Abstract
The HOX/HOM superfamily of homeodomain proteins controls cell fate and segmental embryonic patterning by a mechanism that is conserved in all metazoans. The linear arrangement of the Hox genes on the chromosome correlates with the spatial distribution of HOX protein expression along the anterior–posterior axis of the embryo. Most HOX proteins bind DNA cooperatively with members of the PBC family of TALE-type homeodomain proteins, which includes human Pbx1. Cooperative DNA binding between HOX and PBC proteins requires a residue N-terminal to the HOX homeodomain termed the hexapeptide, which differs significantly in sequence between anterior- and posterior-regulating HOX proteins. We report here the 1.9-Å-resolution structure of a posterior HOX protein, HoxA9, complexed with Pbx1 and DNA, which reveals that the posterior Hox hexapeptide adopts an altered conformation as compared with that seen in previously determined anterior HOX/PBC structures. The additional nonspecific interactions and altered DNA conformation in this structure account for the stronger DNA-binding affinity and altered specificity observed for posterior HOX proteins when compared with anterior HOX proteins. DNA-binding studies of wild-type and mutant HoxA9 and HoxB1 show residues in the N-terminal arm of the homeodomains are critical for proper DNA sequence recognition despite lack of direct contact by these residues to the DNA bases. These results help shed light on the mechanism of transcriptional regulation by HOX proteins and show how DNA-binding proteins may use indirect contacts to determine sequence specificity.
Keywords: HoxA9, Pbx1, Hox, posterior prevalence, DNA binding
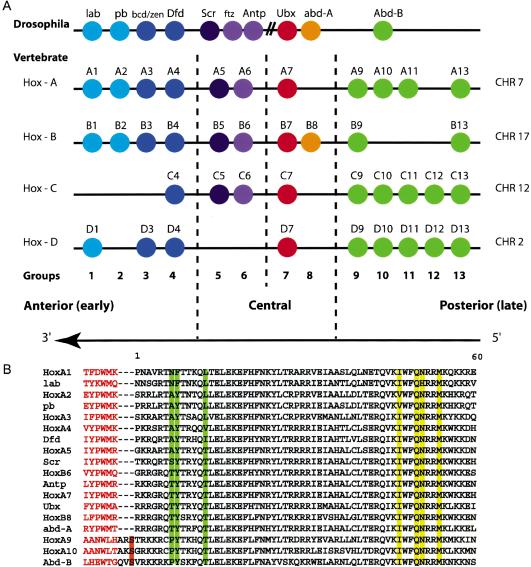
The large family of Hox transcription factors has critical roles in early morphological development and later cell differentiation (McGinnis and Krumlauf 1992; Krumlauf 1994; Lawrence et al. 1996; Magli et al. 1997). Humans have 39 Hox genes arranged in four clusters (HoxA–HoxD). The linear arrangement of genes within each cluster facilitates controlled spatial and temporal expression along the anterior–posterior axis of the body (Fig. 1A). Expression of Hox genes in the wrong segment at the wrong time often results in homeotic transformation of that segment (Lamka et al. 1992; Morgan et al. 1992; Duboule 1994a). Hox homeodomains contain identical DNA-base-contacting residues (Fig. 1B) and have very similar DNA sequence specificity, generally preferring a core DNA-binding site containing the sequence 5′-TTNAT-3′, whose third base depends on the particular Hox protein (Laughon 1991). Cooperative DNA binding with other protein cofactors, such as the PBC family of homeodomain proteins, is thought to enhance the specificity of Hox proteins and thereby allow them to carry out distinct functions (Chan et al. 1994; Chang et al. 1995; Mann and Chan 1996). The PBC family, which includes human Pbx1 and Drosophila Exd, belongs to a class of proteins that contains the TALE (three-amino-acid loop extension)-type homeodomain, so named because of a three-amino-acid insertion between homeodomain residues 23 and 24 in the loop between helices 1 and 2 (Mann 1995; Mann and Chan 1996; Bürglin 1997). A subset of vertebrate Hox proteins interacts with Pbx1 via a conserved six-amino-acid motif, or hexapeptide, enabling cooperative DNA binding (Chang et al. 1995; Neuteboom et al. 1995; Peltenburg and Murre 1996; Shen et al. 1996, 1997; Passner et al. 1999; Piper et al. 1999).
Figure 1.
(A) Arrangement of the Hox genes of the Drosophila and vertebrate clusters. The arrangement 5′ to 3′ is conserved across these species. Members of the same paralog group are colored identically. Abd-B-like Hox genes are shown in green. (B) Alignment of the homeodomain sequences in groups 1–10 of Drosophila and vertebrates shows extensive sequence conservation. Broken lines indicate where linkers of variable lengths would be. The three-amino-acid linkers of the Abd-B-like Hox proteins are shown with the conserved Ser –1 highlighted in orange. The hexapeptides are shown in red text, and the DNA recognition residues are highlighted in yellow. Positions 7, 8, and 13 of the homeodomains are highlighted in green.
Hox proteins required for anterior development are expressed earliest and are restricted to anterior domains. The more posterior proteins are expressed later in development and in more posterior domains (Duboule 1994b; Krumlauf 1994). This character is critical to their in vivo function. In general, when posterior proteins are expressed ectopically in anterior domains, those domains are transformed to have posterior characteristics. However, when anterior Hox proteins are expressed in posterior domains, transformation to an anterior phenotype is not observed. This phenomenon has been referred to as posterior prevalence. Posterior prevalence has been postulated to occur through competition for binding sites, or competition for cofactors, because the effect occurs without changes in the level of expression of endogenous Hox genes, and in some cases transformations are dose-dependent (Cribbs et al. 1995). Competition for binding sites has in the past seemed unlikely because of varying binding-site preference among anterior and posterior Hox proteins. Anterior Hox proteins prefer a G at the second position of the 5′-TNAT/CNNN-3′ half-site, middle Hox proteins accept a G or A, whereas the more posterior proteins bind G, A, or T. A preference for C in the fourth position is exhibited by the most posterior Hox proteins (Shen et al. 1997). Given that Hox proteins contain the same four helix-3 residues that make direct DNA base contacts in the major groove, this specificity has been postulated to be determined by residues in the N-terminal arm through interactions with the minor groove of DNA (Ekker et al. 1994) and modulated by the interaction with PBC proteins (Chang et al. 1996; Passner et al. 1999; Piper et al. 1999).
The structures of two Hox/Hom-C–PBC–DNA ternary complexes were previously determined (Passner et al. 1999; Piper et al. 1999). HoxB1–Pbx1–DNA is an example of an anterior-regulating vertebrate Hox protein complex bound to a sequence containing the anterior Hox core binding site, 5′-TTGAT-3′ (Piper et al. 1999). The Ultrabithorax (Ubx)–EXD–DNA structure containing the core DNA-binding site, 5′-TTTAT-3′, is an example of a central–posterior Hox protein complex, because Ubx is the Drosophila protein homologous to proteins in paralog group 7 (Fig. 1; Passner et al. 1999). The Hox protein in each structure interacts with the PBC protein using its hexapeptide motif, which has the sequence TFDWM in the case of HoxB1 (Piper et al. 1999) and FYPWM in the case of Ubx (Passner et al. 1999; Piper et al. 1999). In both cases, the hexapeptide motif forms a 310-helix that packs against the PBC homeodomain and inserts the conserved tryptophan into the hexapeptide binding pocket. The Hox linker region and the first four amino acids of the homeodomain are disordered in both structures (Passner et al. 1999; Piper et al. 1999).
This work describes the structure of the HoxA9 and Pbx1 homeodomains bound to a DNA fragment containing the sequence 5′-ATGATTTACGAC-3′. This sequence was identified as a high-affinity binding site for HoxA9–Pbx1 (Shen et al. 1997) and differs in the Hox–Pbx core recognition sequence by 1 bp from an in vivo binding site (Taylor 1998; Pan et al. 2001). HoxA9 is a posterior-regulating Hox protein required for proper limb development in mammals and is implicated as a factor in the induction of Acute Myeloid Leukemias (AMLs; Kroon et al. 1998; Lawrence et al. 1999; Thorsteinsdottir et al. 2001). The structure described here is the first containing an Abd-B-like Hox homeodomain, so named for their similarity to Drosophila Abdominal-B (Abd-B). Members of this class of HOX proteins contain a variant hexapeptide (AANWLH) and a three-amino-acid linker, which is the shortest found among Hox proteins. In contrast to previous structures, all the residues in the homeodomain arm and linker are well ordered in the structure. Strikingly, the divergent HoxA9 hexapeptide adopts a notably different conformation as compared with that found in the other two known structures, while preserving the previously observed location of the tryptophan within the Pbx pocket. Comparisons with the two other Hox–PBC–DNA structures suggest that variation in DNA backbone interactions may be correlated with differences in minor groove compression, which in turn affects contacts with the variant base. These observations suggest that site specificity may be determined to some extent by residues within the globular portion of the homeodomain that contact the backbone, rather than the base pairs. We show through DNA-binding studies that mutation of residues that contact the DNA backbone in a region where the minor groove is compressed reduces DNA-binding specificity. These results suggest that in the context of HoxA9, these residues influence binding-site specificity either by inducing a DNA distortion or by favoring a particular backbone conformation. We find that Pbx1 does not influence binding-site specificity with HoxA9 but does increase the site specificity of HoxB1. This suggests that Pbx1 only influences the specificity of a subset of the Hox proteins with which it binds DNA. We also show that HoxA9 binds its high-affinity binding site 20-fold more tightly than HoxB1 binds its high-affinity site, consistent with the structural observation that HoxA9 forms 60% more contacts with the DNA than does HoxB1. This provides a basis for increasing DNA binding affinity anterior to posterior. Based on these findings, we present a plausible model for posterior prevalence that explains how Hox proteins may recognize their target sites and compete for DNA sites of more anterior Hox proteins.
Results
Overview of the complex
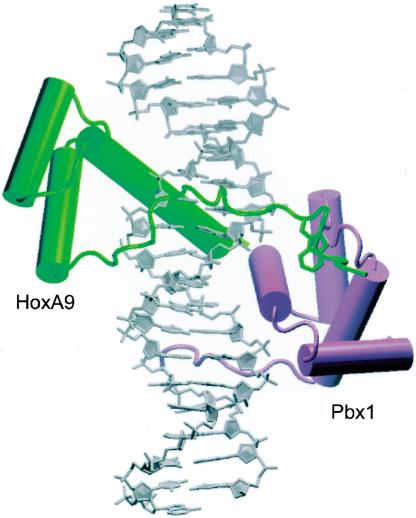
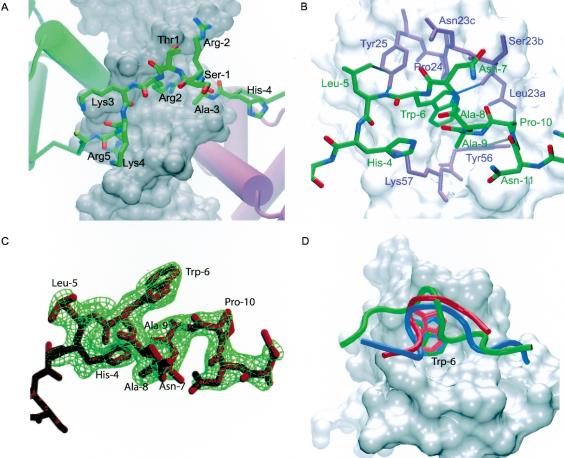
The 1.9-Å-resolution ternary complex structure (see Fig. 2) contains HoxA9 and Pbx1 bound to a 20-bp DNA duplex containing the consensus HoxA9–Pbx1-binding site, 5′-ATGATTTACGAC-3′, as identified by PCR-mediated site selection assays (Shen et al. 1997). The model of the complex reported here contains residues –12 to 64 of HoxA9, 1–70 of Pbx1, the full 20-bp DNA duplex, and 247 water molecules (see Fig. 1B for explanation of protein numbering scheme). Two residues on the N terminus and two residues on the C terminus are disordered in the HoxA9 model, and 14 residues on the C terminus are disordered in the Pbx1 model.
Figure 2.
The HoxA9–Pbx1–DNA complex. HoxA9 is shown in green, Pbx1 in purple. The linker and the hexapeptide of HoxA9 spans the minor groove (front), and the conserved tryptophan is inserted in the Pbx1 binding pocket. This figure and subsequent structure graphics were made using VMD.
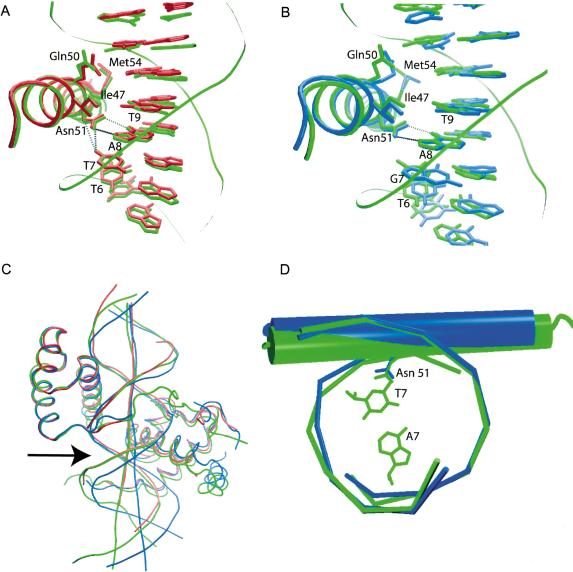
The Hox and Pbx homeodomains are arranged head-to-tail, contacting overlapping sites on nearly opposite faces of the DNA (Fig. 2). The third helix of each homeodomain lies in the major groove and contacts the core bases of the DNA recognition site, while the N-terminal arm of each homeodomain makes contacts in the minor groove (Figs. 2, 4, below). The DNA is bent around the HoxA9 recognition helix by 20.2° and to a lesser extent (11.0°) around that of Pbx1. The orientation of HoxA9 relative to Pbx1 is identical to that seen in both of the previously solved Hox–PBC–DNA complexes (Passner et al. 1999; Piper et al. 1999). The three α-helices of the HoxA9 homeodomain align closely with those of HoxB1 and Ubx, with a root mean square difference (RMSD) in main-chain atom positions of 0.52 Å2 for HoxB1 and 0.48 Å2 for Ubx (Fig. 5A,B, below). One difference in the present structure is that the recognition helix of HoxA9 is longer than that of HoxB1 by three residues.
Figure 4.
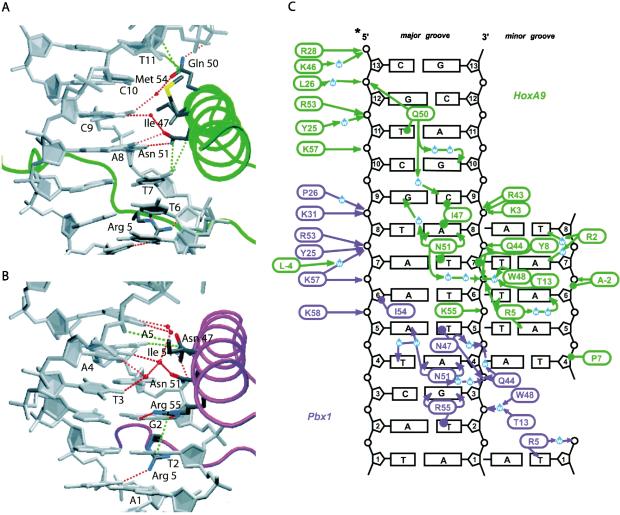
DNA contacts made by HoxA9 (A) and Pbx1 (B). Minor groove interactions are made by Arg 5 in the back for both homeodomains. Hydrophobic interactions (green) and hydrogen bonds (red) are shown as broken lines. (C) Schematic of HoxA9 (green) and Pbx1 (purple) DNA interactions. Hydrogen bonds are indicated with arrows, and van der Waals interactions are shown as lines terminating in circles. Minor groove interactions are on the left side of the figure. The HoxA9/Pbx1 site is 5′-A1T2G3A4T5T6T7A8C9G10A11C12-3′.
Figure 5.
Alignment of the third helix of HoxA9 (green) with Ubx (A; red) and HoxB1 (B; blue). The interaction of Asn 51 with T7 is conserved in Ubx, but may not be made in HoxB1. (C) Alignment of the full homeodomain of Ubx (red) and HoxB1 (blue) with HoxA9 (green) shows that the DNA in both the HoxA9 and Ubx complexes contains a bend (arrow) relative to HoxB1. (D) Alignment of the DNA in the region of the Hox binding sites of HoxA9 (green) and HoxB1 (blue) shows a shift of the HoxA9 recognition helix and Asn 51 into the major groove relative to HoxB1.
Pbx1 is a TALE-type homeodomain with a fourth helix at the C terminus (Fig. 2). The Pbx1 homeodomain in the present structure is virtually identical to that observed in the previously solved complexes (RMSD of 0.30 Å for the superposition of the first three helices of Pbx1 with that in the complex with HoxB1, and 0.52 Å for Pbx1 in the present structure superimposed on Exd in complex with Ubx), except that helix 4 is shorter than that seen in the HoxB1/Pbx1/DNA structure (Piper et al. 1999). The residues comprising this helix were not included in the protein fragment in the Ubx/Exd structure (Passner et al. 1999).
The HoxA9 hexapeptide has a unique conformation
In contrast with previous structures of Hox proteins (Billeter et al. 1993; Fraenkel et al. 1998; Passner et al. 1999; Piper et al. 1999), the full N-terminal arm and linker joining the hexapeptide with the homeodomain of HoxA9 are well ordered (Fig. 3A). The longer linker regions of HoxB1 (18 residues) and Ubx (7 residues) are disordered in structures of those proteins, as are the first four residues of their respective N-terminal arms. The three-amino-acid linker of HoxA9 forms a type 1 β-turn between residues –2 and 2 (Figs. 1B, 3A). As a result, residue Thr 1 and the conserved Ser –1 are forced outward, away from the DNA (Fig. 3A). Despite its short length, the three amino acids comprising the linker are sufficient to span the minor groove to allow the HoxA9 hexapeptide to interact with Pbx1.
Figure 3.
The linker and the hexapeptide of HoxA9. (A) Detailed view of the N-terminal arm and the linker region of the HoxA9 homeodomain spanning the minor groove. The linker region is composed of Ala –3, Arg –2, and Ser –1. Arg 2 makes water-mediated contacts in the minor groove, and Lys 4 and Ala –3 contact the sugar-phosphate backbone. (B) The HoxA9 hexapeptide (green) in the Pbx1 hexapeptide-binding pocket (purple residues and white surface). Hydrophobic interactions and hydrogen bonds between HoxA9 and Pbx1 are shown in gray and blue, respectively. The TALE insertion in the Pbx1 homeodomain is composed of Leu 23a, Ser 23b, and Asn 23c. (C) Simulated annealing omit map contoured at 3 σ for residues –12 to –4 of HoxA9. The hexapeptide residues as well as additional residues in the N terminus are shown. (D) Comparison of the hexapeptide backbone conformations of HoxA9 (green), HoxB1 (blue), and Ubx (red). The PBC homeodomains of the HoxB1 and Ubx complexes were aligned with the Pbx1 homeodomain (white) of the HoxA9 complex to overlay the peptides. The conserved tryptophan is labeled as Trp –6.
The structure of the HoxA9 hexapeptide is markedly different from that in the HoxB1 and Ubx complex structures (Fig. 3D; Passner et al. 1999; Piper et al. 1999). The hexapeptide of HoxA9 forms two type 1 β-turns (Fig. 3B,C) that include the entire hexapeptide as well as the two neighboring N-terminal residues. The sixth hexapeptide residue, His –4, makes a hydrogen bond with its second indole nitrogen and the carbonyl oxygen of Ala –9. A hydrogen bond is also observed between the peptide backbone carbonyl oxygen of Pro –10 to the Trp –6 indole nitrogen (Fig. 3B,C). Ala –9 packs over the hydrophobic tryptophan residue. In contrast, the hexapeptides of Ubx and HoxB1 adopt an entirely different conformation, forming a 310-helix in which a methionine packs over the conserved tryptophan (Fig. 3D; Passner et al. 1999; Piper et al. 1999). With the exception of the identical position of the conserved tryptophan residue within the hexapeptide-binding pocket of Pbx1, there is no relation between the structure or binding of the HoxA9 hexapeptide and that of HoxB1 or Ubx (Fig. 3D).
A mixture of van der Waals and hydrogen-bond contacts stabilizes the interaction between HoxA9 and Pbx1 (Fig. 3B). The conserved Trp –6 of the HoxA9 hexapeptide inserts into the hexapeptide-binding pocket of Pbx1 formed by the three-amino-acid loop insertion between helices 1 and 2, Leu 23a, Ser 23b, and Asn 23c, and the C-terminal end of helix 3 (Fig. 3B), and is covered by Ala –8 instead of Met as seen in the other two structures (Passner et al. 1999; Piper et al. 1999). A stabilizing hydrogen bond is formed between the indole nitrogen of Trp –6 and the backbone carbonyl of Pbx1 residue Leu 23a as is seen in previous Hox/PBC/DNA structures. His –4 of HoxA9 forms a hydrogen bond with the terminal amide of Lys 57 and is also within optimal van der Waals contact distance of the aliphatic portion of the Lys 57 side chain. Leu –5 makes van der Waals contact with the aromatic ring of Tyr 25, while the backbone amide of Leu –5 forms a hydrogen bond to the Tyr 25 hydroxyl. In addition, the peptide carbonyl of Leu –5 forms a water-mediated hydrogen bond with the sugar-phosphate backbone of the DNA. The interaction between HoxA9 and Pbx1 buries 742 Å2, which is 100 Å2 less than that between HoxB1 and Pbx1 or Ubx and Exd.
Minor groove contacts by HoxA9 induce minor groove compression and DNA bending
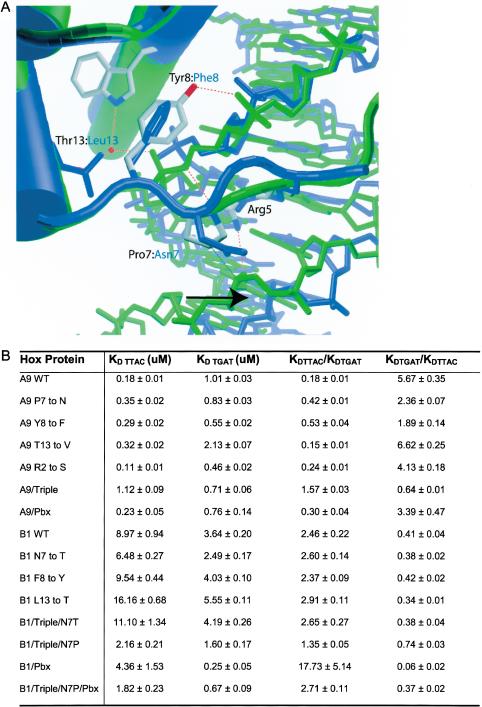
Contacts made by HoxA9 in the minor groove induce deformations in the DNA that promote bending around the homeodomain recognition helix. A series of contacts mediated by HoxA9 N-terminal arm residues Pro 7 and Tyr 8, and helix-1 residue Thr 13 result in significant minor groove compression (Fig. 4A,C). The result is a 20.2° bend of the DNA around the HoxA9 recognition helix, as compared with a 10.0° bend around the HoxB1 helix (Fig. 5C). Tyr 8 makes van der Waals contact with the sugar-phosphate backbone between T6 and T7 (Fig. 6A). In addition, Tyr 8 hydrogen-bonds with a phosphate oxygen via its hydroxyl group. A second hydrogen bond is observed between the peptide backbone amide nitrogen of Tyr 8 and the sugar-phosphate backbone between T6 and T7 (Fig. 6A). Combined with a van der Waals contact from Pro 7 to the DNA backbone on the other strand of the minor groove between T4 and A5, these interactions result in a bridge that compresses the minor groove. The hydrophobic interaction from residue 7 is preserved in the structure of Ubx, and the same DNA minor groove compression is observed, whereas the HoxB1 complex structure contains no interaction between residue 7 (Asn) and the DNA and therefore no minor groove compression. In addition, Thr 13 may help to stabilize the groove compression by hydrogen-bonding with a water that mediates a hydrogen bond between the indole nitrogen of Trp 48 and the sugar-phosphate backbone in the HoxA9 and Ubx structures (Fig. 6A). HoxB1, in contrast, contains a leucine at this position and cannot participate in the same hydrogen-bonding network.
Figure 6.
DNA-binding studies of HoxA9 and HoxB1. (A) Residues 7, 8, and 13 of HoxA9 contact the sugar-phosphate backbone and allow Arg 5 to make contact across the minor groove in HoxA9, whereas in HoxB1 no such contacts are observed. (B) Table of equilibrium dissociation constants measured by gel retardation assays using wild-type and mutant HoxA9 and HoxB1 on DNA containing the consensus binding sites for HoxA9 (TTTAC) and HoxB1 (TTGAT).
Major groove interactions of the HoxA9/Pbx1 complex
Contacts with the DNA major groove are mediated by helix-3 residues of HoxA9 with the HoxA9 site 5′-T5T6T7A8C9G10A11C12-3′ (see Fig. 4A,C). With respect to the HoxA9-binding site, T5 and T6 are specified through interactions with Arg 5. A8 is specified through a bidentate interaction with Asn 51, conserved among Hox homeodomains, and an interaction of its base pair with Arg 5 in the minor groove. Because of the DNA bending induced in the minor groove, Asn 51 is also able to form van der Waals interactions with T7, the variant Hox core base. Contacts on the minor groove side between A7* and Arg 2 further select for this base pair. Ile 47 and Glu 50 interact with C9 and the interaction of G9* with Asn 51 completes DNA recognition of that position. Ile 47 also adopts an alternate conformation that aligns well with that observed in the structure of the Ubx complex. The Ubx complex contains a T at position 9 of the DNA, and HoxA9 is able to bind both a T and a C at this position (Shen et al. 1997; data not shown). G10 is specified through interaction of C10* with Met 54. Glu 50, thought to be a critical residue for determining specificity at bases 10 and 11, hydrogen-bonds with T11* and G12* (Fig. 4A,C).
Of the three Hox structures that have been determined—HoxB1/Pbx1–DNA (Piper et al. 1999), HoxA9/Pbx1–DNA, and Ubx/exd–DNA (Passner et al. 1999)— HoxA9 forms the greatest number of interactions with the DNA and HoxB1 forms the least (Fig. 5A,B). The overall number of indirect DNA contacts is greater in the HoxA9 structure than in either of the other complexes (Fig. 4C). Overall, HoxA9 binding to DNA is more similar to Ubx than to HoxB1. Interactions of Pbx1 with DNA in this structure are the same as those observed in previous structures (see Fig. 4B,C). In the major groove, Asn 47 makes a van der Waals interaction with the T5 of the consensus site (see Fig. 4B,C). Asn 51 forms bidentate hydrogen-bond interactions with the A4, while Arg 55 makes bidentate hydrogen-bond interactions with the G3 and a van der Waals interaction with the T2. In the minor groove, Arg 5 makes a hydrogen bond with the T1.
Residues 7, 8, and 13 of HoxA9 and HoxB1 influence Hox DNA site recognition
To investigate how N-terminal arm contacts influence DNA specificity of Hox proteins, the DNA-binding specificity of wild-type HoxA9 and HoxB1 were measured and compared with HoxA9 and HoxB1 mutated at positions 7, 8, and 13. The dissociation equilibrium constants (KD) for wild-type HoxA9 and HoxB1 were measured using labeled oligonucleotides containing the sequence TTTAC (HoxA9 site) or TTGAT (HoxB1 site). Previous studies (Shen et al. 1997) showed that the third base in this site is differently recognized by HoxA9 and HoxB1, but HoxA9 can bind equally well to sites containing either a C or a T at the fifth position (Shen et al. 1997; data not shown). As expected, HoxA9 prefers the HoxA9 site, and HoxB1 prefers the HoxB1 site (Fig. 6B).
The degree of specificity can be expressed as the ratio of the KD of binding to TTGAT to the KD of binding to TTTAC (KD (TTGAT)/KD (TTTAC)). For HoxA9 only, that ratio is 5.67 ± 0.35, indicating that HoxA9 prefers TTTAC sixfold more than TTGAT. The HoxB1 ratio is 0.41 ± 0.04, indicating HoxB1 prefers TTGAT 2.5-fold more than TTTAC. Interestingly, Pbx1 does not improve the specificity of HoxA9, but renders HoxB1 significantly more specific for the HoxB1 site (Fig. 6B). When binding DNA with Pbx1, HoxA9 shows less discrimination between the sites, showing only a 3.5-fold preference for the HoxA9 site, whereas HoxB1 shows significantly more discrimination, with a 18-fold preference for the HoxB1 site (Fig. 6B). Overall, HoxA9 binds to its high-affinity DNA site (TTTAC) 20-fold more tightly than HoxB1 binds to its high-affinity DNA site (TTGAT) [KD (TTTAC) = 0.18 ± 0.01 μM for HoxA9, KD (TTGAT) = 3.64 ± 0.24 μM for HoxB1; Fig. 6B]. Although HoxA9 prefers TTTAC, HoxA9 binds TTGAT three times tighter than HoxB1 binds TTGAT on its own (KD (TTGAT) = 1.01 ± 0.03 μM for HoxA9). However, with Pbx1, HoxA9 binds the HoxB1 site twofold less tightly than HoxB1 (Fig. 6B).
Mutations were made in the HoxA9 homeodomain residues 7 and 8 to those found in HoxB1. These mutations, Pro 7 to Asn and Tyr 8 to Phe, decreased HoxA9 preference for the HoxA9 site versus the HoxB1 site. Both mutants had a twofold preference for the HoxA9 site (2.36 ± 0.07 for Pro 7 to Asn, 1.89 ± 0.14 for Tyr 8 to Phe), which reflects a threefold decrease in specificity compared with the wild-type protein (Fig. 6). The substitution of HoxA9 residue Thr 13 with Val 1, which tests the importance of the water-mediated hydrogen bond between Thr 13 and the sugar-phosphate backbone, does not decrease specificity, however, and has a slightly increased ratio of binding (6.62 ± 0.25; Fig. 6).
Single mutations of the corresponding residues in the HoxB1 homeodomain did not, however, have the effect of decreasing the preference for TTGAT over TTTAC when HoxB1 binds DNA on its own. All three single mutants containing the mutations Asn 7 to Thr, Phe 8 to Tyr, and Leu 13 to Thr have ratios of binding similar to wild type (0.38 ± 0.02 for Asn 7 to Thr, 0.42 ± 0.02 for Phe 8 to Tyr, 0.34 ± 0.01 for Leu 13 to Thr and 0.38 ± 0.04 for the triple mutant; Fig. 6B). This indicates that these residues have little effect on binding-site specificity in the context of the HoxB1 protein on its own.
To determine the effect of mutating all three residues, residues 7, 8, and 13 in HoxA9 were mutated to those present at these positions in HoxB1 and vice versa. Most significantly, the HoxA9 triple mutant (Pro 7 to Asn, Tyr 8 to Phe, and Thr 13 to Leu) exhibits a reversal of binding-site specificity (0.64 ± 0.02), indicating a slight preference for TTGAT over TTTAC (Fig. 6B). These N-terminal arm and helix-1 residues are therefore able to influence site specificity significantly in the context of the HoxA9 homeodomain, as predicted from structural observations. The HoxB1 triple mutant (Asn 7 to Pro, Phe 8 to Tyr, and Leu 13 to Thr) exhibits a modest twofold decrease in preference for TTGAT over TTTAC, with a ratio of 0.74 ± 0.03 (Fig. 6B). Because the preference for TTGAT over TTTAC is significantly increased for HoxB1 in the presence of Pbx1, the HoxB1 triple mutant was tested in the presence of Pbx1. There is a significant decrease in preference for TTGAT over TTTAC in the presence of Pbx1 for the HoxB1 triple mutant, with a ratio of 0.38 ± 0.02, compared with 0.06 ± 0.02 for the HoxB1 wild-type protein in the presence of Pbx1 (Fig. 6B). Therefore, the triple mutations act together to decrease the preference for the HoxB1 binding site, and also serve to decrease the effect of Pbx on increasing preference of HoxB1 for TTGAT. These results also support the hypothesis, based on structure, that residues 7, 8, and 13 play a role in determining binding-site preference.
Discussion
The HoxA9–Pbx1–DNA structure helps illuminate several questions regarding the mechanism of transcriptional regulation by Hox proteins. Comparison of this structure with the other two known Hox ternary complex structures provides new insights into the molecular determinants of Hox recognition of DNA sequence and the role that protein partners play. A striking observation is that the total number of nonspecific interactions with DNA is greater the more posterior the Hox protein is, with the number increased by 60% in HoxA9 and 30% in Ubx as compared with HoxB1 (Fig. 4C; Passner et al. 1999; Piper et al. 1999). This increased number of contacts accounts for the overall increase in DNA-binding affinity observed for HoxA9 as compared with HoxB1. Despite differences in binding-site preference, the number of direct base contacts does not differ significantly. This increase in nonspecific DNA-binding contacts in the more posterior proteins may be an important part of a mechanism that allows posterior Hox proteins to compete with anterior Hox proteins at anterior Hox binding sites.
Hox DNA-binding site specificity
The determinants for recognition at the variant Hox core position revealed by the present structure highlight the importance of both backbone and base contacts in determining DNA sequence specificity (Fig. 4, 5, 6). HoxB1 prefers a G, whereas both HoxA9 and Ubx prefer a T at that position. HoxA9 and Ubx make clear van der Waals contact with the methyl group at that residue (Fig. 5A). This van der Waals interaction between Asn 51 and the variant base, not observed in the HoxB1 complex structure, is made possible by a DNA distortion that allows the variant base and Asn 51 to be positioned closer together (Fig. 5). Optimal contacts with the DNA distortion are a result of interactions of residues 7, 8, and 13 in the homeodomain N-terminal arm of HoxA9 and Ubx with the sugar-phosphate backbone and in the minor groove of the DNA (Fig. 6). These interactions likely distort the DNA relative to the HoxB1 DNA, which allows Asn 51 access to this interaction in the case of HoxA9 and Ubx. It is also possible that the DNA distortion is, in part, caused by the single base difference between the HoxA9 and HoxB1 DNA-binding sites. In either case, the contacts formed by homeodomain residues 7, 8, and 13 depend on the observed conformation of the DNA backbone. Ubx, which also prefers a T at position 7, also binds DNA that contains a similar set of distortions in the DNA (Passner et al. 1999). Our binding studies of HoxA9 containing mutations at residues 7, 8, and 13 confirm that these residues are capable of influencing DNA-binding specificity in HoxA9 (Fig. 6). Because these residues vary among paralog groups, their influence on contacts made by Asn 51 could account for differing Hox specificities. However, single mutations of these residues in HoxB1 had no effect on DNA-binding specificity, indicating that, in the case of HoxB1, there are other factors responsible for DNA-binding selectivity of the variant base pair. The triple mutant of HoxB1 in which all three residues at positions 7, 8, and 13 were changed to the residues present at these positions of the HoxA9 homeodomain (i.e., Asn 7 to Pro, Phe 8 to Tyr, and Leu 13 to Thr) showed only a twofold decrease in preference for TTGAT over TTTAC, but in the presence of Pbx1, there is a decrease in preference for TTGAT over TTTAC by about sixfold, indicating that Pbx1 does have significant influence on the specificity decision of the mutated HoxB1 homeodomain (Fig. 6B,C). The varying effect of Pbx1 on the DNA-binding specificity of different Hox proteins remains to be explained.
The variant HoxA9 hexapeptide and three-amino-acid linker
The structure of the HoxA9/Pbx/DNA complex shows that Hox hexapeptides can have surprisingly large variations in conformation and sequence, yet still bind specifically to their partner proteins. The HoxA9 hexapeptide is significantly diverged in sequence from the consensus sequence for Hox paralog groups 1 through 8. We find that the conformation of the HoxA9 hexapeptide is significantly different from that observed for HoxB1 and Ubx (Fig. 5B–D). However, the position of the invariant tryptophan is well conserved (Fig. 5D), suggesting that the tryptophan may be the principal determinant for binding to the pocket found in Pbx1 and EXD. Other divergent hexapeptide sequences (Fig. 1B) may therefore be expected to bind to Pbx1 or Exd with their tryptophan residues in the conserved orientation observed here, but there is clearly not a unique structure that a hexapeptide must adopt to bind Pbx1/Exd.
Unlike previous structures, the residues joining the hexapeptide to the Hox homeodomain are well ordered in the HoxA9/Pbx–DNA structure. The three-amino-acid linker, present in all Abd-B-like Hox proteins that interact with PBC proteins, lies in the minor groove but does not appear to contribute to DNA specificity. Ala –3, at the N terminus of the linker, does make DNA contacts to the sugar-phosphate backbone of the DNA, however, and may provide added DNA-binding energy. The conserved Ser –1 is projected away from the DNA along with Thr 1 in the linker. Because this Ser is a protein kinase C phosphorylation consensus site and several homeodomains are shown to be regulated by phosphorylation (Jaffe et al. 1997; Berry and Gehring 2000; Eklund et al. 2000; Yaron et al. 2001), there is a possibility that this residue is conserved because it is required for phosphorylation-mediated regulation.
In light of the extensive interactions between the HoxA9 hexapeptide and Pbx1, as well as the contacts between the N-terminal arm of HoxA9 with the DNA minor groove, it is surprising that insignificant cooperativity was observed in our assays of HoxA9–Pbx1 binding. The HoxA9 hexapeptide motif was previously shown to be important for transformation of hematopoietic cells (Schnabel et al. 2000). In particular, a point mutation substituting the tryptophan in the hexapeptide with alanine abrogated the ability of HoxA9 to immortalize primary myeloid progenitors. HoxA10 and HoxB9, two posterior homeodomain proteins that contain divergent hexapaptide motifs nearly identical to that in HoxA9 (AANWLT in HoxA10 and SANWLH in HoxB9 versus AANWLH in HoxA9) have been shown to bind DNA cooperatively with Pbx1 (Chang et al. 1996; Shen et al. 1997). In myeloid nuclear extracts, HoxA9 is found in complex with Pbx2 and with Meis1, a homeodomain protein that binds tightly to Pbx1 or Pbx2 via a separate N-terminal domain. It is possible that the HoxA9 proteins used in the present structure–function studies exhibit cooperative binding on DNA sequences different from those used in the studies here. The lack of cooperativity seen with the oligonucleotides used here suggests that HoxA9 on its own may form additional interactions with the DNA that are disrupted when it forms a ternary complex with Pbx1.
A model for posterior prevalence
Our results suggest a plausible mechanism for Hox protein binding and site recognition that explains posterior prevalence. A central role is played by the N-terminal arm residues and homeodomain residue 13, which differ between anterior and posterior homeodomain proteins. These residues facilitate binding of posterior homeodomain proteins to posterior DNA sequences. The additional ability of posterior Hox proteins, such as HoxA9, to bind to anterior Hox sites may be caused by a “default” mode of binding mediated by direct base contacts between helix 3 residues Ile 47, Glu 50, and Asn 51 (Figs. 4, 6). Anterior Hox proteins, such as HoxB1, are only capable of this default mode of binding to the anterior sequence, ATGATTGAT, because of sequence differences at key residues 7, 8, and 13. As a result, a posterior Hox protein expressed in any anterior domain can compete for the anterior Hox binding site, whereas the reverse cannot occur. In the case of two Hox proteins that bind similar binding sites, for example Hox proteins from paralog groups 6 and 9, the competition will be based on the precise individual binding affinities, which may account for dosage-dependent effects seen in some cases (Cribbs et al. 1995). This model would account for posterior prevalence, and the requirement for tight spatial and temporal control in a precise anterior-to-posterior sequence as is observed for Hox protein expression.
Materials and methods
Protein–DNA complex purification and preparation
A DNA fragment encoding residues 190–271 of murine HoxA9 containing the hexapeptide, the linker, and the homeodomain (murine and human HoxA9 are identical in amino acid sequence in this fragment) was subcloned from a cDNA vector (M. Cleary) into Pet-28a+ (Novagen) with a stop codon after residue 271 to prevent expression of the plasmid-derived histidine tag (HoxA9–Pet28a), and transformed into Escherichia coli BL21-DE3 Codon+-RIL cells (Stratagene). The cells were grown at 37°C in LB broth to mid-log phase, and expression was induced with 1 mM IPTG. The cells were harvested by centrifugation and stored at–80°C until lysis. Cells were lysed in a microfluidizer in buffer containing 25 mM sodium citrate (pH 5.5), 200 mM NaCl, 1 mM DTT, and 1 mM EDTA. The lysate was centrifuged at 17,000 rpm in an SS-34 rotor (Sorvall). The supernatant was filtered and separated by cation exchange chromatography on an SP Sepharose column (Pharmacia) followed by chromatography on a Mono S column (Pharmacia). The protein was eluted from both columns with a 0.2–1 M NaCl gradient in the same buffer conditions used for lysis, yielding HoxA9 protein that was >95% pure. Further purification to >99% homogeneity was achieved by loading the peak fractions onto a Vydac C4 reverse-phase column and eluting the protein with a 0%–100% acetonitrile gradient in 0.1% trifluoroacetic acid (TFA). Purified protein was then dialyzed against storage buffer containing 25 mM HEPES (pH 7.5), 100 mM NaCl, 1 mM EDTA, and 1 mM DTT, concentrated to ∼10 mg/mL and stored frozen at –80°C until needed. Pbx1 was expressed and purified as described in Piper et al. (1999).
The DNA oligonucleotides used for crystallization containing complementary single strands with the sequences 5′-ACTCTATGATTTACGACGCT-3′ and 5′-TAGCGTCGTAAATCATAGAG-3′ used for crystallization were synthesized on an Applied Biosystems DNA Synthesizer using the phosphoramidite method. Single-stranded oligonucleotides were purified in two steps. Oligonucleotides with the trityl group were purified by reverse phase HPLC over a Dynamax Pure DNA column (Varian) developed with a 0%–100% acetonitrile gradient in 0.1 M triethylamine acetate (TEAA; pH 7). The purified oligonucleotides were dialyzed against 10 mM triethylamine bicarbonate (pH 7), lyophilized, and resuspended in 0.1 M TEAA. The oligonucleotides were loaded onto the Pure DNA column again, and the trityl group was cleaved on the column with 0.5% TFA. The DNA was then eluted with a 0%–100% acetonitrile gradient in 0.1 M TEAA (pH 7). The purified single DNA strands were dialyzed against 10 mM triethylamine bicarbonate (pH 7), mixed in an equimolar ratio, heated to 70°C for 10 min, and annealed overnight at room temperature to produce a double-stranded 20-bp DNA fragment with a single 5′-base overhang at each end.
HoxA9, Pbx1, and DNA were mixed in a molar ratio of 1:1:1.2 and dialyzed into 10 mM HEPES (pH 7.5), 1 mM DTT, and 1 mM EDTA. Crystals were grown by the hanging-drop method by mixing 1 μL of well solution with 1 μL of complex and allowing it to equilibrate at 20°C. Crystals grew in 11% PEG 1000, 100 mM Tris (pH 8.0), and 4 mM cobaltic hexamine to an average size of 600 × 400 × 50 μm. The crystals were transferred into cryoprotectant containing 16% PEG 1000, 100 mM Tris (pH 8.0), 5 mM cobaltic hexamine, and 25% glycerol, and were frozen under a liquid nitrogen stream at –180°C for data collection.
Structure determination
Diffraction data to 1.9 Å were recorded at Beamline X24A, National Synchotron Light Source with a CCD detector and processed using Denzo and Scalepack (Rossmann and van Beek 1999; see Table 1 for data statistics). The space group was C2221, with unit cell dimensions a = 60.77 Å, b = 114.85 Å, c = 108.17 Å, α = β = γ = 90°. The structure was solved by molecular replacement with AMoRe (Navaza 2001), using as a search model the HoxB1/Pbx1/DNA structure (PDB code 1B72) with the HoxB1 hexapeptide deleted (Piper et al. 1999). Rigid-body and simulated annealing refinement was done using CNS (Brünger et al. 1998) with application of a bulk-solvent correction. Xtalview (McRee 1999) was used to build in the correct HoxA9 residues and DNA bases. Multiple rounds of rebuilding, positional refinement, and group B factor refinement were done using 2Fo – Fc, Fo – Fc, and simulated annealing omit maps generated by CNS and displayed with Xtalview. The hexapeptide and linker region, absent in the search model, were built into difference density maps generated by a model containing only the HoxA9 homeodomain. The model for the hexapeptide and linker region was confirmed by generating simulated annealing omit maps in which the linker or hexapeptide had been omitted and examining the fit of the model. Waters were placed in 3 σ peaks of the Fo – Fc maps using Xtalview and those that refined with a B-factor of <70 Å2 were retained. The final model was refined to an Rfree of 26.9% and an Rcryst of 23.4% using data from 20 Å to 1.9 Å, with 9.5% of the reflections excluded for the Rfree calculation. Model statistics are provided in Table 1. The model contains residues 192–268 of HoxA9, 233–316 of Pbx1, and all the DNA residues. This corresponds to homeodomain residues –12 to 66 of HoxA9 and 1–84 of Pbx1 in canonical homeodomain numbering scheme. DNA parameters were analyzed using the program Curves (Lavery and Sklenar 1988).
Table 1.
Data collection and refinement statistics
| Completeness (%) (outer shell) | 95.7 (94.1) |
| Redundancy | 4.3 (3.8) |
| Overall I/σ(I) (outer shell) | 22.3 (4.9) |
| Rsym (%) (outer shell) | 6.5 (26.0) |
| Refinement | |
| Resolution range (Å) | 20-1.9 |
| Rcryst/Rfree (%) | 23.4/26.9 (F > 0 σ) |
| Rfree reflections (%) | 2873 (9.5) |
| RMSD | Bonds 0.010 Å angles 1.43° |
Mutagenesis
Mutations were made to HoxA9 using the QuikChange Site-Directed Mutagenesis kit (Stratagene) with HoxA9–Pet28a as a template. Single mutants changing Pro 7 (homeodomain numbering, see Fig. 1B) to Asn, Tyr 8 to Phe, and Thr 13 to Val were made using the complimentary oligonucleotides 5′-CGGAAGAAGCGATGCAACTACACAAAACACCAG-3′ and 5′-CTG GTGTTTTGTGTAGTTGCATCGCTTCTTCCG-3′, 5′-AAGA AGCGATGCCCTTTCACAAAACACCAGACG-3′ and 5′-CGTCTGGTGTTTTGTGAAGGTGCATCGCTTCTT-3′, and 5′-TACACAAAACACCAGGTGCTGGAACTGCTGGAGAAG-3′ and 5′-CTTCTCCAGTTCCAGCACCTGGTGTTTTGTGTA-3′, respectively. The triple mutant was made from a double-mutant plasmid containing the Pro 7 to Asn and the Thr 13 to Val mutations using the oligonucleotides 5′-AAGAAGCGATGCAACTTCACAAAACACCAGGTG-3′ and 5′-CACCTGGTGTTTTGTGAAGTTGCATCGCTTCTT-3′ to change Tyr 8 to Phe. Mutations in HoxB1 were made using a Pet-11d (Novagen) plasmid containing HoxB1 residues 171–266. Single mutants changing Asn 7 (homeodomain numbering) to Thr, Phe 8 to Tyr, and Leu 13 to Thr were made using the complementary oligonucleotides 5′-AGTGGCCTCCGCACCACCTTCACCACAAGGCAG-3′ and 5′-CTGCCTTGTGGTGAAGGTGGTGCGGAGGCCACT-3′, 5′-GGCCTCCGCACCAACTACACCACAAGGCAGCTG-3′ and 5′-CAGCTGCCTTGTGGTGTAGTTGGTGCGGAGGCC-3′, and 5′-TTCACCACAAGGCAGACCACAGAACTGGAAAAG-3′ and 5′-CTTTTCCAGTTCTGTGGTCTGCCTTGTGGTGAA-3′, respectively. The triple mutant was made from a double-mutant plasmid containing the Asn 7 to Thr and the Leu 13 to Thr mutations using the oligonucleotides 5′-GGCCTCCGCACCACCTACACCACAAGGCAGACC-3′ and 5′-GGTCTGCCTTGTGGTGTAGGTGGTGCGGAGGCC-3′ to change Phe 8 to Tyr.
The mutant proteins were expressed as described above for HoxA9. Cells were lysed in a microfluidizer in buffer containing 25 mM sodium citrate (pH 5.5), 200 mM NaCl, 1 mM DTT, and 1 mM EDTA. The lysates were centrifuged at 17,000 rpm in an SS-34 rotor (Sorvall). The supernatant was filtered and separated by cation exchange chromatography on a 5 mL Hi-Trap SP-HP column (Pharmacia). The mutant proteins were eluted from the column with a 0.2–1 M NaCl gradient in the same buffer conditions as used for lysis, yielding >90% pure protein. The mutant proteins were concentrated to ∼10 mg/mL, dialyzed into buffer containing 20 mM sodium HEPES (pH 7.5), 100 mM NaCl, 1 mM DTT, and 1 mM EDTA and stored frozen at –80°C until use. The concentration for each protein was calculated using absorbance at 280 nm and calculated extinction coefficients. HoxB1 wild-type protein was purified as described in Piper et al. (1999).
DNA-binding assays
Gel mobility shift assays were performed using 10 twofold dilutions of each protein spanning the concentration range 1.5 × 10–5 M to 3 × 10–8 M. Some of these points were discarded in final calculations because of inaccuracy in data collection. Pbx1 protein at a concentration of 10–8 M was present in assays of Hox–Pbx1 binding to DNA. Double-stranded DNA oligonucleotides containing the sequences 5′-ACTCTATGATTTACGACGCT-3′ (HoxA9 site, TTTAC) and 5′-ACTCTATGATTGATCGGCTG-3′ (HoxB1 site, TTGAT) were labeled at the 3′ end with [α-32P]ATP and Klenow fragment (New England Biolabs). The DNA-binding reactions were carried out in 10 mM Tris (pH 7.5), 75 mM NaCl, 6% glycerol, 1 mM DTT, 1 mM EDTA, 6.7 ng/μL dIdC, 6.7 ng/μL BSA, titrating concentrations of protein, and labeled DNA at ∼0.6 nM. The reactions, done in triplicate, were incubated at room temperature for 30 min and separated using precast DNA retardation 6% 0.5 × TBE polyacrylamide gels (Invitrogen) at a constant voltage of 90 V for 50 min. The gels were dried and placed on a phosphorimaging screen (Kodak) overnight. The screen was read using a phophorimager (Molecular Dynamics), and the bands representing the shifted bands and the free DNA were analyzed for counts using ImageQuant version 2. For each lane, the fraction bound was calculated using the formula: fraction bound = 1 – (free DNA counts)/(free DNA counts + shifted DNA counts). The data were analyzed using the program Prism to calculate the equilibrium dissociation constant (KD) from nonlinear hyperbolic curve fit of plots of fraction bound versus concentration (M) assuming single-site binding. The ratio of the KD of HoxB1 site (TTGAT) binding to the KD of HoxA9 site (TTTAC) binding was taken as a measure of degree of site specificity.
Acknowledgments
We thank M. Cleary for expression vectors, C. Garvie and R. Campbell for help in structure determination, S. Muhammad for technical advice and assistance, and other lab members for helpful discussion. We thank C. Ogata for assistance at Beamline X4A of the National Synchotron Light Source at Brookhaven National Laboratory. This work was supported by grants MCB-9808412 and MCB-0220191 from the National Science Foundation. The PDB ID code for the structure is 1PUF.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1103303.
References
- Berry, M. and Gehring, W. 2000. Phosphorylation status of the SCR homeodomain determines its functional activity: Essential role for protein phosphatase 2A,B′. EMBO J. 19: 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter, M., Qian, Y.Q., Otting, G., Muller, M., Gehring, W., and Wuthrich, K. 1993. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain–DNA complex. J. Mol. Biol. 234: 1084–1093. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 (Pt 5): 905–921. [DOI] [PubMed] [Google Scholar]
- Bürglin, T.R. 1997. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25: 4173–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S.K., Jaffe, L., Capovilla, M., Botas, J., and Mann, R.S. 1994. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell 78: 603–615. [DOI] [PubMed] [Google Scholar]
- Chang, C.P., Shen, W.F., Rozenfeld, S., Lawrence, H.J., Largman, C., and Cleary, M.L. 1995. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes & Dev. 9: 663–674. [DOI] [PubMed] [Google Scholar]
- Chang, C.P., Brocchieri, L., Shen, W.F., Largman, C., and Cleary, M.L. 1996. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol. Cell. Biol. 16: 1734–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs, D.L., Benassayag, C., Randazzo, F.M., and Kaufman, T.C. 1995. Levels of homeotic protein function can determine developmental identity: Evidence from low-level expression of the Drosophila homeotic gene proboscipedia under Hsp70 control. EMBO J. 14: 767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule, D. 1994a. Temporal colinearity and the phylotypic progression: A basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Development Suppl: 135–142. [PubMed]
- Duboule, D., ed. 1994b. Guidebook to homeobox genes, pp. 575–576. Oxford University Press, Oxford.
- Ekker, S.C., Jackson, D.G., von Kessler, D.P., Sun, B.I., Young, K.E., and Beachy, P.A. 1994. The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. EMBO J. 13: 3551–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund, E.A., Jalava, A., and Kakar, R. 2000. Tyrosine phosphorylation of HoxA10 decreases DNA binding and transcriptional repression during interferon γ-induced differentiation of myeloid leukemia cell lines. J. Biol. Chem. 275: 20117–20126. [DOI] [PubMed] [Google Scholar]
- Fraenkel, E., Rould, M.A., Chambers, K.A., and Pabo, C.O. 1998. Engrailed homeodomain–DNA complex at 2.2 Å resolution: A detailed view of the interface and comparison with other engrailed structures. J. Mol. Biol. 284: 351–361. [DOI] [PubMed] [Google Scholar]
- Jaffe, L., Ryoo, H.D., and Mann, R.S. 1997. A role for phosphorylation by casein kinase II in modulating Antennapedia activity in Drosophila. Genes & Dev. 11: 1327–1340. [DOI] [PubMed] [Google Scholar]
- Kroon, E., Krosl, J., Thorsteinsdottir, U., Baban, S., Buchberg, A.M., and Sauvageau, G. 1998. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17: 3714–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf, R. 1994. Hox genes in vertebrate development. Cell 78: 191–201. [DOI] [PubMed] [Google Scholar]
- Lamka, M.L., Boulet, A.M., and Sakonju, S. 1992. Ectopic expression of UBX and ABD-B proteins during Drosophila embryogenesis: Competition, not a functional hierarchy, explains phenotypic suppression. Development 116: 841–854. [DOI] [PubMed] [Google Scholar]
- Laughon, A. 1991. DNA binding specificity of homeodomains. Biochemistry 30: 11357–11367. [DOI] [PubMed] [Google Scholar]
- Lavery, R. and Sklenar, H. 1988. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 6: 63–91. [DOI] [PubMed] [Google Scholar]
- Lawrence, H.J., Sauvageau, G., Humphries, R.K., and Largman, C. 1996. The role of HOX homeobox genes in normal and leukemic hematopoiesis. Stem Cells 14: 281–291. [DOI] [PubMed] [Google Scholar]
- Lawrence, H.J., Rozenfeld, S., Cruz, C., Matsukuma, K., Kwong, A., Komuves, L., Buchberg, A.M., and Largman, C. 1999. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia 13: 1993–1999. [DOI] [PubMed] [Google Scholar]
- Magli, M.C., Largman, C., and Lawrence, H.J. 1997. Effects of HOX homeobox genes in blood cell differentiation. J. Cell Physiol. 173: 168–177. [DOI] [PubMed] [Google Scholar]
- Mann, R.S. 1995. The specificity of homeotic gene function. Bioessays 17: 855–863. [DOI] [PubMed] [Google Scholar]
- Mann, R.S. and Chan, S.K. 1996. Extra specificity from extradenticle: The partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 12: 258–262. [DOI] [PubMed] [Google Scholar]
- McGinnis, W. and Krumlauf, R. 1992. Homeobox genes and axial patterning. Cell 68: 283–302. [DOI] [PubMed] [Google Scholar]
- McRee, D.E. 1999. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125: 156–165. [DOI] [PubMed] [Google Scholar]
- Morgan, B.A., Izpisua-Belmonte, J.C., Duboule, D., and Tabin, C.J. 1992. Targeted misexpression of Hox-4.6 in the avian limb bud causes apparent homeotic transformations. Nature 358: 236–239. [DOI] [PubMed] [Google Scholar]
- Navaza, J. 2001. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 57: 1367–1372. [DOI] [PubMed] [Google Scholar]
- Neuteboom, S.T., Peltenburg, L.T., van Dijk, M.A., and Murre, C. 1995. The hexapeptide LFPWMR in Hoxb-8 is required for cooperative DNA binding with Pbx1 and Pbx2 proteins. Proc. Natl. Acad. Sci. 92: 9166–9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, L., Xie, Y., Black, T.A., Jones, C.A., Pruitt, S.C., and Gross, K.W. 2001. An Abd-B class HOX.PBX recognition sequence is required for expression from the mouse Ren1c gene. J. Biol. Chem. 276: 32489–32494. [DOI] [PubMed] [Google Scholar]
- Passner, J.M., Ryoo, H.D., Shen, L., Mann, R.S., and Aggarwal, A.K. 1999. Structure of a DNA-bound Ultrabithorax–Extradenticle homeodomain complex. Nature 397: 714–719. [DOI] [PubMed] [Google Scholar]
- Peltenburg, L.T. and Murre, C. 1996. Engrailed and Hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. EMBO J. 15: 3385–3393. [PMC free article] [PubMed] [Google Scholar]
- Piper, D.E., Batchelor, A.H., Chang, C.P., Cleary, M.L., and Wolberger, C. 1999. Structure of a HoxB1–Pbx1 heterodimer bound to DNA: Role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96: 587–597. [DOI] [PubMed] [Google Scholar]
- Rossmann, M.G. and van Beek, C.G. 1999. Data processing. Acta Crystallogr. D Biol. Crystallogr. 55 (Pt 10): 1631–1640. [DOI] [PubMed] [Google Scholar]
- Schnabel, C.A., Jacobs, Y., and Cleary, M.L. 2000. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene 19: 608–616. [DOI] [PubMed] [Google Scholar]
- Shen, W.F., Chang, C.P., Rozenfeld, S., Sauvageau, G., Humphries, R.K., Lu, M., Lawrence, H.J., Cleary, M.L., and Largman, C. 1996. Hox homeodomain proteins exhibit selective complex stabilities with Pbx and DNA. Nucleic Acids Res. 24: 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W.F., Rozenfeld, S., Lawrence, H.J., and Largman, C. 1997. The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. J. Biol. Chem. 272: 8198–8206. [DOI] [PubMed] [Google Scholar]
- Taylor, H.S. 1998. A regulatory element of the empty spiracles homeobox gene is composed of three distinct conserved regions that bind regulatory proteins. Mol. Reprod. Dev. 49: 246–253. [DOI] [PubMed] [Google Scholar]
- Thorsteinsdottir, U., Kroon, E., Jerome, L., Blasi, F., and Sauvageau, G. 2001. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol. Cell. Biol. 21: 224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron, Y., McAdara, J.K., Lynch, M., Hughes, E., and Gasson, J.C. 2001. Identification of novel functional regions important for the activity of HOXB7 in mammalian cells. J. Immunol. 166: 5058–5067. [DOI] [PubMed] [Google Scholar]