Abstract
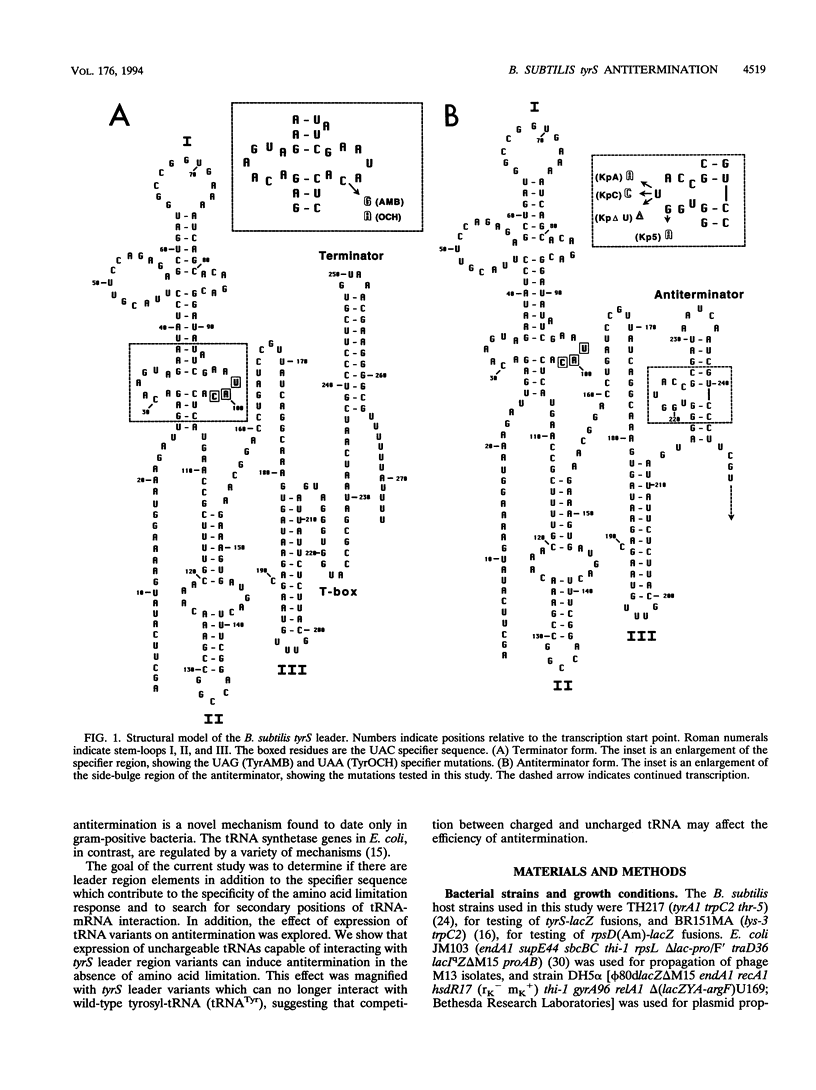
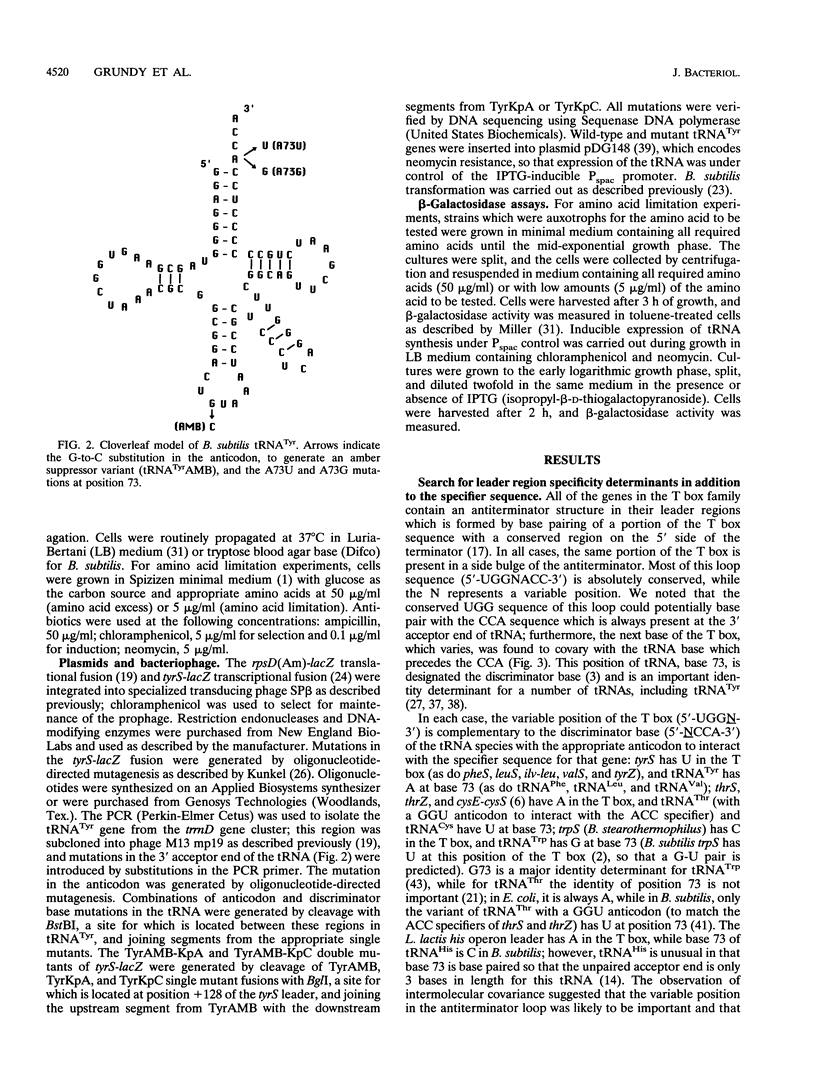
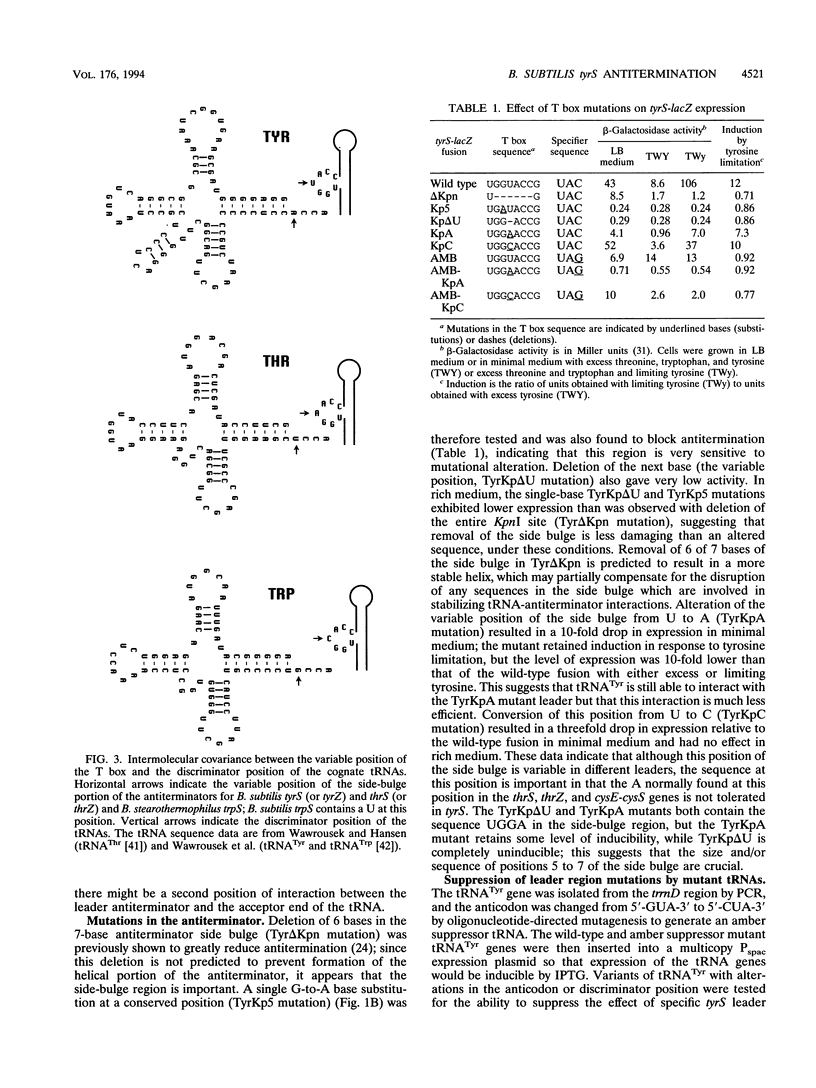
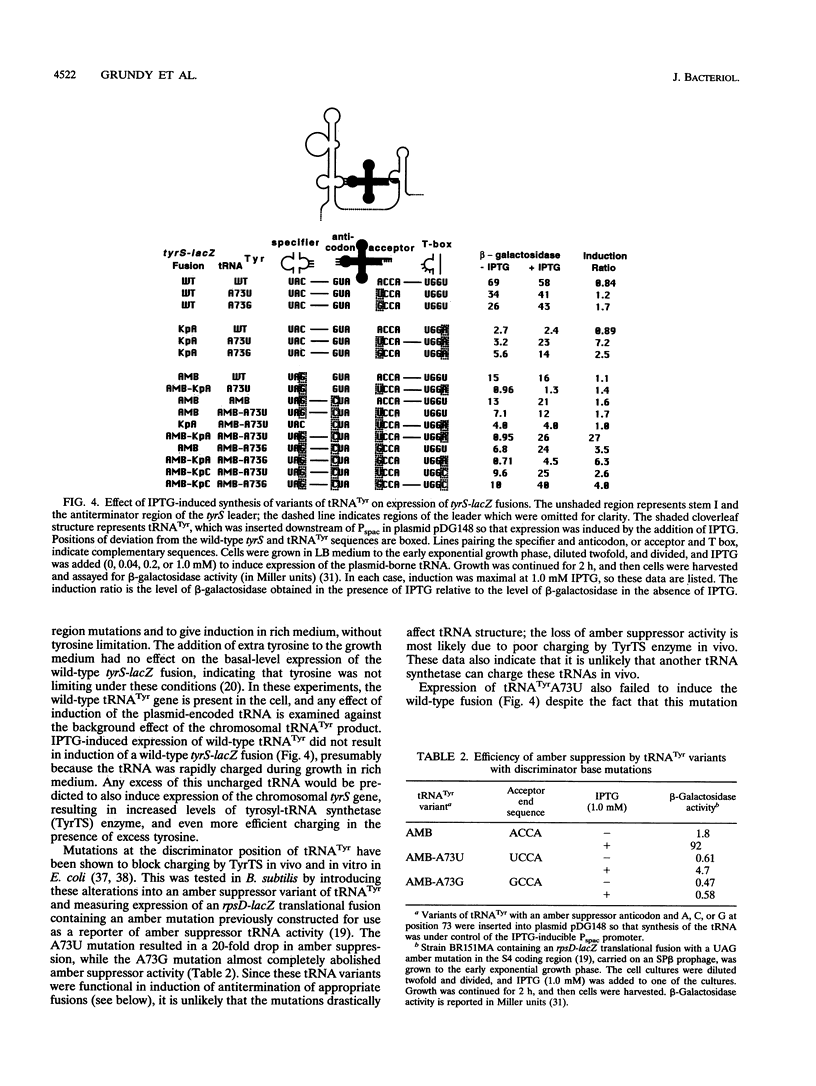
The Bacillus subtilis tyrS gene is a member of a group of gram-positive aminoacyl-tRNA synthetase and amino acid biosynthesis genes which are regulated by transcription antitermination. Each gene in the group is specifically induced by limitation for the appropriate amino acid. This response is mediated by interaction of the cognate tRNA with the mRNA leader region to promote formation of an antiterminator structure. The tRNA interacts with the leader by codon-anticodon pairing at a position designated the specifier sequence which is upstream of the antiterminator. In this study, an additional site of possible contact between the tRNA and the leader was identified through covariation of leader mRNA and tRNA sequences. Mutations in the acceptor end of tRNA(Tyr) could suppress mutations in the side bulge of the antiterminator, in a pattern consistent with base pairing. This base pairing may thereby directly affect the formation and/or function of the antiterminator. The discriminator position of the tRNA, an important identity determinant for a number of tRNAs, including tRNA(Tyr), was shown to act as a second specificity determinant for assuring response to the appropriate tRNA. Furthermore, overproduction of an unchargeable variant of tRNA(Tyr) resulted in antitermination in the absence of limitation for tyrosine, supporting the proposal that uncharged tRNA is the effector in this system.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K. C., Wong J. T. Cloning and nucleotide sequence of the structural gene coding for Bacillus subtilis tryptophanyl-tRNA synthetase. Gene. 1988 Dec 20;73(2):537–543. doi: 10.1016/0378-1119(88)90518-5. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Seno T., Söll G. Is there a discriminator site in transfer RNA? Proc Natl Acad Sci U S A. 1972 Oct;69(10):3063–3067. doi: 10.1073/pnas.69.10.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debarbouille M., Arnaud M., Fouet A., Klier A., Rapoport G. The sacT gene regulating the sacPA operon in Bacillus subtilis shares strong homology with transcriptional antiterminators. J Bacteriol. 1990 Jul;172(7):3966–3973. doi: 10.1128/jb.172.7.3966-3973.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron N., Putzer H., Grunberg-Manago M. Expression of both Bacillus subtilis threonyl-tRNA synthetase genes is autogenously regulated. J Bacteriol. 1994 Jan;176(2):486–494. doi: 10.1128/jb.176.2.486-494.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon J. J., Chopin M. C., Ehrlich S. D. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992 Oct;174(20):6580–6589. doi: 10.1128/jb.174.20.6580-6589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollnick P. Regulation of the Bacillus subtilis trp operon by an RNA-binding protein. Mol Microbiol. 1994 Mar;11(6):991–997. doi: 10.1111/j.1365-2958.1994.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Goodman H. M., Abelson J., Landy A., Brenner S., Smith J. D. Amber suppression: a nucleotide change in the anticodon of a tyrosine transfer RNA. Nature. 1968 Mar 16;217(5133):1019–1024. doi: 10.1038/2171019a0. [DOI] [PubMed] [Google Scholar]
- Grandoni J. A., Fulmer S. B., Brizzio V., Zahler S. A., Calvo J. M. Regions of the Bacillus subtilis ilv-leu operon involved in regulation by leucine. J Bacteriol. 1993 Dec;175(23):7581–7593. doi: 10.1128/jb.175.23.7581-7593.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandoni J. A., Zahler S. A., Calvo J. M. Transcriptional regulation of the ilv-leu operon of Bacillus subtilis. J Bacteriol. 1992 May;174(10):3212–3219. doi: 10.1128/jb.174.10.3212-3219.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy F. J., Henkin T. M. Conservation of a transcription antitermination mechanism in aminoacyl-tRNA synthetase and amino acid biosynthesis genes in gram-positive bacteria. J Mol Biol. 1994 Jan 14;235(2):798–804. doi: 10.1006/jmbi.1994.1038. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Henkin T. M. Inducible amber suppressor for Bacillus subtilis. J Bacteriol. 1994 Apr;176(7):2108–2110. doi: 10.1128/jb.176.7.2108-2110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy F. J., Henkin T. M. The rpsD gene, encoding ribosomal protein S4, is autogenously regulated in Bacillus subtilis. J Bacteriol. 1991 Aug;173(15):4595–4602. doi: 10.1128/jb.173.15.4595-4602.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy F. J., Henkin T. M. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993 Aug 13;74(3):475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- Hasegawa T., Miyano M., Himeno H., Sano Y., Kimura K., Shimizu M. Identity determinants of E. coli threonine tRNA. Biochem Biophys Res Commun. 1992 Apr 15;184(1):478–484. doi: 10.1016/0006-291x(92)91219-g. [DOI] [PubMed] [Google Scholar]
- Henkin T. M., Chambliss G. H. Genetic mapping of a mutation causing an alteration in Bacillus subtilis ribosomal protein S4. Mol Gen Genet. 1984;193(2):364–369. doi: 10.1007/BF00330694. [DOI] [PubMed] [Google Scholar]
- Henkin T. M., Glass B. L., Grundy F. J. Analysis of the Bacillus subtilis tyrS gene: conservation of a regulatory sequence in multiple tRNA synthetase genes. J Bacteriol. 1992 Feb;174(4):1299–1306. doi: 10.1128/jb.174.4.1299-1306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski H., Goldman E. Quantities of individual aminoacyl-tRNA families and their turnover in Escherichia coli. J Bacteriol. 1984 Jun;158(3):769–776. doi: 10.1128/jb.158.3.769-776.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouze E., Bedouelle H. Structural and kinetic bases for the recognition of tRNATyr by tyrosyl-tRNA synthetase. J Mol Biol. 1989 Feb 20;205(4):729–735. doi: 10.1016/0022-2836(89)90317-3. [DOI] [PubMed] [Google Scholar]
- Lapointe J., Duplain L., Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1 in vitro. J Bacteriol. 1986 Jan;165(1):88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mulbry W. W., Ambulos N. P., Jr, Lovett P. S. Bacillus subtilis mutant allele sup-3 causes lysine insertion at ochre codons: use of sup-3 in studies of translational attenuation. J Bacteriol. 1989 Oct;171(10):5322–5324. doi: 10.1128/jb.171.10.5322-5324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M., Willis N. M., Bossi L., Gesteland R. F., Atkins J. F. Functional tRNAs with altered 3' ends. EMBO J. 1993 Jun;12(6):2559–2566. doi: 10.1002/j.1460-2075.1993.tb05911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguiza J. A., Malumbres M., Eriani G., Pisabarro A., Mateos L. M., Martin F., Martín J. F. A gene encoding arginyl-tRNA synthetase is located in the upstream region of the lysA gene in Brevibacterium lactofermentum: regulation of argS-lysA cluster expression by arginine. J Bacteriol. 1993 Nov;175(22):7356–7362. doi: 10.1128/jb.175.22.7356-7362.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putzer H., Gendron N., Grunberg-Manago M. Co-ordinate expression of the two threonyl-tRNA synthetase genes in Bacillus subtilis: control by transcriptional antitermination involving a conserved regulatory sequence. EMBO J. 1992 Aug;11(8):3117–3127. doi: 10.1002/j.1460-2075.1992.tb05384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuven N. B., Deutscher M. P. Substitution of the 3' terminal adenosine residue of transfer RNA in vivo. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4350–4353. doi: 10.1073/pnas.90.10.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman J. M., Rogers K., Rogers M. J., Söll D. Synthetase competition and tRNA context determine the in vivo identify of tRNA discriminator mutants. J Mol Biol. 1992 Dec 20;228(4):1055–1062. doi: 10.1016/0022-2836(92)90314-a. [DOI] [PubMed] [Google Scholar]
- Sherman J. M., Rogers M. J., Söll D. Competition of aminoacyl-tRNA synthetases for tRNA ensures the accuracy of aminoacylation. Nucleic Acids Res. 1992 Jun 11;20(11):2847–2852. doi: 10.1093/nar/20.11.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P., Bonamy C., Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988 Mar 11;52(5):697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- Swanson R., Hoben P., Sumner-Smith M., Uemura H., Watson L., Söll D. Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science. 1988 Dec 16;242(4885):1548–1551. doi: 10.1126/science.3144042. [DOI] [PubMed] [Google Scholar]
- Wawrousek E. F., Hansen J. N. Structure and organization of a cluster of sic tRNA genes in the space between tandem ribosomal RNA gene sets in Bacillus subtilis. J Biol Chem. 1983 Jan 10;258(1):291–298. [PubMed] [Google Scholar]
- Wawrousek E. F., Narasimhan N., Hansen J. N. Two large clusters with thirty-seven transfer RNA genes adjacent to ribosomal RNA gene sets in Bacillus subtilis. Sequence and organization of trrnD and trrnE gene clusters. J Biol Chem. 1984 Mar 25;259(6):3694–3702. [PubMed] [Google Scholar]
- Xue H., Shen W., Giegé R., Wong J. T. Identity elements of tRNA(Trp). Identification and evolutionary conservation. J Biol Chem. 1993 May 5;268(13):9316–9322. [PubMed] [Google Scholar]



