotes and crossed to produce Fancd2 mutants and controls in the syngeneic 129S4, the congenic C57BL/6J, and a mixed 129S4, C57BL/6J strain background (Fig. 1C).
Figure 1.
Targeted disruption of murine Fancd2–/– gene. (A) Schematic of targeting vector, wild-type Fancd2 locus, targeted allele with neo–ura cassette, and targeted allele without neo–ura cassette. Homologous integration of the targeting plasmid replaces Fancd2 exons 26 and 27with a neo–ura cassette flanked by lox sites. The binding site for the probe is indicated by a black bar. (B) Southern blot analysis using genomic DNA from targeted ES cell clones digested with XbaI. Homologous integration of the targeting plasmid eliminates an XbaI site in intron 26, resulting in a nontargeted band at 3.6 kb and a wild-type band at 8.3 kb. (C) PCR genotyping using genomic DNA from wild-type, heterozygous, and mutant tail as templates. Mutant Fancd2 alleles with and without the neo–ura cassette are shown. The mutant band is 556 bp (with neo–ura) or 459 bp (without neo–ura). The wild-type band is 303 bp. (D) Western blot with anti-Fancd2 antibody on protein lysate from Fancd2 mutant, Fanca/c double mutant, wild-type, and Fanca mutant testes. Fancd2-S (155 kD) and Fancd2-L (162 kD) are visible in wild-type extracts. Only Fancd2-S is detectable in Fanca/c mutant and Fanca mutant extracts. No signal is detectable in Fand2 mutant extract. (E) Western blot with anti-Fancd2 antibody on protein lysate from wild-type and Fancd2 mutant MEFs either untreated or irradiated with 10 Gy. Fancd2 (155/162 kD) is detected in wild-type extracts but absent from Fancd2 mutant extracts. The absence of a band at 88 kD, corresponding to the predicted size of truncated Fancd2, is indicated.
To confirm that the targeted allele of Fancd2 was a null mutation, reverse transcriptase PCR (RT–PCR) was performed using mRNA generated from Fancd2 mutant and wild-type testes. A product of the predicted size was amplified from wild-type testes, and a product lacking exons 26 and 27 of Fancd2 was amplified from Fancd2 mutant testes, indicating that some mRNA is transcribed from the mutant allele (data not shown). We next performed Western blots on protein extract prepared from mouse bone marrow and testes and mouse embryonic fibroblasts (MEFs) using an N terminus-specific antibody [kindly provided by Alan D'Andrea (Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA); Fig. 1D,E; data not shown]. Both isoforms of Fancd2 (Fancd2-S, 156 kD and Fancd2-L, 162 kD) were detectable in wild-type testes extract. Only Fancd2-S was observed in extract from Fanca mutant and Fanca/c double mutant testes. Full-length Fancd2 was also observed in Western blots using protein extract from wild-type MEFs and wild-type bone marrow (Fig. 1E; data not shown). Importantly, no full-length Fancd2 or any bands of the predicted truncated protein's molecular weight (∼88 kD) were observed in protein extract from testes, bone marrow, or MEFs. Thus, we conclude that the targeted allele of Fancd2 is a null mutation at the protein level.
Fancd2 knockouts display phenotypes similar to Fanca, Fancc, and Fancg knockouts
Upon gross examination, Fancd2 mutants on the 129S4 background were indistinguishable from their littermates. However, Fancd2–/– mice on the C57BL/6J background were smaller than controls, similar to Fancc mice (M. Buchwald, pers. comm.). Mutants were consistently smaller than controls at each data point analyzed (see Supplementary Fig. 1). At day 5, mutant males weighed 2.5 ± 0.7g (n = 9), and nonmutant control males weighed 3.1 ± 0.4 g (n = 23; p < 0.05, t-test), indicating delayed development in utero. At day 40 (adulthood), mutant males weighed 18.3 ± 1.2 g (n = 3), and nonmutant control males weighed 21.2 ± 1.0 g (n = 19; p < 0.01, t-test). A similar result was found for female mutants and controls. We conclude that Fancd2 mutant mice dis play a developmental delay in utero as well as postnatally.
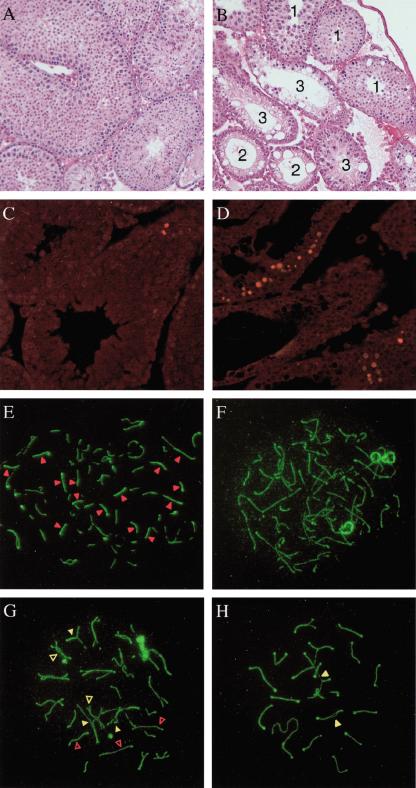
Individual testes from adult (age 2–6 mo) Fancd2 mutant and control animals on the 129S4 background were also examined. The average weight per testis from Fancd2 mutants (23 ± 4 mg; n = 20) was smaller than that of controls (90 ± 19 mg; n = 13; p < 0.001, one-way ANOVA). Interestingly, the testicular weight of Fancd2 mutants was also significantly reduced compared with Fanca (78 ± 13 mg; n = 14; p < 0.001, one-way ANOVA) and Fancc mutants (56 ± 15 mg; n = 14; p < 0.001, one way ANOVA) on the same strain background (Noll et al. 2002). Despite the weight difference, histological examination of adult Fancd2 mutant testes showed a similar mosaic pattern of normal and abnormal tubules. This characteristic pattern consists of apparently normal tubules, abnormal tubules that lack all germ cells, and abnormal tubules that contain germ cells but have increased numbers of apoptotic pachytene spermatocytes along with a relative paucity of diplotene spermatocytes and spermatids (Fig. 2A–D).
Figure 2.
Germ-cell and meiotic pairing abnormalities in Fancd2–/– mice. (A,B) H&E staining of testis sections from 7-week-old wild-type (A) or Fancd2–/– (B) littermates. All stages of spermatogenesis are seen in the wild-type adult testes. Fancd2–/– testes show a mosaic pattern of normal-appearing tubules (tubule 1), tubules with vacuolated Sertoli cell cytoplasm and no germ cells (tubule 2) and tubules with vacuolated Sertoli cell cytoplasm and decreased numbers of spermatocytes and spermatids (tubule 3). Magnification, 20×. (C,D) TUNEL staining of testes sections from 7-week-old wild-type (C) and Fancd2–/– (D) littermates. Occasional cells were labeled in wild-type tubules near the basement membrane, whereas Fancd2–/– testes contain tubules with multiple numbers of apoptotic spermatocytes. The nuclei of apoptotic cells are TUNEL-labeled with Cy3-dCTP and fluoresce red. Magnification, 20×. (E–H) Meiotic pairing abnormalities in Fancd2–/– spermatocytes detected by indirect immunofluorescence staining for Scp3 (green) and Rad51 (red). (E) Zygotene wild-type nucleus. (F) Zygotene Fancd2–/– nucleus with unusually long unpaired axial elements. (G) Late zygotene Fancd2–/– nucleus with multiple synaptic abnormalities. (H) Pachytene Fancd2–/– nucleus with unpaired (open arrowheads) and mispaired homologs (closed arrowheads). Magnification, 100×.
Ovaries from adult females were also abnormal and contained a markedly reduced number of developing follicles (data not shown.) Importantly, there was a lack of germ cells in testes of newborn Fancd2 mutants (data not shown), as previously also reported for Fancc mice (Whitney et al. 1996). This finding, together with the observed deficiency of spermatogonia in adult males (Fig. 2B), provides evidence that the FA pathway may be important for the survival of early germ cells. Alternatively, the phenotype might also be caused indirectly as a result of an inability to repair DNA damage that occurs constitutively.
The excess of apoptotic spermatocytes seen in some Fancd2 tubules suggested that loss of Fancd2 function may lead to aberrant germ-cell development during meiosis I. To follow meiotic events, we examined surface spreads of mouse spermatocytes using polyclonal antibodies against Rad51 and against Scp3, a component of the axial elements of unsynapsed chromosomes and of synaptonemal complexes that form between homologous chromosomes (Dobson et al. 1994). Fancd2–/– mice display the same meiotic pairing abnormalities as those seen in other FA mice (S. Meyn, unpubl.). The earliest substages of meiotic prophase (leptonema and early zygonema) appear normal in Fancd2–/– spermatocytes, as monitored by Scp3 and Rad51 staining. However, there was an increased frequency of synaptic abnormalities beginning in mid-zygonema. Synapsis between homologs is normally initiated while axial elements are still forming. As a result, unpaired axial elements in wild-type zyogtene spermatocytes are typically short, with nearly full-length but unpaired axial elements uncommon (Fig. 2E). Unpaired axial elements in most zygotene Fancd2–/– nuclei tended to be longer than normal (Fig. 2F), and some zygotene Fancd2–/– nuclei contained multiple full-length axial elements that had aligned with their homologs but failed to synapse properly (Fig. 2G). The majority of Fancd2–/– pachytene nuclei appeared to be normal. However, there was an excess of Fancd2–/– pachytene nuclei with autosomal axial elements that had failed to synapse and/or with nonhomologs that had attempted to pair (Fig. 2H). These observations suggest that, although the FA pathway is not absolutely required for meiosis I, it acts to promote efficient and accurate pairing of homologs.
Fancd2 knockouts display phenotypes not observed in previous mouse models of FA
Fancd2 mutants also had phenotypes not previously observed in Fanca, Fancc, or Fancg knockout mice. Pups were born in near-expected Mendelian ratios from crosses of heterozygous 129S4 breeders; however, a significant deviation from the expected ratios was observed on the C57BL/6J background strain (data not shown). Only 16.5% (50/303), versus the expected 25%, of the pups present at the time of genotyping (postnatal day 5) were mutant (p < 0.005, χ2 test). Although perinatal lethality has not been previously reported for FA knockout mice, it has been described in mice homozygous for a C terminus truncating mutation in Brca2 in which an ∼33% reduction of mutant animals was noted (McAllister et al. 2002). The lack of this phenotype in the 129S4 strain suggests the presence of a modifier locus in either the 129S4 or C57BL/6J strain, which affects the penetrance of the perinatal lethality phenotype.
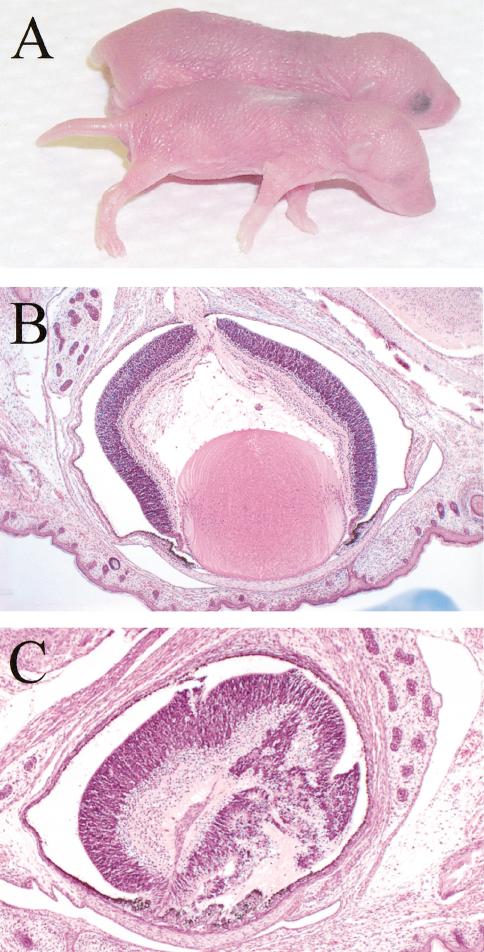
An increased incidence of eye abnormalities was observed for Fancd2 mutants in the C57BL/6J background (Fig. 3), with 32 of 41 (78%) Fancd2 mutants examined having microphthalmia. Consistent with previous reports, nonmutants in the C57BL/6J population also showed a low incidence (3.2%) of microphthalmia (Kalter 1968). To determine if the phenotype was caused by altered expression of nearby genes, also referred to as position effects (Olson et al. 1996), and not the Fancd2 mutation itself, the neo–ura cassette was removed by breeding to transgenic mice expressing Cre recombinase (see Materials and Methods). Consistent with our previous finding, 9 of 19 (47%) Fancd2–/– pups with the allele lacking the neo–ura cassette but none of the controls had microphthalmia. This confirmed that Fancd2 is capable of modifying the microphthalmia predisposition of C57BL/6J mice.
Figure 3.
Fancd2 mutants are predisposed to microphthalmia. (A) Fancd2 mutants are smaller than wild-type controls at postnatal day 1. Microphthalmia is visible in Fancd2 mutants. (B) Histology of eye from a wild-type control pup (postnatal day 1). Magnification, 50×. (C) Representative histology of eye from a Fancd2–/– pup (postnatal day 1) shows a disorganized retina with lack of a distinct chamber or lens development. Magnification, 100×.
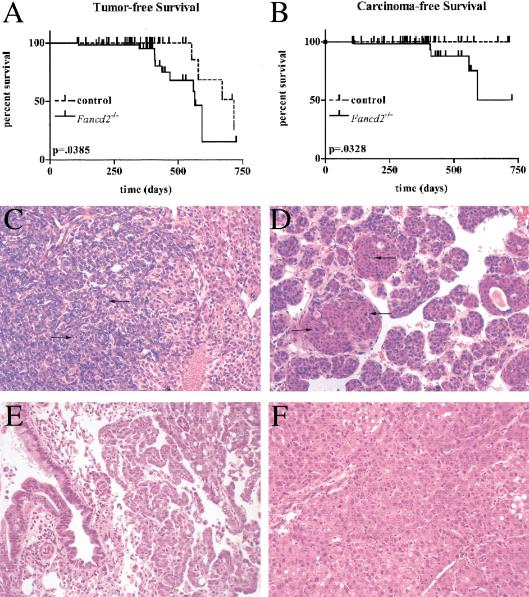
To determine whether Fancd2 mutants are tumorprone, a cohort of animals in the mixed and 129S4 backgrounds were monitored long-term. Animals were killed if abnormal growths were obvious or if they appeared otherwise unhealthy. A final sacrifice of all animals occurred at 18–24 mo for those in the mixed background and 7–17 mo for those in the 129S4 background. To date, 10 of 54 Fancd2 mutants, 3 of 28 Fancd2 heterozygote controls (p > 0.05), and 1 of 14 wild-type controls (p > 0.05) have developed adenomas (p = 0.0385 for Fancd2 mutant vs. controls) or carcinomas (p = 0.0328 for Fancd2 mutant vs. controls) at between 3 and 23 mo of age (Fig. 4A,B). Tumors occurred in multiple different epithelial cell types (summarized in Table 1). Paraffin-embedded sections of these tumors were stained with H&E (Fig. 4C–F) and classified by histological examination. Abnormal cell proliferation was confirmed by immunohistochemistry with an antibody to phosphorylated histone H3, a marker of mitosis (data not shown). The increased incidence of tumors in Fancd2 mutants but not other FA mice demonstrates that complete loss of Fancd2 function is necessary for the presentation of a cancer phenotype. The occurrence of hepatic adenomas in three of the heterozygous controls suggests that Fancd2 heterozygotes may also be at an increased risk for carcinogenesis. However, these data are not statistically significant, and a larger study will be necessary to demonstrate this definitively. Importantly, although some controls developed hepatic adenomas, no carcinomas were observed in these mice.
Figure 4.
Histology of carcinomas and survival curves for Fancd2 mutants and controls. Tumor-free (A; both adenoma and carcinoma) and carcinoma-free (B; only carcinoma) survival curves are shown for the combination of mixed and 129S4 populations. P values were determined by Logrank test. (C–F) H&E-stained paraffin-embedded sections of tumors from Fancd2–/– animals. Arrows indicate mitotic cells with condensed chromatin. Magnification, 200×. (C) Ovarian adenocarcinoma with large acini lined by tall columnar cells having abundant eosinophilic cytoplasm and highly pleomorphic hyperchromatic nuclei (129S4, 3 mo). (D) Mammary adenocarcinoma in which large clusters of mitotically active and pleomorphic cells were found, ranging from columnar cells to nests of small, undifferentiated cells in clusters (mixed, 13 mo). (E) Lung adenocarcinoma of broncho-alveolar origin, with rows of columnar epithelial cells having a high nucleus-to-cytoplasm ratio lining septa, sometimes forming papillae or filling air spaces (129S4, 13 mo). (F) Hepatocellular carcinoma is multinodular and well differentiated. The neoplastic cells vary from being smaller than normal hepatocytes to 2×–3× larger (mixed background, 18 mo).
Table 1.
Increased incidence of tumors of epithelial cell origin in Fancd2 mutant mice
| Genotype | Background | Sex | Age at death | Tumor type |
|---|---|---|---|---|
| Fancd2-/- | Mixed | Female | 14 mo | Ovarian adenoma |
| Fancd2-/- | Mixed | Male | 15 mo | Hepatic adenoma |
| Fancd2-/- | Mixed | Male | 19 mo | Hepatic adenomaa |
| Fancd2-/- | Mixed | Male | 19 mo | Adenoma of fundic mucosaa |
| Fancd2-/- | Mixed | Male | 18 mo | Gastric adenoma |
| Fancd2-/- | Mixed | Male | 13 mo | Facial seborrheic keratosis |
| Fancd2-/- | Mixed | Female | 13 mo | Mammary adenocarcinoma |
| Fancd2-/- | Mixed | Male | 19 mo | B-cell lymphomab |
| Fancd2-/- | Mixed | Male | 19 mo | Hepatocellular carcinomab |
| Fancd2-/- | Mixed | Male | 19 mo | Bronchoalveolar carcinomab |
| Fancd2-/- | Mixed | Male | 19 mo | Bronchoalveolar carcinomab |
| Fancd2-/- | Mixed | Male | 18 mo | Hepatocellular carcinoma |
| Fancd2-/- | 129S4 | Male | 13 mo | Lung adenocarcinoma |
| Fancd2-/- | 129S4 | Female | 3 mo | Ovarian adenocarcinoma |
| Fancd2-/+ | Mixed | Male | 23 mo | Hepatic adenoma |
| Fancd2-/+ | Mixed | Female | 22 mo | Ovarian cystadenoma |
| Fancd2-/+ | Mixed | Female | 18 mo | Hepatic adenoma |
| Wild type | Mixed | Male | 19 mo | Hepatic adenoma |
Hepatic adenoma and adenoma of fundic mucosa occurred in the same Fancd2 mutant.
B-cell lymphoma and three primary tumors occurred in the same Fancd2 mutant.
Fancd2 and Fancc knockouts share a similar DNA damage response profile
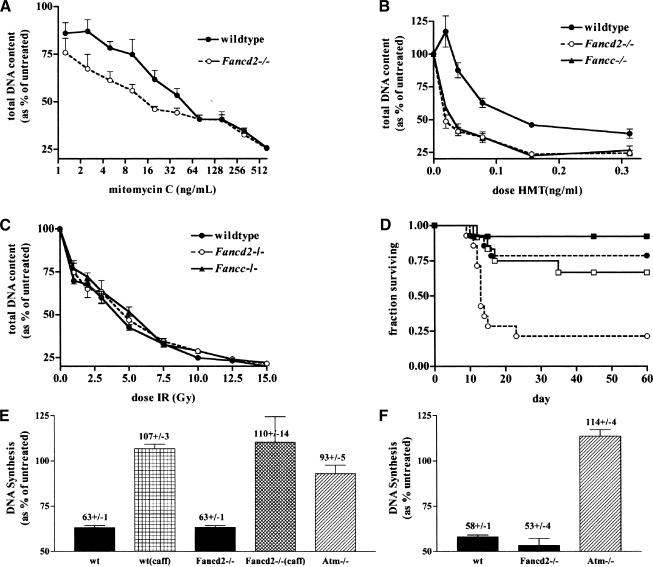
FANCD2 acts downstream of the FA nuclear complex, and thus it may act as a convergence point between multiple upstream DNA damage response pathways (Garcia-Higuera et al. 2001). Furthermore, FANCD2 is phosphorylated by ATM following IR (Taniguchi et al. 2002b). These observations suggested that cells lacking FANCD2 might be more sensitive to DNA damage, particularly IR, than cells lacking FANCA, FANCC, or FANCG. To test this hypothesis, primary MEFs were prepared, and their sensitivity to various DNA-damaging agents was measured in cell growth assays. Cells were treated with increasing doses of mitomycin C (MMC), IR, or the photoactivated DNA cross-linking agent, 4′-hydroxymethyl-4,5′, 8-trimethylpsoralen (HMT; Sigma) plus ultraviolet light (PUVA). As expected, Fancd2 mutant cells displayed a heightened sensitivity to MMC-induced DNA damage as compared with wild-type controls (Fig. 5A). Fancd2 and Fancc mutant cells were equally sensitive to PUVA-induced ICLs (Fig. 5B). However, neither Fancc nor Fancd2 mutant fibroblasts showed a differential sensitivity to IR when compared with wild-type controls (Fig. 5C).
Figure 5.
DNA damage sensitivity and S-phase checkpoint in Fancd2–/– MEFs and mice. Primary MEFs were challenged with increasing doses of mitomycin C (A), 4′-hydroxymethyl-4,5′,8-trimethylpsoralen (HMT) plus UVA (PUVA; B), or ionizing radiation (C). (•) Wild-type control MEFs; (○) Fancd2 mutant MEFs; (▴) Fancc mutant MEFs. Experiments were performed in quadruplicate. Each data point represents the mean ± standard error of the mean (S.E.M.). (D) Survival curve of control (▪, •) and Fancd2 mutant (□, ○) mice following a dose of 9 (solid line) and 10 (broken line) Gy of whole-body irradiation. (E,F) Primary Fancd2–/– MEFs have an intact S-phase checkpoint. DNA synthesis was measured 30 min after treatment with 10 Gy of IR (E) or 0.5 ng/mL H-methoxy-tripsoralen (F) and is represented as the percentage of untreated controls. As a control, cells were also pretreated with 10 mM caffeine (caff). Experiments were performed in quintuplet. Error bars indicate the S.E.M.
To further investigate the role of Fancd2 in the response to IR, we also exposed Fancd2 mutants and controls to whole-body irradiation at doses of 9, 10, and 11 Gy (Fig. 5D; data not shown). We hypothesized that although neither Fancd2 nor Fancc seems to function in response to IR in fibroblasts, a role for Fancd2 may be revealed in whole animals. At a dose of 10 Gy, 11 of 14 controls survived, whereas only 3 of 14 Fancd2 mutants survived (p < 0.005, Logrank test). No Fancd2 mutants survived following a dose of 11 Gy, whereas 4 of 6 control animals survived. Thus, the LD50 for Fancd2 mutants was estimated to be 9.5 Gy compared with 11 Gy for nonmutant littermate controls (proportional difference ∼1.15), which is identical to that previously reported for Fancc mutants (proportional difference of ∼1.2; Noll et al. 2001). Although the Fancd2 mutants had a moderate in vivo IR sensitivity similar to that of Fancc mutants, the lack of a differential sensitivity of Fancd2 mutant MEFs or a major in vivo sensitivity of whole animals to irradiation supports the argument against a significant role for Fancd2 in cellular responses to IR.
Primary Fancd2 mutant fibroblasts have an intact ATM-dependent S-phase checkpoint
Recent experiments have demonstrated that FANCD2 is phosphorylated at Ser 222 by the DNA damage signaling kinase, ATM. Using mutant cDNA constructs of human FANCD2, it was shown that this modification was necessary for the arrest of DNA synthesis after DNA damage, an ATM-dependent function (Taniguchi et al. 2002b). In addition, others have reported a defect in S-phase arrest in response to PUVA in multiple FA cell lines (Centurion et al. 2000; Sala-Trepat et al. 2000). These findings prompted the investigation of the status of the checkpoint in primary Fancd2 mutant MEFs.
Primary (passage 2) Fancd2 mutant and wild-type MEFs, and primary (passage 3) Atm mutant ear fibroblasts were treated with 10 Gy of IR, and their ability to inhibit DNA synthesis was measured. Whereas Fancd2 mutant and wild-type cells inhibited DNA synthesis equally, Atm mutant cells did not (Fig. 5E). As a control, both Fancd2 mutant and wild-type cells were then preincubated in 10 mM caffeine, an inhibitor of ATM (Sarkaria et al. 1999). Inhibition of ATM in these cells was capable of inducing a radioresistant DNA synthesis phenotype similar to that of Atm mutant cells (Fig. 5E).
Furthermore, both Fancd2 mutant and wild-type cells were equally capable of inhibiting DNA synthesis following treatment with 0.5 ng/mL of PUVA, whereas Atm mutant cells were not and continued to incorporate DNA following damage (Fig. 5F). Together, these experiments demonstrate that primary Fancd2 mutant MEFs have an intact ATM-dependant S-phase checkpoint.
Expression and interaction of Rad51 and Brca2 is normal in Fancd2 mutants
Cells harboring mutations in genes implicated in control of homologous recombination repair of DNA damage, including BRCA1, BRCA2, and RAD51, are sensitive to DNA interstrand cross-linking agents (Yu et al. 2000; Moynahan et al. 2001a). After treatment with ICLs, these cells show gross rearrangements involving nonhomologous chromosomes, a phenotype identical to cells from all FA complementation groups. RAD51 functions in the process of homologous recombination (Baumann et al. 1996). BRCA2 interacts directly with RAD51 (Moynahan et al. 2001b), and cells lacking wild-type BRCA2 have increased levels of error-prone homology-directed repair of DNA double-strand breaks, suggesting that BRCA2 may function in error-free repair (Tutt et al. 2001). As Fancd2 acts downstream of the FA complex, it was hypothesized that it may function to modulate the stability of Rad51 and Brca2 or their interaction.
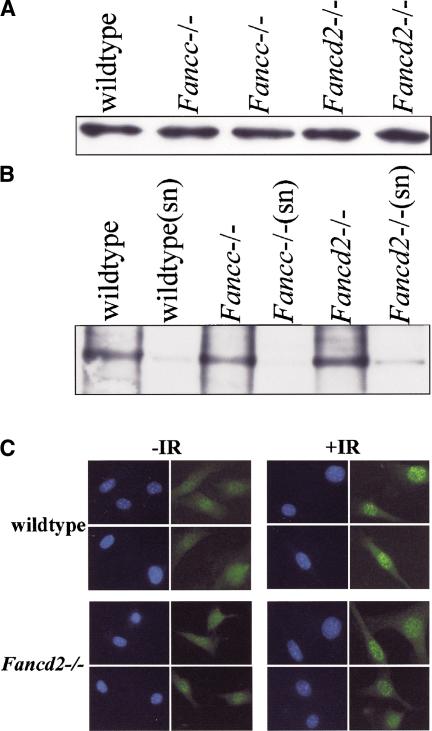
To investigate this hypothesis, immunoblots with testicular lysates from Fancd2 mutant, Fancc mutant, and control mice were prepared. Immunoblotting revealed no significant differences in the steady-state level of Rad51 (Fig. 6A). Immunoprecipitation of Brca2 from these lysates probed with the same Brca2 antibody also showed no significant differences in expression of Brca2 (data not shown).
Figure 6.
Interaction of Rad51/Brca2 and Rad51 foci formation. (A) Whole-cell lysates from mouse testis of the indicated genotypes were separated by SDS-PAGE and probed with an antibody against Rad51. (B) Anti-Brca2 antibodies were used to immunoprecipitate protein complexes. Complexes were separated by SDS-PAGE and probed with an antibody against Rad51. “sn” indicates supernatant from immunocomplexes to compare bound versus unbound fraction. (C) Rad51 foci formation in wild-type and Fancd2–/– MEFs either untreated (–IR) or following 12 Gy of ionizing radiation (+IR). Cell nuclei were counterstained with DAPI (blue).
We next asked whether Fancd2 plays a role in modulating the interaction between Rad51 and BRCA2. Immunoprecipitation of Brca2 was performed on whole-cell lysates from Fancd2 mutant, Fancc mutant, and wild-type mouse testes. An anti-Brca2 antibody but not control IgG was capable of immunoprecipitating Rad51 from whole-cell lysates of testis from Fancd2 mutant, Fancc mutant, and wild-type animals (Fig. 6B). We conclude that the protein levels and interaction of Rad51 and Brca2 are normal in Fancd2 mutant and Fancc mutant mice.
Finally, we performed immunocytochemistry experiments designed to examine whether Rad51 foci form normally in the nuclei of Fancd2-null fibroblasts following DNA damage (Fig. 6C). Primary wild-type and Fancd2–/– MEFs were scored for the presence of Rad51 foci. An equal number of untreated wild-type (36%) and Fancd2–/– (32%) MEFs contained >5 foci per nucleus. Similarly, no difference in the level of Rad51 foci was observed between wild-type (51%) and Fancd2–/– (58%) MEFs 6 h after treatment with ionizing radiation. We find that Rad51 foci formation is unaltered in Fancd2-null fibroblasts when compared with wild-type controls.
Discussion
The phenotype of human Fanconi anemia patients indicates that the FA pathway is important for normal embryonic development, the maintenance of genomic stability, and the preservation of several types of stem cells (Auerbach et al. 2001). The recent discovery that the breast cancer gene, BRCA2, is an FA gene (Howlett et al. 2002) has connected this rare disorder to a common form of cancer. Nonetheless, the precise function(s) of the FA/BRCA pathway remains unknown (D'Andrea and Grompe 2003). We and others have previously created strains of mice with targeted deletions of the murine Fanca, Fancc, and Fancg genes, all components of the FA nuclear complex. These mutants have very similar phenotypes (Chen et al. 1996; Whitney et al. 1996; Cheng et al. 2000; Yang et al. 2001; Koomen et al. 2002; Noll et al. 2002), supporting a model in which the components of the complex participate in a common function. To determine whether Fancd2 participates only in this function in vivo or has additional roles, we generated a strain of mice with a null allele in this gene.
Earlier studies have suggested that FANCD2 may have unique roles that are distinct from the other FA proteins. First, it acts downstream of the FA nuclear complex and is the target of the monoubiquitination mediated by the complex (Garcia-Higuera et al. 2001; Timmers et al. 2001). Hence, the integrity of the FA nuclear complex is not perturbed in FA-D2 cells, unlike in other complementation groups (Garcia-Higuera et al. 1999; de Winter et al. 2000c). Second, FANCD2 is the only FA protein known to form nuclear foci after DNA damage and to colocalize with the repair proteins BRCA1 and RAD51 (Garcia-Higuera et al. 2001; Taniguchi et al. 2002a). Finally, FANCD2 is directly phosphorylated by ATM in response to IR but not in response to ICLs (Nakanishi et al. 2002; Taniguchi et al. 2002b). Other FA proteins are not known to be targets of the ATM kinase.
Despite these biochemical differences, human FA-D2 patients do not differ significantly clinically from FA patients belonging to other complementation groups (Timmers et al. 2001). However, FA-D2 is a rare complementation group, and all human patients reported to date have at least one mutant allele that could have some residual function, and, therefore, the phenotype of a true FANCD2-null mutation remained uncertain. The Fancd2 mutant mice reported here display all the features of previously reported strains of FA knockout mice, but also have important differences.
Fancd2, ionizing radiation, and the ATM signaling pathway
Ser 222 of human FANCD2 is phosphorylated by ATM in response to IR, and this posttranslational modification has been correlated to increased IR sensitivity in immortalized fibroblasts (Taniguchi et al. 2002b). In addition, phosphorylation of Ser 222 was associated with radiation-induced arrest of DNA synthesis. Therefore, to test whether Fancd2 played an important role in the response to IR, the survival of isogenic, primary cells from Fancc mutant and Fancd2 mutant cells was compared. Interestingly, Fancd2-null cells were not differentially sensitive to IR or ICLs compared with Fancc-null cells. Furthermore, although mild hypersensitivity to IR was observed in vivo (LD50 = ∼9.5 Gy), this sensitivity was also not greater than that observed for Fancc mutant mice (Noll et al. 2001).
In addition, and in contrast to Atm mutants, Fancd2-deficient cells did not display radiation-resistant DNA synthesis. Therefore, in murine primary cells, Fancd2 is not necessary for the ATM-mediated S-phase arrest following IR. Overall, the phenotype of Fancd2 mutant mice is significantly different from that of Atm-null mice, which display marked radiation sensitivity, immune deficiency, and hematologic tumors (Xu et al. 1996). Together, our observations suggest that Fancd2 does not play a significant role in ATM-mediated physiologic responses to IR. Perhaps human FANCD2 and murine Fancd2 differ in this aspect of their function, although the proteins are highly homologous and Ser 222 is conserved between the species. Alternatively, Fancd2 may play a role as a mediator of ATM function in only some specific circumstances, for example, T-antigen immortalized fibroblasts.
Novel phenotypes in Fancd2 knockout mice
Although Fancd2 mutant mice do not differ measurably from Fancc mutants in terms of their response to DNA damage, they have some features not seen in Fanca, Fancc, or Fancg mutant animals. These phenotypes include microphthalmia, perinatal lethality, more severe hypogonadism, and tumor development. Normal function of the FA nuclear complex is necessary for the monoubiquitination of FANCD2 in response to DNA damage or DNA replication (Garcia-Higuera et al. 2001; Taniguchi et al. 2002a). Therefore, two basic models exist to explain the divergence of phenotypes between mutants in Fancd2 and nuclear complex genes. First, nonubiquitinated Fancd2 protein, as present in Fanca, Fancc, and Fancg knockout mice, may have some residual activity in the function common to the FA pathway. In this model, knockouts of the nuclear complex genes would be similar to hypomorphic mutations of the Fancd2 gene. The phenotypes would be the result of deficiency in the same biochemical function, but they would be more severe in Fancd2 mutant mice. In the second model, FANCD2 is a multifunctional protein with a domain that functions in the FA pathway and other domains that mediate unrelated functions. The additional phenotypes of Fancd2 mutants would then be caused by deficiency of these additional functions.
Most of the features observed in Fancd2-null mice can be interpreted as more severe manifestations of the qualitatively similar defects seen in other FA knockouts. This is most obvious with the germ-cell defects. Although testicular weight is clearly more affected in Fancd2 mutant mice, the histology is similar to other FA models. Similarly, we have observed microphthalmia in the mutant offspring of some Fancc breeding pairs (M. Grompe, unpubl.), indicating that this is not a truly novel phenotype but a more complete manifestation of a defect common to all FA mice. The increased susceptibility of FA cells to apoptotic cell death after DNA damage could provide a common explanation for the observed germ-cell deficiency, microphthalmia, small size, and perinatal lethality. In the case of these developmental defects, the cells would be responding to spontaneous DNA damage. Overall, the phenotype of Fancd2 mutant mice does not convincingly establish the existence of additional functional domains in the Fancd2 protein.
Fancd2, Brca2, and carcinomas
Although human patients with FA develop a variety of cancers, tumors have never been reported in the other FA knockout mice even when followed to very late ages. In contrast, it is well established that mice with a truncating mutation in Brca2 are at an increased risk of a variety of neoplasms (Connor et al. 1997; McAllister et al. 2002). Interestingly, the tumor spectrum of the Brca2 hypomorphic mice was comparable to that seen in Fancd2 mutants (McAllister et al. 2002). In both cases, a predisposition toward epithelial cancers was seen, and the age of onset was similar. Brca2 hypomorphic mice also have other similarities such as an FA-like germ-cell defect, small size, and perinatal lethality. Taken together, this extensive phenotypic overlap is consistent with the hypothesis that the C-terminal domain of Brca2 functions in the same pathway as Fancd2.
Human BRCA2 is known to interact with and modulate the activity of RAD51, a central player in homologous recombination, and Brca2 mutant cells are known to have defects in error-free recombination (Moynahan et al. 2001b; Tutt et al. 2001). Therefore, the identification of Brca2 as an FA gene supports a role for the FA pathway in recombination DNA repair. This hypothesis is strengthened by the observation that chromosomes in the pachytene stage of meiosis mispair, as described here. One potential function for the FA pathway could be to control the interaction between Rad51 and Brca2 and thereby modulate homologous recombination events at sites of DNA damage. However, Western blot analysis of Fancd2 mutant tissues showed normal Rad51 levels as well as normal interaction of the Brca2 and Rad51 proteins as determined by coimmunoprecipitation. Furthermore, Rad51 foci formation following DNA damage was normal in Fancd2–/– MEFs. Thus, our data do not support a simple model in which the FA pathway controls the stability/rate of Brca2/Rad51 interaction.
In humans, heterozygosity for inactivating BRCA2 germ-line mutations is associated with breast, ovarian, and pancreatic cancer, all tumors of epithelial origin. It is unclear at present why these tissues are particularly affected, but it is interesting to note that epithelial tumors in rodents have been associated with telomere shortening (Artandi et al. 2000; Chang et al. 2001). Human FA patients are known to display significantly shortened telomeres in hematopoietic cells (Leteurtre et al. 1999; Adelfalk et al. 2001; Brummendorf et al. 2001; Hanson et al. 2001), and it is therefore interesting to speculate that the FA/BRCA pathway could be involved in telomere maintenance in some tissues.
The high incidence of epithelial tumors observed in Fancd2 mutants here also raises the issue whether FANCD2 could be an important gene in some human epithelial cancers, especially of the breast and ovary. Importantly, some cases of familial ovarian cancer have been linked precisely to the region where FANCD2 resides in the human genome (Sekine et al. 2001; Simsir et al. 2001; Zhang and Xu 2002). Future studies of human patients with these disorders will be needed to determine whether FA genes play a role in sporadic or inherited human cancers.
Materials and methods
Targeting vector design and generation of Fancd2-deficient mice
A λ phage library of mouse cDNA (Lambda FIX II-Stratagene) was screened with a 3′ 3.0-kb RACE fragment of mouse Fancd2 cDNA. DNA was purified from positive phage clones, and a 7.9-kb BamHI fragment of genomic DNA was subcloned into the Saccharomyces cerevisiae/Escherichia coli shuttle vector, YCplac22-TK (Gietz and Sugino 1988), to generate YCplac22-TK + BamHI fragment. This 7.9-kb fragment was further subcloned, sequenced, and found to contain exons 21–28 of the mouse Fancd2 gene. The following primer pairs were used to amplify sequence flanking exons 26 and 27, and the PCR products were cloned into the plasmid pRAY-1 (Storck et al. 1996) to generate pRAY-1+ flanking arms: 5′-TCAGCCTCACATGGA GTTTAACG-3′/5′-GTGTGGACACTAACCTCACTCGC-3′ and 5′-TTTCCTGTCCTCATCTGCG-3′/5′-TGCAAAGAGAGAAA TGACTCAAG-3′. pRAY-1+ flanking arms and the YCplac22-TK + BamHI fragment were cotransfected into yeast strain MW3317-21A (–trp, –ura) and selected in –ura/–trp dropout media. Clones were screened for a recombination event between the two plasmids that replaced mouse exons 26 and 27with the neo–ura cassette from pRAY-1. YCplac22-TK + BamHI (del exon26/27) was linearized and injected into 129S4 ES cells. gDNA was isolated from clones that were doubly resistant to G418 and gancyclovir, digested with XbaI, and Southern blotted using a 333-bp probe that binds 266–599 bp 3′ of the BamHI site marking the 3′-most end of genomic mouse sequence in the targeting vector (see Fig. 1B). The probe was amplified from total mouse genomic DNA using the following PCR primers pairs: 5′-GAGTTGTTCTTTTGTATGCCCGTC-3′/5′-GTCTTGTATCTGCTTTCCTCTGCC-3′, and cloned into pCR-Blunt II-TOPO (Invitrogen). The probes were digested out with EcoRI. Four highly chimeric males were produced from blastocyst injections of correctly targeted ES cells.
Animal husbandry
Chimeric animals were crossed to C57BL/6J mice, and homozygous Fancd2 knockout mice on a mixed 129S4, C57BL/6J background were generated. Founder animals were also back-crossed for six generations to pure C57BL/6J mice to generate a population of homozygous Fancd2 knockout mice on the C57BL/6J background. A strain of congenic 129S4 Fancd2 mutants was generated by breeding a germ-line chimera with 129S4 females. To generate Fancd2 mutants that lacked the neo–ura knockout cassette, a Fancd2 heterozygous male was crossed to a female mouse in a mixed C57BL/6J, DBA background containing the transgene encoding Cre recombinase driven by a testes-specific promoter [Jackson Labs strain #3466, B6;D2-TgN(Sycp1-Cre)4Min].
PCR genotyping
Fifty nanograms of gDNA was prepared from toes as previously described (Miller et al. 1988) and used as a template in PCR (Mullis and Faloona 1987) to genotype mice: a forward primer, MG968 (5′-TCAGCCTCACATGGAGTTTAACG-3′) and two reverse primers, MG1007(5′-AATTCGCCAATGACAAGAC GC-3′; amplifies a mutant band of 556 bp) and MG1008 (5′-CAGGGATGAAAGGGTCTTACGC-3′; amplifies a wild-type band of 303 bp). The reaction conditions were 94°C for 3 min; 37 cycles of 94°C for 25 sec, 48°C for 25 sec, and 72°C for 35 sec; and a final extension at 72°C for 2 min. For detection of the Fancd2 mutant allele lacking the neo–ura cassette, the same reaction was used but primer MG1007 was replaced with primer MG1280 (5′-GCTACACAGCATTGCCCATAAAG-3′; amplifies a mutant band of 459 bp).
Histology
Tissues fixed in 10% phosphate-buffered formalin for 24 h (pH 7.4) were dehydrated in 100% EtOH and embedded in paraffin wax at 58°C. Then 4-μm sections were rehydrated and stained with hematoxylin and eosin (H&E).
Meiotic chromosome staining
Surface spreads of pachytene and diplotene spermatocytes from male mice between the ages of 16 and 35 d old were prepared as described by Peters et al. (1997). A polyclonal goat antibody to the mouse Scp3 protein was used to visualize axial elements and synaptonemal complexes in the meiotic preparations (Plug et al. 1997). The anti-Rad51 rabbit polyclonal antibody (Ab-1) was purchased from Oncogene Research Products. Antibody incubation and detection procedures were a modification of the protocol of Moens et al. (1987) as described by Keegan et al. (1996). Combinations of donkey anti-rabbit and donkey anti-goat IgG secondary antibodies conjugated to either FITC or TRITC were used for detection (Jackson ImmunoResearch Laboratories). All preparations were counterstained with 4′,6′-diamino-2-phenylindole (DAPI, Sigma) and mounted in a DABCO (Sigma) antifade solution. The preparations were examined on a Zeiss Axioplan-2 microscope (63× Plan Apochromat and 100× Plan Apochromat oil-immersion objectives). Each fluorochrome (FITC, TRITC, and DAPI) image was captured separately as a 1344 × 1024-pixel, 12-bit grayscale source image via OpenLab software (Improvision) controlling a cooled-CCD camera (Hamamatsu Orca ER), and the separate grayscale images were cropped, 24-bit pseudocolored, and merged using Adobe Photoshop.
Cell growth assays
Primary (passage 2) mouse embryonic fibroblasts (MEFs) were prepared from pregnant mice at between 12 and 15 d postcoitus and seeded in quadruplicate at a density of 500 cells/well in 96-well plates. Cells were grown in Dulbecco's modified Eagle's medium (DMEM; GIBCO-BRL) supplemented with 15% fetal bovine serum (FBS; HyClone) and 1× penicillin/streptomycin (GIBCO-BRL). Cells were treated with increasing doses of mitomycin C (Sigma; 0–640 ng/mL) or 4′-hydroxymethyl-4,5′,8-trimethylpsoralen (HMT; Sigma) plus UVA (PUVA; 0–0.5 ng/mL), as previously described (Akkari et al. 2000). Ionizing radiation was administered at a dose of 145.9 rad/min from a 137Cs source. Following treatment, cells were allowed to grow at 37°C for 5 d in 5% CO2. Plates were frozen at –80°C. Total DNA was quantified using CyQuant (Molecular Probes) and compared with untreated controls as an indication of cell growth as previously described (Cheng et al. 2000). Quantification of DNA using CyQuant was performed using a Fluorstar plate reader (B&L Systems) according to the manufacturer's instructions.
Radioresistant DNA synthesis (RDS) assay
The RDS assay was performed as previously described with modifications (Painter and Young 1980; Taniguchi et al. 2002b). Primary MEFs (wild-type and Fancd2, p2) or ear (Atm, p3) fibroblasts were seeded at a cell density of 75,000 per 60-mm dish. Cells were preincubated in media containing 10 nCi/mL thymidine [methyl-14C] (NEN Life Sciences, catalog #NEC568) for 24 h to control for total DNA content and then incubated for 24 h in nonradioactive media. Following DNA damage, cells were incubated at 37°C for 30 min with 5% CO2 to allow arrest of DNA synthesis and then incubated in media containing 2.5 μCi/mL thymidine [methyl-3H] (NEN Life Sciences, catalog #NET027) for 3 h. Cells were postincubated at 37°C for 30 min with 5% CO2 in nonradioactive media to remove unincorporated thymidine pools, washed three times in ice-cold phosphate-buffered saline (PBS), and fixed at room temperature in 90% methanol. Cells were scraped onto Whatman QF/C filters and air-dried for 10 min. Radioactivity was measured using a Beckman LS6500 scintillation counter. DNA synthesis is calculated from the ratio of 3H:14C cpm corrected for channel crossover and is represented as a percentage of untreated controls. Experiments were performed in quintuplet. The data shown are the mean ± standard error of the mean (S.E.M.).
Immunoblotting of Brca2, Rad51, and Fancd2
Preparation of whole-cell lysates: Testes from 4–6-week-old mice belonging to complementation groups A, C, D2, and control mice or MEFs were homogenized with an Eppendorf homogenizer containing 500 μL of lysis buffer (1× PBS, 1% Nonidet P-450, 0.5% sodium deoxycholate, and 0.2% SDS) including 20 μg/mL phenylmethylsulfonyl fluoride, 1 μg/mL leupeptin, and 1 μg of aprotinin. The suspension was passed through a 27-gauge needle a few times to shear the DNA and centrifuged at 15,000g and 4°C for 20 min to remove the debris. The supernatant was stored at –80°C. Total protein from each of the samples was analyzed in triplicate using the Lowry assay (Lowry et al. 1951). For immunoprecipitation and Western blotting of Brca2, mouse monoclonal Brca2 (Ab-1) antibody (Oncogene Research Products) was used for the human cells, and a rabbit polyclonal anti-Brca2 (H-300) antibody was used for the mouse samples. Anti-Rad51 antibody (Santa Cruz Biotechnology, Inc.) was used on the mouse testicular samples for both immunoprecipitation (IP) and Western blotting.
Immunoblot analysis of Rad51: Testicular lysates were resolved on pre-cast Nupage 10%–12% Bis-Tris gels (Invitrogen, Life Technologies). Then 100 μg of lysate was denatured by heating at 70°C for 10 min after the addition of an appropriate amount of 4× Nupage sample buffer and 10× reducing agent. The proteins were transferred to Immobilon-P Transfer membranes (Millipore Corporation), and Rad51 was detected using a 1:500 dilution of anti-Rad51 antibody (Santa Cruz Biotechnology, Inc.), a 1:2000 dilution of anti-rabbit secondary antibody (Amersham Life Science), and the ECL detection system (Amersham Life Science).
Immunoblot analysis of Fancd2: Testicular lysates were resolved on precast Nupage 3%–8% gradient gels (Invitrogen Life Technologies). MEF lysates were resolved on a 5% Tris-glycine gel. One-hundred micrograms of the lysates was denatured by heating at 70°C for 10 min after the addition of appropriate amount of 4× Nupage sample buffer and 10× reducing agent. The proteins were transferred to Immobilon-P Transfer membranes (Millipore Corp.) and Fancd2 was detected using a 1:5000 (testes) or 1:1000 (MEFs) dilution of anti-FANCD2 polyclonal antibody and 1:10,000 dilution of anti-rabbit secondary antibody (Amersham Life Science) and the ECL system (Amersham Life Science).
Immunoprecipitation of Rad51 and Brca2 from testicular lysates
Preparation of Protein A Sepharose: The media (Amersham Life Science) was washed three times with the lysis buffer described above and centrifuged at 12,000g for 20 sec. The supernatant was discarded between washes. A 50% slurry was prepared by mixing equal volumes of media and lysis buffer, and was stored at 4°C.
Cell lysis: For Brca2, VU423 (Howlett et al. 2002) and HeLa cells were used as controls. The culture medium was removed, and the cells were washed twice with ice-cold PBS. The dishes were placed on ice, and the cells were collected with a cell scraper. Tissues were lysed in 100 μL of lysis buffer and incubated on ice for 10–15 min with occasional shaking. The cells were further disrupted by passage through a 21-gauge needle. The lysates were centrifuged at 12,000g and 4°C for 10 min to remove particulate matter, and the supernatant was transferred to a fresh tube.
Immunoprecipitation: For immunoprecipitation, 1 mg (1 mL) of whole-cell or tissue extract was transferred to a 2-mL centrifuge tube. The lysate was precleared by adding 1 μg of the appropriate normal mouse, rat, rabbit, or goat IgG together with 20 μL of Protein A Sepharose conjugate. Incubation was done at 4°C for 1 h and pelleted by centrifugation at 2500 rpm and 4°C for 5 min. The precleared supernatant was transferred to a fresh Eppendorf tube.
Coupling of antigen to the antibody: The above supernatant was aliquoted to 500 μL, and the respective primary antibodies (anti-Rad51, H-300 for mouse Brca2 and Ab-1 for human BRCA2) were added to a final concentration of 10 μg/mL and mixed on a rotary shaker at 4°C for 1 h.
Precipitation of the immune complexes: Fifty microliters of Protein A Sepharose 4 Fast Flow suspension (50% slurry, described above) was added to the antigen–antibody complex and mixed on a rotary shaker at 4°C for 1 h. The complex was centrifuged at 12,000g for 20 sec, and both the pellet and the supernatant were saved to compare bound versus unbound protein. The pellet was washed three times with 1 mL of lysis buffer and once with wash buffer (50 mM Tris at pH 8.0).
Dissociation and analysis: Final pellets were suspended in 30 μL of 4× Nupage sample buffer (Invitrogen) containing 1× DTT. The samples were heated to 95°C for 3 min and centrifuged at 12,000g for 20 sec to remove the beads. The supernatant was removed and analyzed by immunoblot as described above.
Rad51 foci formation immunocytochemistry
Primary wild-type and Fancd2–/– MEFs were seeded on 18-mm coverslips and grown in DMEM + 15% FBS. Cells (untreated or treated with 12 Gy of ionizing radiation) were fixed 6 h after treatment with 4% paraformaldehyde at room temperature for 10 min and permeabilized with PBS + 0.5% Triton X-100 for 5 min. Cells were blocked in PBS-Tween 20 (0.25%) + 2% BSA at 4°C overnight. Anti-human RAD51 antibody (Oncogene) was used at a dilution of 1:1000 in PBS-Tween 20 (0.25%) + 2% BSA at room temperature for 1 h. Cells were washed four times for 5 min in PBS-Tween 20 (0.25%). Anti-rabbit-Cy3-conjugated secondary antibody (Upstate Biotechnology) was used at a dilution of 1:2000 in PBS-Tween 20 (0.25%) at room temperature for 1 h. Cells were washed four times for 5 min in PBS-Tween 20 (0.25%). Coverslips were mounted in a DAPI-containing antifade solution (Molecular Probes) and analyzed by fluorescence microscopy. Images were captured using Openlab software. The results are the mean of two independent experiments in which 200 cells were counted per experiment. Cells were scored positive if they contained >5 foci per nucleus.
Acknowledgments
We thank Alan D'Andrea for providing Fancd2 antibodies. This work was supported by NHLBI Program Project Grant 1PO1HL48546 (M.G.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1103403.
References
- Adelfalk, C., Lorenz, M., Serra, V., von Zglinicki, T., Hirsch-Kauffmann, M., and Schweiger, M. 2001. Accelerated telomere shortening in Fanconi anemia fibroblasts—A longitudinal study. FEBS Lett. 506: 22–26. [DOI] [PubMed] [Google Scholar]
- Akkari, Y.M., Bateman, R.L., Reifsteck, C.A., Olson, S.B., and Grompe, M. 2000. DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell Biol. 20: 8283–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter, B.P. 1993. Fanconi's anaemia and its variability. Br. J. Haematol. 85: 9–14. [DOI] [PubMed] [Google Scholar]
- Artandi, S.E., Chang, S., Lee, S.L., Alson, S., Gottlieb, G.J., Chin, L., and DePinho, R.A. 2000. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406: 641–645. [DOI] [PubMed] [Google Scholar]
- Auerbach, A.D., Buchwald, M., and Joenje, H. 2001. Fanconi anemia. In The Metabolic and molecular basis of inherited disease (ed. C.R. Scriver et al.), pp. 753–783. McGraw-Hill, New York.
- Baumann, P., Benson, F.E., and West, S.C. 1996. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87: 757–766. [DOI] [PubMed] [Google Scholar]
- Brummendorf, T.H., Rufer, N., Holyoake, T.L., Maciejewski, J., Barnett, M.J., Eaves, C.J., Eaves, A.C., Young, N., and Lansdorp, P.M. 2001. Telomere length dynamics in normal individuals and in patients with hematopoietic stem cell-associated disorders. Ann. NY Acad. Sci. 938: 293–304. [DOI] [PubMed] [Google Scholar]
- Centurion, S.A., Kuo, H.R., and Lambert, W.C. 2000. Damage-resistant DNA synthesis in Fanconi anemia cells treated with a DNA cross-linking agent. Exp. Cell Res. 260: 216–221. [DOI] [PubMed] [Google Scholar]
- Chang, S., Khoo, C., and DePinho, R.A. 2001. Modeling chromosomal instability and epithelial carcinogenesis in the telomerase-deficient mouse. Semin. Cancer Biol. 11: 227–239. [DOI] [PubMed] [Google Scholar]
- Chen, M., Tomkins, D.J., Auerbach, W., McKerlie, C., Youssoufian, H., Liu, L., Gan, O., Carreau, M., Auerbach, A., Groves, T., et al. 1996. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat. Genet. 12: 448–451. [DOI] [PubMed] [Google Scholar]
- Cheng, N.C., van de Vrugt, H.J., van der Valk, M.A., Oostra, A.B., Krimpenfort, P., de Vries, Y., Joenje, H., Berns, A., and Arwert, F. 2000. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum. Mol. Genet. 9: 1805–1811. [DOI] [PubMed] [Google Scholar]
- Connor, F., Bertwistle, D., Mee, P.J., Ross, G.M., Swift, S., Grigorieva, E., Tybulewicz, V.L., and Ashworth, A. 1997. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat. Genet. 17: 423–430. [DOI] [PubMed] [Google Scholar]
- D'Andrea, A.D. and Grompe, M. 2003. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 3: 23–34. [DOI] [PubMed] [Google Scholar]
- Davies, A.A., Masson, J.Y., McIlwraith, M.J., Stasiak, A.Z., Stasiak, A., Venkitaraman, A.R., and West, S.C. 2001. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7: 273–282. [DOI] [PubMed] [Google Scholar]
- de Winter, J.P., Waisfisz, Q., Rooimans, M.A., van Berkel, C.G., Bosnoyan-Collins, L., Alon, N., Carreau, M., Bender, O., Demuth, I., Schindler, D., et al. 1998. The Fanconi anaemia group G gene FANCG is identical with XRCC9. Nat. Genet. 20: 281–283. [DOI] [PubMed] [Google Scholar]
- de Winter, J.P., Leveille, F., van Berkel, C.G., Rooimans, M.A., van Der Weel, L., Steltenpool, J., Demuth, I., Morgan, N.V., Alon, N., Bosnoyan-Collins, L., et al. 2000a. Isolation of a cDNA representing the Fanconi anemia complementation group E gene. Am. J. Hum. Genet. 67: 1306–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winter, J.P., Rooimans, M.A., van Der Weel, L., van Berkel, C.G., Alon, N., Bosnoyan-Collins, L., de Groot, J., Zhi, Y., Waisfisz, Q., Pronk, J.C., et al. 2000b. The Fanconi anaemia gene FANCF encodes a novel protein with homology to ROM. Nat. Genet. 24: 15–16. [DOI] [PubMed] [Google Scholar]
- de Winter, J.P., van der Weel, L., de Groot, J., Stone, S., Waisfisz, Q., Arwert, F., Scheper, R.J., Kruyt, F.A., Hoatlin, M.E., and Joenje, H. 2000c. The Fanconi anemia protein FANCF forms a nuclear complex with FANCA, FANCC and FANCG. Hum. Mol. Genet. 9: 2665–2674. [DOI] [PubMed] [Google Scholar]
- Dobson, M.J., Pearlman, R.E., Karaiskakis, A., Spyropoulos, B., and Moens, P.B. 1994. Synaptonemal complex proteins: Occurrence, epitope mapping and chromosome disjunction. J. Cell Sci. 107 (Pt 10): 2749–2760. [DOI] [PubMed] [Google Scholar]
- Fanconi, G. 1967. Familial constitutional panmyelocytopathy, Fanconi's anemia (F.A.). I. Clinical aspects. Semin. Hematol. 4: 233–240. [PubMed] [Google Scholar]
- Garcia-Higuera, I., Kuang, Y., Naf, D., Wasik, J., and D'Andrea, A.D. 1999. Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol. Cell. Biol. 19: 4866–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera, I., Taniguchi, T., Ganesan, S., Meyn, M.S., Timmers, C., Hejna, J., Grompe, M., and D'Andrea, A.D. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7: 249–262. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D. and Sugino, A. 1988. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534. [DOI] [PubMed] [Google Scholar]
- Hanson, H., Mathew, C.J., Docherty, Z., and Mackie Ogilvie, C. 2001. Telomere shortening in Fanconi anaemia demonstrated by a direct FISH approach. Cytogenet. Cell Genet. 93: 203–206. [DOI] [PubMed] [Google Scholar]
- Howlett, N.G., Taniguchi, T., Olson, S., Cox, B., Waisfisz, Q., De Die-Smulders, C., Persky, N., Grompe, M., Joenje, H., Pals, G., et al. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297: 606–609. [DOI] [PubMed] [Google Scholar]
- Joenje, H., Oostra, A.B., Wijker, M., di Summa, F.M., van Berkel, C.G., Rooimans, M.A., Ebell, W., van Weel, M., Pronk, J.C., Buchwald, M., et al. 1997. Evidence for at least eight Fanconi anemia genes. Am. J. Hum. Genet. 61: 940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalter, H. 1968. Sporadic congenital malformations of newborn inbred mice. Teratology 1: 193–199. [DOI] [PubMed] [Google Scholar]
- Keegan, K.S., Holtzman, D.A., Plug, A.W., Christenson, E.R., Brainerd, E.E., Flaggs, G., Bentley, N.J., Taylor, E.M., Meyn, M.S., Moss, S.B., et al. 1996. The Atr and Atm protein kinases associate with different sites along meiotically pairing chromosomes. Genes & Dev. 10: 2423–2437. [DOI] [PubMed] [Google Scholar]
- Koomen, M., Cheng, N.C., van de Vrugt, H.J., Godthelp, B.C., van der Valk, M.A., Oostra, A.B., Zdzienicka, M.Z., Joenje, H., and Arwert, F. 2002. Reduced fertility and hypersensitivity to mitomycin C characterize Fancg/Xrcc9 null mice. Hum. Mol. Genet. 11: 273–281. [DOI] [PubMed] [Google Scholar]
- Kupfer, G.M., Naf, D., Suliman, A., Pulsipher, M., and D'Andrea, A.D. 1997. The Fanconi anaemia proteins, FAA and FAC, interact to form a nuclear complex. Nat. Genet. 17: 487–490. [DOI] [PubMed] [Google Scholar]
- Leteurtre, F., Li, X., Guardiola, P., Le Roux, G., Sergere, J.C., Richard, P., Carosella, E.D., and Gluckman, E. 1999. Accelerated telomere shortening and telomerase activation in Fanconi's anaemia. Br. J. Haematol. 105: 883–893. [DOI] [PubMed] [Google Scholar]
- Lo Ten Foe, J.R., Rooimans, M.A., Bosnoyan-Collins, L., Alon, N., Wijker, M., Parker, L., Lightfoot, J., Carreau, M., Callen, D.F., Savoia, A., et al. 1996. Expression cloning of a cDNA for the major Fanconi anaemia gene, FAA. Nat. Genet. 14: 320–323. [DOI] [PubMed] [Google Scholar]
- Lowry, O.H., Rosenbrough, N.J., Farr, A.L., and Randall, R.J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193: 265–275. [PubMed] [Google Scholar]
- Marmorstein, L.Y., Ouchi, T., and Aaronson, S.A. 1998. The BRCA2 gene product functionally interacts with p53 and RAD51. Proc. Natl. Acad. Sci. 95: 13869–13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister, K.A., Bennett, L.M., Houle, C.D., Ward, T., Malphurs, J., Collins, N.K., Cachafeiro, C., Haseman, J., Goulding, E.H., Bunch, D., et al. 2002. Cancer susceptibility of mice with a homozygous deletion in the COOH-terminal domain of the Brca2 gene. Cancer Res. 62: 990–994. [PubMed] [Google Scholar]
- Medhurst, A.L., Huber, P.A., Waisfisz, Q., de Winter, J.P., and Mathew, C.G. 2001. Direct interactions of the five known Fanconi anaemia proteins suggest a common functional pathway. Hum. Mol. Genet. 10: 423–429. [DOI] [PubMed] [Google Scholar]
- Miller, S.A., Dykes, D.D., and Polesky, H.F. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens, P.B., Heyting, C., Dietrich, A.J., van Raamsdonk, W., and Chen, Q. 1987. Synaptonemal complex antigen location and conservation. J. Cell Biol. 105: 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan, M.E., Cui, T.Y., and Jasin, M. 2001a. Homology-directed DNA repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 61: 4842–4850. [PubMed] [Google Scholar]
- Moynahan, M.E., Pierce, A.J., and Jasin, M. 2001b. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7: 263–272. [DOI] [PubMed] [Google Scholar]
- Mullis, K.B. and Faloona, F.A. 1987. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 155: 335–350. [DOI] [PubMed] [Google Scholar]
- Nakanishi, K., Taniguchi, T., Ranganathan, V., New, H.V., Moreau, L.A., Stotsky, M., Mathew, C.G., Kastan, M.B., Weaver, D.T., and D'Andrea, A.D. 2002. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat. Cell Biol. 4: 913–920. [DOI] [PubMed] [Google Scholar]
- Noll, M., Bateman, R.L., D'Andrea, A.D., and Grompe, M. 2001. Preclinical protocol for in vivo selection of hematopoietic stem cells corrected by gene therapy in Fanconi anemia group C. Mol. Ther. 3: 14–23. [DOI] [PubMed] [Google Scholar]
- Noll, M., Battaile, K.P., Bateman, R., Lax, T.P., Rathbun, K., Reifsteck, C., Bagby, G., Finegold, M., Olson, S., and Grompe, M. 2002. Fanconi anemia group A and C double-mutant mice. Functional evidence for a multi-protein Fanconi anemia complex. Exp. Hematol. 30: 679–688. [DOI] [PubMed] [Google Scholar]
- Olson, E.N., Arnold, H.H., Rigby, P.W., and Wold, B.J. 1996. Know your neighbors: Three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell 85: 1–4. [DOI] [PubMed] [Google Scholar]
- Painter, R.B. and Young, B.R. 1980. Radiosensitivity in ataxiatelangiectasia: A new explanation. Proc. Natl. Acad. Sci. 77: 7315–7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, A.H., Plug, A.W., van Vugt, M.J., and de Boer, P. 1997. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5: 66–68. [DOI] [PubMed] [Google Scholar]
- Plug, A.W., Clairmont, C.A., Sapi, E., Ashley, T., and Sweasy, J.B. 1997. Evidence for a role for DNA polymerase beta in mammalian meiosis. Proc. Natl. Acad. Sci. 94: 1327–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Trepat, M., Rouillard, D., Escarceller, M., Laquerbe, A., Moustacchi, E., and Papadopoulo, D. 2000. Arrest of S-phase progression is impaired in Fanconi anemia cells. Exp. Cell Res. 260: 208–215. [DOI] [PubMed] [Google Scholar]
- Sarkaria, J.N., Busby, E.C., Tibbetts, R.S., Roos, P., Taya, Y., Karnitz, L.M., and Abraham, R.T. 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59: 4375–4382. [PubMed] [Google Scholar]
- Sekine, M., Nagata, H., Tsuji, S., Hirai, Y., Fujimoto, S., Hatae, M., Kobayashi, I., Fujii, T., Nagata, I., Ushijima, K., et al. 2001. Localization of a novel susceptibility gene for familial ovarian cancer to chromosome 3p22-p25. Hum. Mol. Genet. 10: 1421–1429. [DOI] [PubMed] [Google Scholar]
- Sharan, S.K., Morimatsu, M., Albrecht, U., Lim, D.S., Regel, E., Dinh, C., Sands, A., Eichele, G., Hasty, P., and Bradley, A. 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386: 804–810. [DOI] [PubMed] [Google Scholar]
- Simsir, A., Palacios, D., Linehan, W.M., Merino, M.J., and Abati, A. 2001. Detection of loss of heterozygosity at chromosome 3p25-26 in primary and metastatic ovarian clear-cell carcinoma: Utilization of microdissection and polymerase chain reaction in archival tissues. Diagn. Cytopathol. 24: 328–332. [DOI] [PubMed] [Google Scholar]
- Storck, T., Kruth, U., Kolhekar, R., Sprengel, R., and Seeburg, P.H. 1996. Rapid construction in yeast of complex targeting vectors for gene manipulation in the mouse. Nucleic Acids Res. 24: 4594–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathdee, C.A., Gavish, H., Shannon, W.R., and Buchwald, M. 1992. Cloning of cDNAs for Fanconi's anaemia by functional complementation. Nature 356: 763–767. [DOI] [PubMed] [Google Scholar]
- Taniguchi, T., Garcia-Higuera, I., Andreassen, P.R., Gregory, R.C., Grompe, M., and D'Andrea, A.D. 2002a. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100: 2414–2420. [DOI] [PubMed] [Google Scholar]
- Taniguchi, T., Garcia-Higuera, I., Xu, B., Andreassen, P.R., Gregory, R.C., Kim, S.T., Lane, W.S., Kastan, M.B., and D'Andrea, A.D. 2002b. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109: 459–472. [DOI] [PubMed] [Google Scholar]
- Timmers, C., Taniguchi, T., Hejna, J., Reifsteck, C., Lucas, L., Bruun, D., Thayer, M., Cox, B., Olson, S., D'Andrea, A.D., et al. 2001. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol. Cell 7: 241–248. [DOI] [PubMed] [Google Scholar]
- Tutt, A., Bertwistle, D., Valentine, J., Gabriel, A., Swift, S., Ross, G., Griffin, C., Thacker, J., and Ashworth, A. 2001. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 20: 4704–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney, M.A., Royle, G., Low, M.J., Kelly, M.A., Axthelm, M.K., Reifsteck, C., Olson, S., Braun, R.E., Heinrich, M.C., Rathbun, R.K., et al. 1996. Germ cell defects and hematopoietic hypersensitivity to γ-interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood 88: 49–58. [PubMed] [Google Scholar]
- Xu, Y., Ashley, T., Brainerd, E.E., Bronson, R.T., Meyn, M.S., and Baltimore, D. 1996. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes & Dev. 10: 2411–2422. [DOI] [PubMed] [Google Scholar]
- Yang, H., Jeffrey, P.D., Miller, J., Kinnucan, E., Sun, Y., Thoma, N.H., Zheng, N., Chen, P.L., Lee, W.H., and Pavletich, N.P. 2002. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297: 1837–1848. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Kuang, Y., De Oca, R.M., Hays, T., Moreau, L., Lu, N., Seed, B., and D'Andrea, A.D. 2001. Targeted disruption of the murine Fanconi anemia gene, Fancg/Xrcc9. Blood 98: 3435–3440. [DOI] [PubMed] [Google Scholar]
- Yu, V.P., Koehler, M., Steinlein, C., Schmid, M., Hanakahi, L.A., van Gool, A.J., West, S.C., and Venkitaraman, A.R. 2000. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes & Dev. 14: 1400–1406. [PMC free article] [PubMed] [Google Scholar]
- Zhang, G.L. and Xu, K.L. 2002. Loss of heterozygosity at chromosome 3p in epithelial ovarian cancer in China. Int. J. Gynecol. Cancer 12: 198–201. [PubMed] [Google Scholar]