Abstract
Programmed gene rearrangements are employed by a variety of microorganisms, including viruses, prokaryotes, and simple eukaryotes, to control gene expression. In most instances in which organisms mediate host evasion by large families of homologous gene cassettes, the mechanism of variation is not thought to involve DNA inversion. Here we report that Campylobacter fetus, a pathogenic Gram-negative bacterium, reassorts a single promoter, controlling surface-layer protein expression, and one or more complete ORFs strictly by DNA inversion. Rearrangements were independent of the distance between sites of inversion. These rearrangements permit variation in protein expression from the large surface-layer protein gene family and suggest an expanding paradigm of programmed DNA rearrangements among microorganisms.
Keywords: surface layer proteins, Campylobacter fetus
One way in which microorganisms can alter their surface properties, allowing a fraction of the population to preadapt to environmental changes, is by varying protein expression through programmed genomic DNA rearrangements (1). Phase, antigenic, or size variation of expressed surface proteins are governed by mechanisms such as transposition and DNA inversion. During transposition, a silent gene is activated by movement to an expression site where it displaces the currently expressed gene. In DNA inversion, a segment of DNA is cut, inverted, and then rejoined by a site-specific recombinase. The invertible DNA segment may contain either a promoter that directs expression of fixed structural genes or structural genes controlled by a fixed promoter. Transposition and inversion differ in both the enzymes used and in the number of genes that can be controlled (many versus two).
Campylobacter fetus, a bacterial pathogen of ungulates and humans, is covered by a paracrystalline surface (S) layer, composed of high-molecular weight S-layer proteins (SLPs), that masks most of the underlying Gram-negative surface features (2). More than 300 bacterial genera that possess S layers have been described (3). The S layer renders C. fetus cells resistant to serum killing by prohibiting the binding of C3b (4), and the SLPs themselves may change, permitting antigenic variation (5, 6). These SLPs are encoded by eight tightly clustered and partially homologous promoterless gene cassettes (7, 8). Since previous studies show that C. fetus can express alternative SLPs (4–6, 9), that there exists only a single promoter for SLP expression which is present on a 6.2-kb invertible element (9), and that the structural genes flanking the promoter are subject to substitution (9), we proposed that both the promoter and the eight structural genes (sapA and its homologs) may rearrange strictly by inversion.
To test the hypothesis that nested DNA inversion can occur in which both the sapA promoter and the complete structural genes are mobile, we created mutant strains in which both SLP gene cassettes bracketing the invertible sapA promoter were disrupted. By selecting for mutants able to survive incubation in normal human serum (NHS), and thus expressing native SLPs, we could identify cells that had exchanged at least one of the bracketing cassettes. Inversion has been previously reported in biological systems involving an invertible promoter and fixed structural genes or vice versa, permitting alternate expression of at most two alternate sets of structural genes (10, 11). By genetic and phenotypic analyses, we now demonstrate that C. fetus uses a novel system of DNA inversion, in which the promoter may invert alone or in concert with one or more of the flanking structural genes, resulting in antigenic variation of the expressed SLP.
MATERIALS AND METHODS
Bacterial Strains and Culture Conditions.
Wild-type S+ (possessing an S-layer) C. fetus strain 23D and spontaneous S− (no S-layer present) mutant strain 23B have been extensively characterized (4, 5, 12, 13). Other C. fetus strains used were defined mutants derived from strain 23D, as described below. Stock cultures were stored and grown as described (8). Media containing 7 units/ml polymyxin B, 10 μg/ml vancomycin, 50 μg/ml naladixic acid, and 10 μg/ml trimethoprim lactate were supplemented for kanamycin- or chloramphenicol-resistant strains, with 30 μg/ml kanamycin or 15 μg/ml chloramphenicol, respectively. Escherichia coli strains used in this study, including DH5a, HB101, and XL1-Blue (Stratagene), were grown in L broth or on L plates (14).
Chemicals and Enzymes.
Isopropyl β-d-thiogalactopyranoside (50 mg/ml) and 5-bromo-4-chloro-3-indolyl β-d-galactoside (28 mg/ml) were purchased from Jersey Lab Supply (Livingston, NJ). Restriction enzymes, T4 DNA ligase, Taq polymerase, and E. coli DNA polymerase large (Klenow) fragment were from Promega and United States Biochemical. Antibiotics were from Sigma, and [α-32P]dATP (650 mCi/mmol; 1 Ci = 37 GBq) was from ICN.
Genetic Techniques.
Chromosomal DNA was prepared from 48-hr plate cultures, as described (9). Plasmids were isolated by the procedure of Birnboim and Doly (15). All other standard molecular genetic techniques were done as described (14).
Construction of Mutant C. fetus Strains.
Mutant C. fetus strains were created by mobilization of donor pKO500 or pKO505 plasmid constructs by conjugal mating as described (9) with sequential selection (see Fig. 1) on media containing 30 mg/ml kanamycin or 15 mg/ml chloramphenicol, or in the presence of 10% NHS as described (16). pKO500 is a suicide hybrid plasmid with the sapA ORF disrupted with a chloramphenicol-resistance gene (cm; ref. 17) located 127 bp into the ORF. pKO505 is a suicide hybrid plasmid with the sapA2 ORF disrupted with a promoterless kanamycin-resistance gene (km; ref. 18) located 127 bp into the ORF.
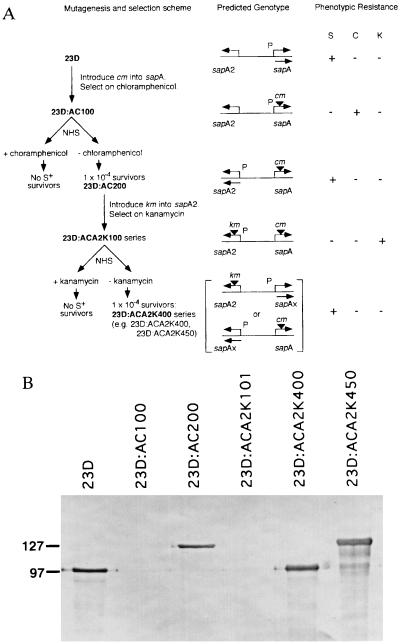
Figure 1.
Schematic representation of serial experiments to use km and cm cassettes to examine SLP gene rearrangement (A). Introduction of cm into strain 23D by marker rescue led to insertion into sapA to create 23D:AC100. This strain was selected on chloramphenicol-containing medium and, as expected, was chloramphenicol (C)-resistant, was S−, and not resistant to serum (S) and kanamycin (K), as previously reported (9). Incubation of 23D:AC100 with NHS selected for survivors (at a frequency of 1 × 10−4; AC200 series) that were S+ and serum-resistant but sensitive to kanamycin and chloramphenicol. Strain 23D:AC200, expressing sapA2, was further mutagenized by introduction of km into sapA2 to create the 23D:ACA2K100 series. These strains were S− and serum- and chloramphenicol-sensitive but kanamycin-resistant. Incubation with NHS selected for survivors (at a frequency of 1 × 10−4) that were S+ and serum-resistant but kanamycin- and chloramphenicol-sensitive (ACA2K400 series). The reciprocal relationship between antibiotic- and serum-resistance suggests that only a single promoter for SLP gene cassettes is present. All ACA2K400 series strains must have an SLP gene cassette other than sapA or sapA2 positioned downstream of the single SLP promoter. The predicted genotypes are depicted. P, sapA promoter; bent arrows, location and direction of transcription of SLP gene cassette; ▾, antibiotic resistance gene insertion. (B) Immunoblot of C. fetus strain 23D and selected mutants into which cm or cm and km were inserted into SLP gene cassettes. As expected, all the serum-resistant strains expressed SLP (of 97 or 127 kDa) recognized by antiserum to conserved C. fetus SLP determinants, whereas there was no expression for strains maintained on either antibiotic.
Bactericidal Assays.
To determine the susceptibilities of the mutant strains to the bactericidal activity of NHS, 10-fold serially diluted cultures (starting from a single colony) were incubated at 37°C for 60 min in the presence of 10% pooled NHS or 10% heat-inactivated NHS as described (9, 16). Wild-type S+ strain 23D and spontaneous S− mutant strain 23B were the serum-resistant and serum-sensitive controls, respectively (9). Cultures then were plated to media containing chloramphenicol, kanamycin, or no antibiotic selection (see Fig. 1A), and following incubation, bacterial colonies were enumerated. Survival rates were determined as the ratio of colony-forming units per milliliter in the presence of NHS or heat-inactivated human serum, or a similar ratio of colony-forming units per milliliter in the presence or absence of the selective antibiotic.
Production of Antiserum to C. fetus SLPs and Immunoblot Analyses.
Antiserum to the 97-kDa SLP of type A strain 82–40LP was raised in adult New Zealand White female rabbits and shows broad recognition of C. fetus SLPs as described (5). To analyze wild-type and transconjugant C. fetus strains for SLP expression, cells were harvested from plates, lysed in sample loading buffer, and examined by SDS/PAGE, and immunoblotting was performed as detailed (5, 13).
Southern Hybridizations and Probes.
C. fetus chromosomal DNA was digested with HincII and processed exactly as described (12). Probes included the gel-purified PCR products specific for the sapA promoter region (9), km, cm, 3′ sapA region (1649–2760 bp), middle sapA1 region (620–1381 bp), and 3′ sapA1 region (1381–2763 bp). Probes were 32P-radiolabeled by primer extension with random hexameric oligonucleotides (19). PCRs were performed as described (9).
PCR.
PCRs were performed as described (9). Amplification of large PCR products was accomplished by denaturing for only 5 sec and extending at 68°C for longer periods, typically 4–7 min. Primers used in this study include the following: km F, 5′-TGTAGAAAAGAGGAAGGAAA-3′; km R, 5′-CTAAAACAATTCATCCAGTA-3′; cm F, 5′-AGTGGATAGATTTATGATATAGTG-3′; cm R, 5′-TTTATTTATTCAGCAAGTCTTG-3′; sapA F middle, 5′-CATCTCTACAGCAGCAAAAG-3′; sapA F 3′, 5′-GCGGAGATAATGTTGTAG- TTGAT G-3′; sapA R, 5′-AACTTTAAGATCTAGCGTACC-3′; sapA1 F middle, 5′-AGGGTACTGATTTAGACGATA-3′; sapA1 F 3′, 5′-GCTGGATTTACAGGAGATTTAACC-3′; sapA1 R 3′ #1, 5′-GTTACTGGTATCAATAACAACATAAGT; sapA1 R 3′ #2, 5′-CTACGTAATCATACTGCTACC-3′.
RESULTS
Construction and Phenotypic Analysis of Mutant Strains.
To test the hypothesis that both the sapA promoter and the complete structural genes can be mobilized by DNA inversion, we created mutant strains in which both SLP-encoding gene cassettes bracketing the invertible sapA promoter were disrupted (Fig. 1A). First, insertion of a chloramphenicol-resistance cassette (cm) into the ORF of the expressed S-layer gene cassette (sapA) ablated SapA expression (Fig. 1B) and rendered the organism (23D:AC100) serum-sensitive. Using the ability of S+ (but not S−) C. fetus strains to survive incubation in NHS (4, 13, 20–22), we then selected for promoter inversion mutants (9) expressing the sapA2 cassette and identified strain 23D:AC200 (Fig. 1). Next, we mutagenized 23D:AC200 by insertion of the kanamycin-resistance cassette (km) to ablate SapA2 expression, creating 23D:ACA2K101 (Figs. 1 and 2D). Incubation of this dually disrupted strain in serum selected for serum-resistant survivors. These were found at a frequency of 1 × 10−4 and, as expected, expressed SLPs of 97 or 127 kDa (Fig. 1B) and were antibiotic-sensitive. Immunoblotting of 26 serum survivors demonstrated that 16 (62%) expressed a 97-kDa SLP (e.g., 23D:ACA2K400) and 10 (38%) expressed a 127-kDa SLP (e.g., 23D:ACA2K450). These results suggested that in each of the survivors there had been exchange of at least one of the bracketing cassettes. None of the mutant strains maintained on antibiotic-containing media produced an SLP band, as revealed by immunoblotting (Fig. 1B), and this confirms our earlier observation that expression of cm or km depends on the single sapA promoter (9). These experiments provided a group of well defined strains with which to examine the genotypic events associated with the observed phenotypic variations.
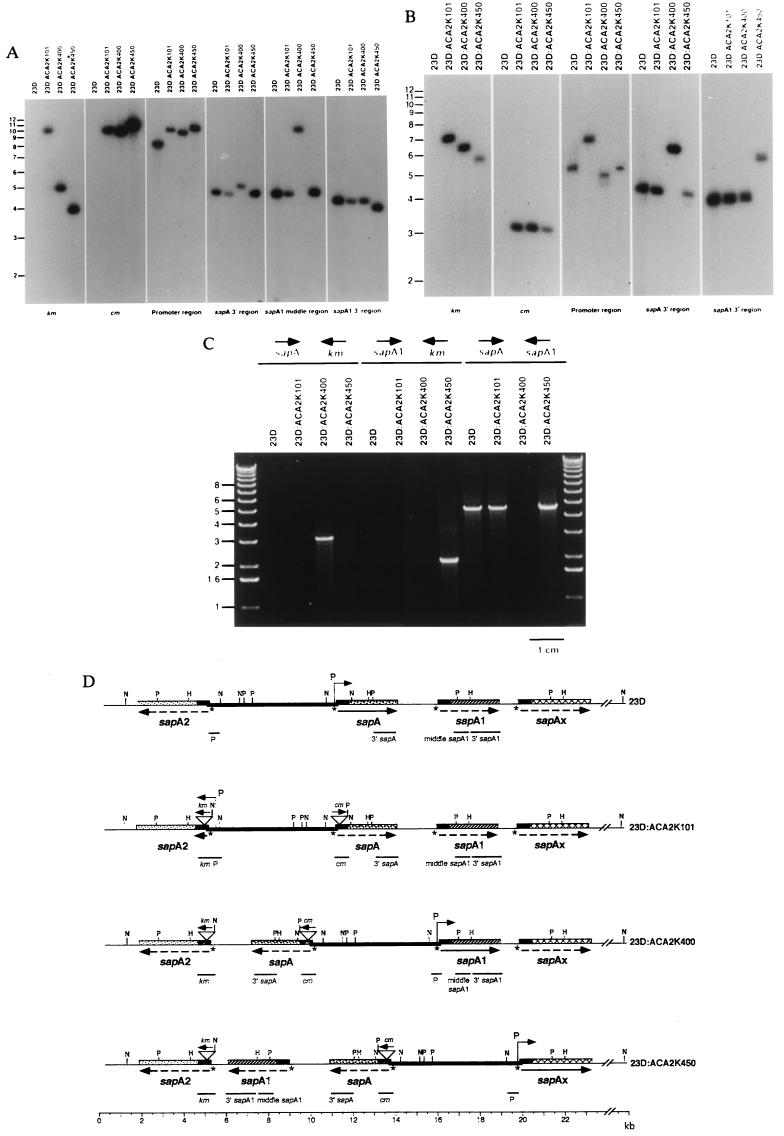
Figure 2.
Southern hybridization of HincII (A) or PstI (B) digestions of chromosomal DNA from C. fetus 23D and ACA2K series mutants using probes to km, cm, the promoter region, the sapA-specific 3′ region, the sapA1-middle region, or the sapA1-specific 3′ region. Each probe hybridized to a single fragment regardless of the phenotype of the C. fetus strain. (C) Mapping of SLP gene cassette arrangement by PCR. PCRs were performed with template chromosomal DNA from strains 23D and ACA2K mutants using sapA-specific 3′ region forward (sapA) and km reverse (km) primers (left four lanes), sapA1-specific 3′ region forward (sapA1) and km primers (center four lanes), or sapA and sapA1-specific 3′ region reverse (sapA1) primers (right four lanes). (D) Cumulative restriction maps of the four strains presented in A–C. The location of the probes as indicated from the hybridizations is shown under the map for each strain. sapAx represents an uncharacterized SLP gene cassette; arrows represent the direction of transcription; solid lines represent expressed genes, dashed lines represent silent genes; P over bent arrows represents the sapA promoter; and the heavy line represents the 6.2-kb invertible promoter-containing element, flanked by opposing SLP gene cassettes. The asterisks represent the palindromic putative recombinase recognition sites (TTAAGGAaTCCTTAA) present in the 5′ conserved region of each SLP gene cassette (7), and restriction sites are indicated: H, HincII; N, NdeI; P, PstI.
Phenotypic Variation Is Associated with Nested DNA Inversions.
To investigate the nature of the recombination event that allowed the single sapA promoter to express a native S-layer gene cassette in the dually disrupted strains, using Southern analyses, we compared these organisms with wild-type C. fetus strain 23D and the S− parental mutant 23D:ACA2K101. The known positions of HincII (Fig. 2A), PstI (Fig. 2B), and NdeI (data not shown) sites located in sapA, sapA1, and sapA2 (8, 9, 23, 24) were used to predict the size of the fragments hybridizing with appropriate probes. The promoter region, the sapA 3′ region, the sapA1 middle region, and the sapA1 3′ region probes hybridized to 8.5-, 4.8-, 4.8-, and 4.3-kb HincII fragments, respectively, in wild-type strain 23D (Fig. 2A). Based on these and previous Southern hybridization and PCR data (9), the location of these genes relative to one another in wild-type strain 23D was defined (Fig. 2D, first line). An identical hybridization pattern was observed for mutant 23D:ACA2K101 with the exception that the km and cm probes hybridized to a 10.4-kb fragment and that the promoter region probe also hybridized to a 10.4-kb fragment (Fig. 2D, second line). These results are entirely consistent with the introduction of the km (0.8-kb) and cm (1.1-kb) markers flanking the promoter. For the mutant strains ACA2K400 and ACA2K450, changes in phenotype were clearly associated with change in probe cohybridization (Fig. 2A). In mutant strain 23D:ACA2K400, the km probe hybridized to the same 5.1-kb fragment as the sapA 3′ region probe, and in strain 23D:ACA2K450, the km probe hybridized to the same 4.0-kb fragment as the sapA1 3′ region probe (Fig. 2A), whereas the promoter region probe hybridized to fragments larger than 9 kb. For each of the mutant strains, the cm and promoter region probes hybridized to fragments of identical sizes (Fig. 2A). These results demonstrate that the ORFs had inverted (Fig. 2D) and reflect the differing locations of each of the SLP gene cassettes in relation to the km marker. The total size of nonoverlapping hybridizing fragments remained constant among the strains (17.6 kb in wild-type and, reflecting addition of the two antibiotic resistance cassettes, 19.5 kb in all mutant strains), indicating that recombination did not involve net duplication or deletion of DNA (Fig. 2D).
Following PstI digestion, as expected, the km probe hybridized to the same 7.3-, 6.5-, and 5.9-kb fragments as the promoter region probe, the sapA 3′ region probe, and the sapA1 3′ region probe in strains ACA2K101, ACA2K400, and ACA2K450, respectively (Fig. 2B). Despite the genotypic differences among the mutant strains, the constancy of the 3.1-kb fragment hybridizing with the cm probe (Fig. 2B) was consistent with the position of the marker downstream of the promoterless end of the 6.2-kb invertible element (Fig. 2D). Hybridization of the promoter region probe to 5.5-, 4.9-, and 5.3-kb fragments in strain 23D and mutants ACA2K400 and ACA2K450, respectively, also reaffirms the model. The sapA 3′ region probe hybridized with a 4.1-kb fragment in strain 23D and mutants ACA2K101 and ACA2K450, but not ACA2K400 (Fig. 2B), consistent with the locations of PstI sites within sapA and sapA1, and the tandem relationship between these two cassettes. Similarly, the sapA1 3′ region probe hybridized with a 4.2-kb fragment in strains 23D, ACA2K101, and ACA2K400 but not ACA2K450 (Fig. 2B), consistent with the tandem relationship between sapA1 and the downstream sapAx in the first three strains. The observed sizes of each of the hybridizing fragments and their cohybridization patterns in PstI (Fig. 2B) or NdeI-digested (not shown) genomic DNA using similar probes were completely consistent with the known restriction sites (8, 9, 23, 24) as depicted in Fig. 2D.
PCR analyses provided independent confirmation for ORF inversion. The 3′ sapA-specific forward primer and a km reverse primer yielded a product for strain 23D:ACA2K400 (Fig. 2C), indicating that sapA had inverted and was located immediately upstream of km. Similarly, for strain 23D:ACA2K450 the data (Fig. 2C) indicate that sapA1 had inverted and was located immediately upstream of km. PCR analyses using the 3′ sapA-specific forward primer and 3′ sapA1-specific reverse primer yielded a 4.3-kb product for all strains except for 23D:ACA2K400 (Fig. 2C, right four lanes), indicating that the tandem relationship of sapA and sapA1 was lost in strain ACA2K400. The data indicate that for mutant strains 23D:ACA2K400 and 23D:ACA2K450, sapA, or both sapA and sapA1, respectively, have inverted in relationship to sapA2 (Fig. 2D, last three lines).
DNA Inversion Events Occur Independently of the Distance Between Recombination Sites.
We next sought to determine the frequency of the DNA inversion events, involving promoter alone or promoter and one or more of the gene cassettes. The mutant strains (Table 1) provided easily definable phenotypes with which to assess the inversion events. We measured these by conducting experiments in which we examined for resistance to a selection that would be lethal (exposure to kanamycin, chloramphenicol, or serum) unless defined inversions allowing expression of genes to overcome the exposure had occurred. Events involving inversion of the promoter-containing element alone, or together with one or two ORFs, occurred at nearly equal frequency (about 1–2.6 × 10−4; Table 1). These data imply that inversions involving two adjacent ORFs occurred in a single event and did not result from two or more sequential inversion events. Recombination occurs at either homologous or palindromic DNA sequences (9). The distance-independence between sites suggests that inversion occurs by a random collision model, as proposed by Gellert and Nash (25).
Table 1.
C. fetus survival after serum or antibiotic selection
| Strain | Relevant phenotype
|
Selection* | No. of experiments | |||||
|---|---|---|---|---|---|---|---|---|
| Susceptibility to
|
Immunoblot presence of S layer, kDa | Survival, ×10−4
|
||||||
| Chloramphenicol | Kanamycin | Serum | Mean | Range | ||||
| 23D:AC100 | R | S | S | — | Serum | 7 | 1.0 | 0.2–3.1 |
| 23D:AC200 | S | S | R | 127 | Chloramphenicol | 6 | 1.2 | 0.1–2.5 |
| 23D:ACA2K101 | S | R | S | — | Chloramphenicol | 12 | 0.7 | 0.2–2.5 |
| 23D:ACA2K101 | S | R | S | — | Serum | 8 | 1.2 | 0.2–1.4 |
| 23D:ACA2K200 | R | S | S | — | Kanamycin | 3 | 0.8 | 0.4–1.1 |
| 23D:ACA2K400 | S | S | R | 97 | Kanamycin | 6 | 1.0 | 0.8–1.4 |
| 23D:ACA2K400 | S | S | R | 97 | Chloramphenicol | 6 | 1.2 | 0.6–2.2 |
| 23D:ACA2K450 | S | S | R | 127 | Kanamycin | 5 | 2.6 | 1.2–4.6 |
| 23D:ACA2K450 | S | S | R | 127 | Chloramphenicol | 6 | 2.5 | 1.0–4.6 |
S, Sensitive; R, resistant.
Selection was performed by plating cells to media supplemented with 15 μg/ml chloramphenicol or 30 μg/ml kanamycin, or incubating cells in 10% NHS for 60 min at 37°C.
DISCUSSION
We have recently demonstrated that SLP expression in C. fetus is based on the single sapA promoter present on an invertible 6.2-kb element, which is bracketed by inverted repeats and oppositely oriented cassettes, sapA and sapA2, that are subject to substitution (9). By creation of appropriate mutants and by use of selection for phenotypic properties, our present experiments demonstrate further that DNA rearrangement can involve inversion of this element in concert with one or more of the tandemly arrayed SLP gene cassettes.
DNA inversion has been believed to involve mutually exclusive promoter or structural gene inversion. Either the promoter inverts relative to fixed structural genes (26, 27) or structural genes invert downstream of a fixed promoter (28–30), permitting expression of two alternate gene copies (10, 11). The system of DNA inversion in C. fetus is novel because it combines the features of each mechanism as both the promoter and the structural genes are mobile, which permits the shuffling of complete genes and their ultimate expression (Fig. 3). Rearrangement of the SLP gene cassettes permits the organism to vary SLP expression and thus surface antigenicity, allowing for evasion of host immune responses (5, 6, 31). This inversion system differs from the recently described Mycoplasma pulmonis vsa gene inversion (32), which rearranges only incomplete coding regions and demonstrates less sequence stability.
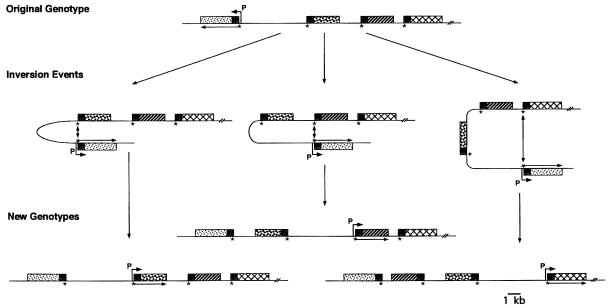
Figure 3.
Proposed model of molecular events involved in SLP gene cassette rearrangement by DNA inversion. DNA inversion between two oppositely oriented cassettes follows DNA strand exchange at the putative recombinase target site (∗) found upstream of each SLP gene cassette within the 5′ conserved region (small shaded box; ref. 8). Patterned boxes represent variable regions of SLP gene cassettes. A 6.2-kb intervening segment is topologically reversed, leading to ordered rearrangement of the SLP gene cassettes. Inversion of DNA segments containing the promoter (P over bent arrow) permits expression of alternate SLP gene cassettes (mRNA, arrow). Illustrated are inversion of the 6.2-kb promoter-containing element alone (Left), and the 6.2-kb element and one (Center) or two (Right) SLP gene cassette ORFs and the resultant genotypes. Each of these genotypes has been observed (Fig. 2D).
Our studies further indicate that inversion occurs randomly between ORFs of opposite orientation independently of the size of the intervening DNA segment. The economy of a simple inversion system may be especially useful for C. fetus, which has a relatively small (1.1-Mb) genome (33); the strict conservation of both coding and noncoding regions related to the sap homologs in type A and type B strains (7) is consistent with the importance of this efficient system. This study expands the paradigms of DNA rearrangement, in which large gene families of complete ORFs can reassort by inversion to vary the surface protein expression of the microbe. Similar mechanisms controlling protein expression may be present in other organisms.
Acknowledgments
We thank Richard Breyer, Gisela Mosig, Stuart Thompson, and Murali Tummuru for helpful discussions and review of the manuscript. This work was supported in part by Grant RO1-AI24145 from the National Institutes of Health and by the Medical Research Service of the Department of Veterans Affairs.
Footnotes
Abbreviations: S, surface; SLP, surface-layer protein; km, kanamycin-resistance gene; cm, chloramphenicol-resistance gene; NHS, normal human serum.
References
- 1.Borst P, Greaves D R. Science. 1982;235:658–667. doi: 10.1126/science.3544215. [DOI] [PubMed] [Google Scholar]
- 2.Fujimoto S, Takade A, Amako K, Blaser M J. Infect Immun. 1991;59:2017–2022. doi: 10.1128/iai.59.6.2017-2022.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beveridge T J, Koval S F. Advances in Paracrystalline Bacterial Surface Layers. New York: Plenum; 1993. [Google Scholar]
- 4.Blaser M J, Smith P F, Repine J E, Joiner K A. J Clin Invest. 1988;81:1434–1444. doi: 10.1172/JCI113474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang E, Garcia M M, Blake M S, Pei Z, Blaser M J. J Bacteriol. 1993;175:4979–4984. doi: 10.1128/jb.175.16.4979-4984.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia M M, Lutze-Wallace C L, Denes A S, Eaglesome M D, Holst E, Blaser M J. J Bacteriol. 1995;177:1976–1980. doi: 10.1128/jb.177.8.1976-1980.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dworkin J, Tummuru M K, Blaser M J. J Biol Chem. 1995;270:15093–15101. doi: 10.1074/jbc.270.25.15093. [DOI] [PubMed] [Google Scholar]
- 8.Dworkin J, Tummuru M K, Blaser M J. J Bacteriol. 1995;177:1734–1741. doi: 10.1128/jb.177.7.1734-1741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dworkin J, Blaser M J. Mol Microbiol. 1996;19:1241–1253. doi: 10.1111/j.1365-2958.1996.tb02469.x. [DOI] [PubMed] [Google Scholar]
- 10.Glasgow A C, Hughes K T, Simon M I. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 637–659. [Google Scholar]
- 11.van de Putte P, Goosen N. Trends Genet. 1992;8:457–462. doi: 10.1016/0168-9525(92)90331-w. [DOI] [PubMed] [Google Scholar]
- 12.Tummuru M K, Blaser M J. J Bacteriol. 1992;174:5916–5922. doi: 10.1128/jb.174.18.5916-5922.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blaser M J, Smith P F, Hopkins J A, Heinzer I, Bryner J H. J Infect Dis. 1987;155:696–705. doi: 10.1093/infdis/155.4.696. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Birnboim H C, Doly J. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaser M J, Smith P F, Kohler P F. J Infect Dis. 1985;151:227–235. doi: 10.1093/infdis/151.2.227. [DOI] [PubMed] [Google Scholar]
- 17.Wang E, Taylor D E. Gene. 1990;94:23–28. doi: 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]
- 18.Trieu-Cout P, Gerbaud G, Lambert T, Courvalin P. EMBO J. 1985;4:3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feinberg A P, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 20.Pei Z, Blaser M J. J Clin Invest. 1990;85:1036–1043. doi: 10.1172/JCI114533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winter A J, McCoy E C, Fullmer C S, Burda K, Bier P J. Infect Immun. 1978;22:963–971. doi: 10.1128/iai.22.3.963-971.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaser M J, Pei Z. J Infect Dis. 1993;167:696–706. doi: 10.1093/infdis/167.2.372. [DOI] [PubMed] [Google Scholar]
- 23.Blaser M J, Gotschlich E C. J Biol Chem. 1990;265:14529–14535. [PubMed] [Google Scholar]
- 24.Tummuru M K R, Blaser M J. Proc Natl Acad Sci USA. 1993;90:7265–7269. doi: 10.1073/pnas.90.15.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gellert M, Nash H. Nature (London) 1987;325:401–404. doi: 10.1038/325401a0. [DOI] [PubMed] [Google Scholar]
- 26.Silverman M, Zieg J, Hilmen M, Simon M. Proc Natl Acad Sci USA. 1979;76:391–395. doi: 10.1073/pnas.76.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abraham J M, Freitag C S, Clements J R, Eisenstein B I. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marrs C F, Ruehl W W, Schoolnik G K, Falkow S. J Bacteriol. 1988;170:3032–3039. doi: 10.1128/jb.170.7.3032-3039.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iida S, Meyer J, Kennedy K E, Arber W. EMBO J. 1982;1:1445–1453. doi: 10.1002/j.1460-2075.1982.tb01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giphart-Gassler M, Plasterk R H A, van de Putte P. Nature (London) 1982;297:339–342. doi: 10.1038/297339a0. [DOI] [PubMed] [Google Scholar]
- 31.Blaser M J, Wang E, Tummuru M K, Washburn R, Fujimoto S, Labigne A. Mol Microbiol. 1994;14:453–462. doi: 10.1111/j.1365-2958.1994.tb02180.x. [DOI] [PubMed] [Google Scholar]
- 32.Bhugra B, Voelker L L, Zou N, Yu H, Dybwig K. Mol Microbiol. 1995;18:703–714. doi: 10.1111/j.1365-2958.1995.mmi_18040703.x. [DOI] [PubMed] [Google Scholar]
- 33.Salama S M, Garcia M M, Taylor D E. Int J Syst Bacteriol. 1992;42:446–450. doi: 10.1099/00207713-42-3-446. [DOI] [PubMed] [Google Scholar]