Abstract
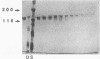
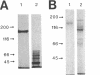
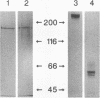
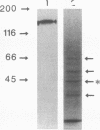
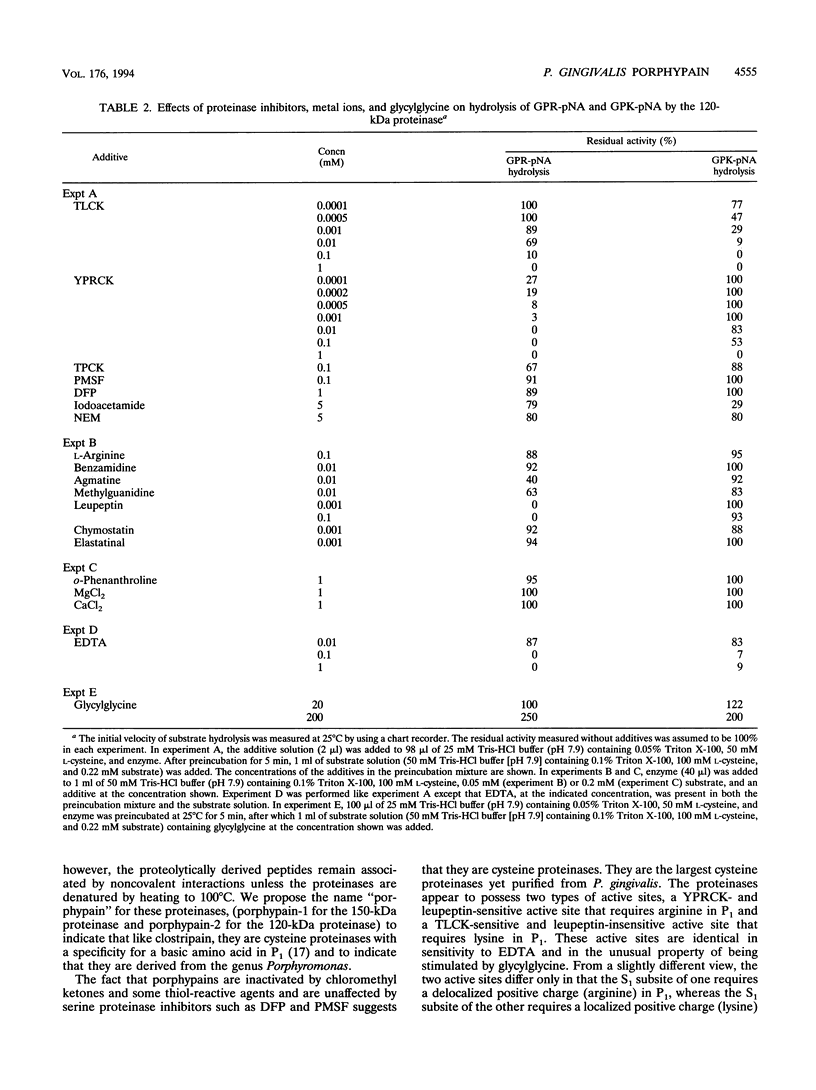
Porphyromonas gingivalis, and organism implicated in the etiology and pathogenesis of human periodontal diseases, produces a variety of potent proteolytic enzymes, and it has been suggested that these enzymes play a direct role in the destruction of periodontal tissues. We now report that two cell-associated cysteine proteinases of P. gingivalis W12, with molecular masses of approximately 150 kDa (porphypain-1) and 120 kDa (porphypain-2), as determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, have been separated and purified to apparent homogeneity. These proteinases appear to be SDS-stable conformational variants of a 180-kDa enzyme, and they are the largest cysteine proteinases yet purified from P. gingivalis. The purified proteinases hydrolyze fibrinogen, tosyl-Gly-L-Pro-L-Arg p-nitroanilide, and tosyl-Gly-L-Pro-L-Lys p-nitroanilide. While hydrolysis of both synthetic substrates by porphypain-1 and -2 requires activation by reducing agents, is inhibited by EDTA, and is stimulated in the presence of derivatives of glycine, the Arg-amidolytic activity is sensitive to leupeptin and H-D-tyrosyl-L-prolyl-L-arginyl chloromethyl ketone, whereas the Lys-amidolytic activity is sensitive to tosyl-L-lysyl chloromethyl ketone and insensitive to leupeptin. These data suggest that porphypains contain two types of active sites. These cell-associated P. gingivalis proteinases may contribute significantly and directly to periodontal tissue destruction.
Full text
PDF
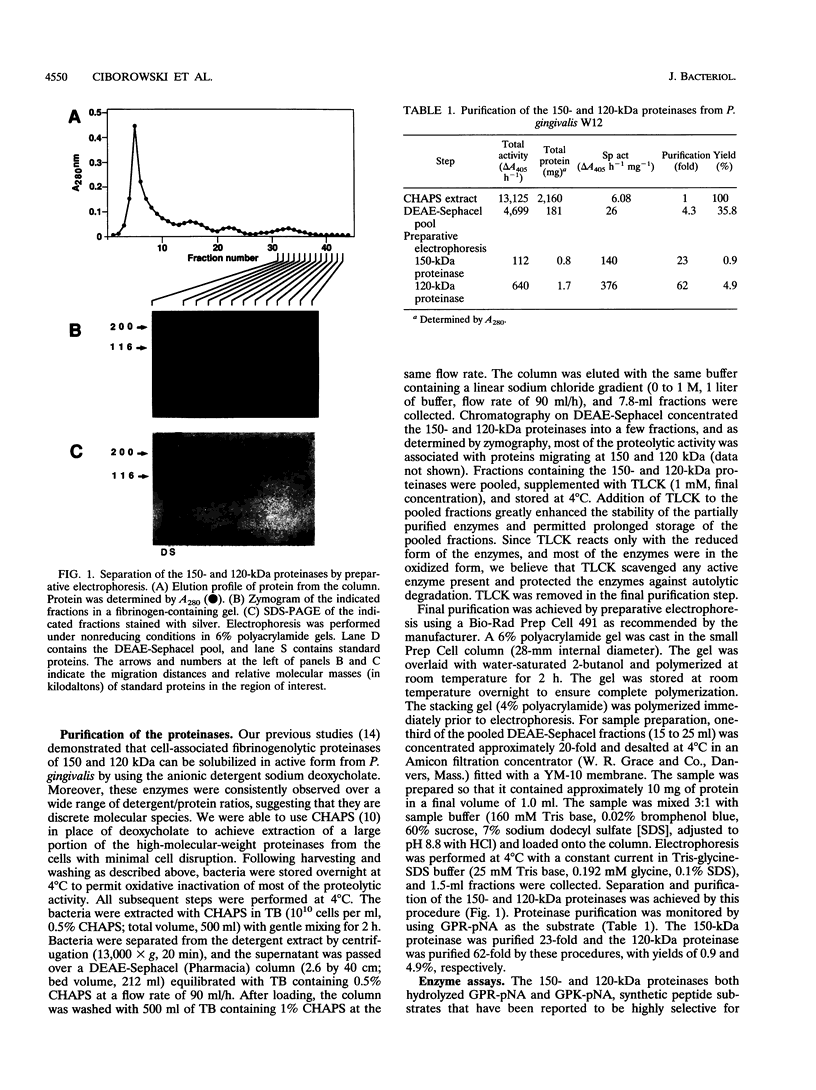


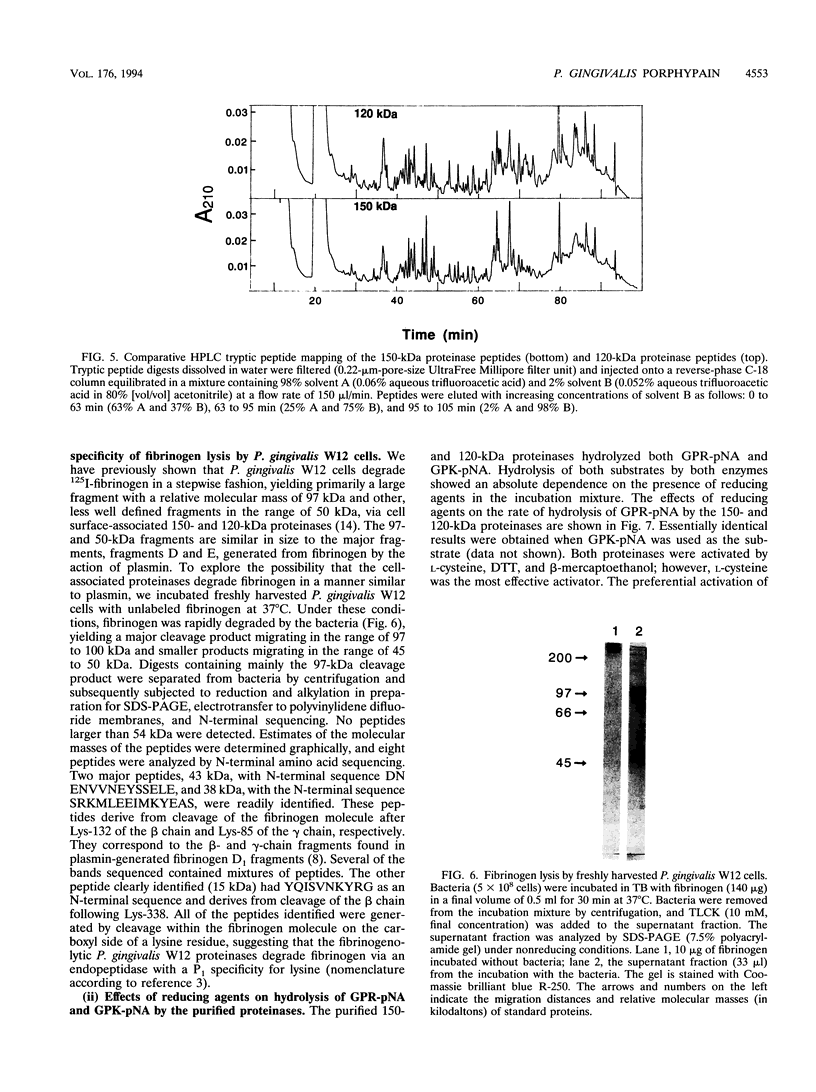
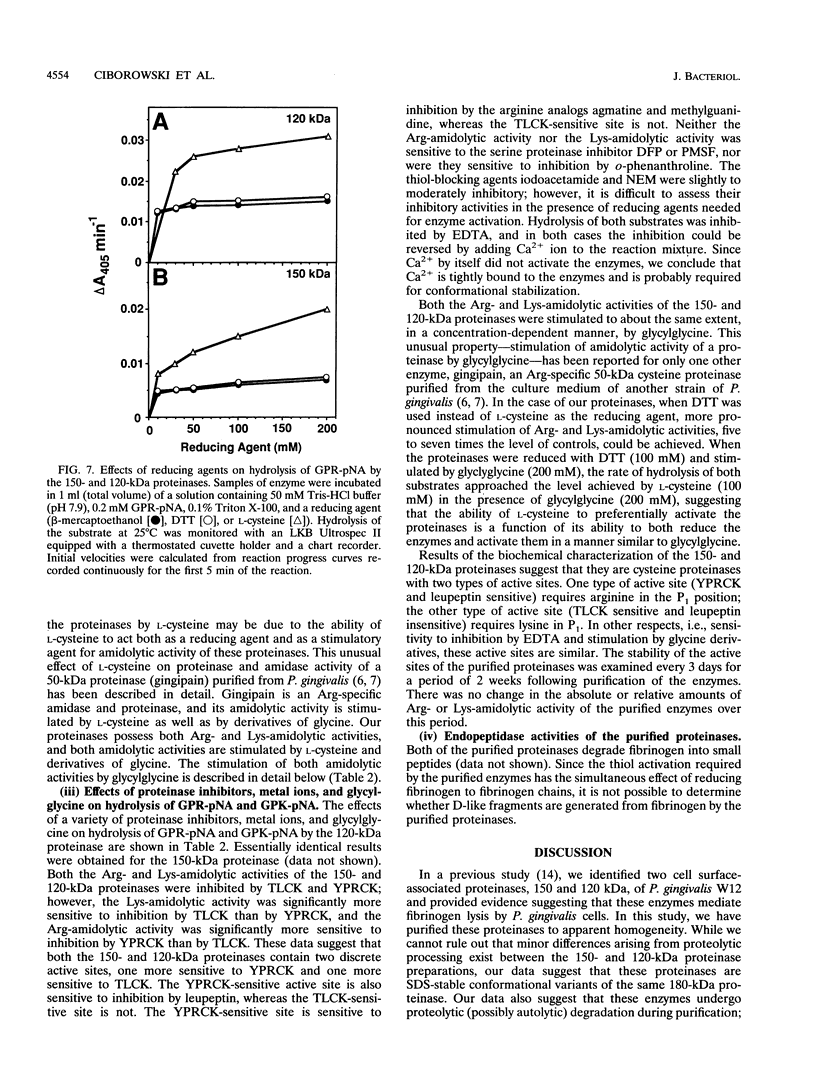


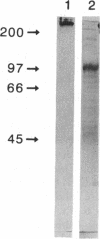
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abiko Y., Hayakawa M., Murai S., Takiguchi H. Glycylprolyl dipeptidylaminopeptidase from Bacteroides gingivalis. J Dent Res. 1985 Feb;64(2):106–111. doi: 10.1177/00220345850640020201. [DOI] [PubMed] [Google Scholar]
- Bedi G. S., Williams T. Purification and characterization of a collagen-degrading protease from Porphyromonas gingivalis. J Biol Chem. 1994 Jan 7;269(1):599–606. [PubMed] [Google Scholar]
- Berger A., Schechter I. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos Trans R Soc Lond B Biol Sci. 1970 Feb 12;257(813):249–264. doi: 10.1098/rstb.1970.0024. [DOI] [PubMed] [Google Scholar]
- Birkedal-Hansen H., Taylor R. E., Zambon J. J., Barwa P. K., Neiders M. E. Characterization of collagenolytic activity from strains of Bacteroides gingivalis. J Periodontal Res. 1988 Jul;23(4):258–264. doi: 10.1111/j.1600-0765.1988.tb01369.x. [DOI] [PubMed] [Google Scholar]
- Bourgeau G., Lapointe H., Péloquin P., Mayrand D. Cloning, expression, and sequencing of a protease gene (tpr) from Porphyromonas gingivalis W83 in Escherichia coli. Infect Immun. 1992 Aug;60(8):3186–3192. doi: 10.1128/iai.60.8.3186-3192.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. X., Potempa J., Polanowski A., Renvert S., Wikström M., Travis J. Stimulation of proteinase and amidase activities in Porphyromonas (Bacteroides) gingivalis by amino acids and dipeptides. Infect Immun. 1991 Aug;59(8):2846–2850. doi: 10.1128/iai.59.8.2846-2850.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Potempa J., Polanowski A., Wikstrom M., Travis J. Purification and characterization of a 50-kDa cysteine proteinase (gingipain) from Porphyromonas gingivalis. J Biol Chem. 1992 Sep 15;267(26):18896–18901. [PubMed] [Google Scholar]
- Doolittle R. F. Fibrinogen and fibrin. Annu Rev Biochem. 1984;53:195–229. doi: 10.1146/annurev.bi.53.070184.001211. [DOI] [PubMed] [Google Scholar]
- Hinode D., Hayashi H., Nakamura R. Purification and characterization of three types of proteases from culture supernatants of Porphyromonas gingivalis. Infect Immun. 1991 Sep;59(9):3060–3068. doi: 10.1128/iai.59.9.3060-3068.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmeland L. M. A nondenaturing zwitterionic detergent for membrane biochemistry: design and synthesis. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6368–6370. doi: 10.1073/pnas.77.11.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S. C., Bramanti T. E. Factors in virulence expression and their role in periodontal disease pathogenesis. Crit Rev Oral Biol Med. 1991;2(2):177–281. doi: 10.1177/10454411910020020301. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lantz M. S., Allen R. D., Duck L. W., Blume J. L., Switalski L. M., Hook M. Identification of Porphyromonas gingivalis components that mediate its interactions with fibronectin. J Bacteriol. 1991 Jul;173(14):4263–4270. doi: 10.1128/jb.173.14.4263-4270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz M. S., Allen R. D., Vail T. A., Switalski L. M., Hook M. Specific cell components of Bacteroides gingivalis mediate binding and degradation of human fibrinogen. J Bacteriol. 1991 Jan;173(2):495–504. doi: 10.1128/jb.173.2.495-504.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottenberg R., Christensen U., Jackson C. M., Coleman P. L. Assay of coagulation proteases using peptide chromogenic and fluorogenic substrates. Methods Enzymol. 1981;80(Pt 100):341–361. doi: 10.1016/s0076-6879(81)80030-4. [DOI] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Van Keuren M. L. Gel protein stains: silver stain. Methods Enzymol. 1984;104:441–447. doi: 10.1016/s0076-6879(84)04111-2. [DOI] [PubMed] [Google Scholar]
- Nilsson T., Carlsson J., Sundqvist G. Inactivation of key factors of the plasma proteinase cascade systems by Bacteroides gingivalis. Infect Immun. 1985 Nov;50(2):467–471. doi: 10.1128/iai.50.2.467-471.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikata M., Kanehira T., Oh H., Tani H., Tazaki M., Kuboki Y. Salivary histatin as an inhibitor of a protease produced by the oral bacterium Bacteroides gingivalis. Biochem Biophys Res Commun. 1991 Jan 31;174(2):625–630. doi: 10.1016/0006-291x(91)91463-m. [DOI] [PubMed] [Google Scholar]
- Nishikata M., Yoshimura F. Characterization of Porphyromonas (bacteroides) gingivalis hemagglutinin as a protease. Biochem Biophys Res Commun. 1991 Jul 15;178(1):336–342. doi: 10.1016/0006-291x(91)91819-x. [DOI] [PubMed] [Google Scholar]
- Otogoto J., Kuramitsu H. K. Isolation and characterization of the Porphyromonas gingivalis prtT gene, coding for protease activity. Infect Immun. 1993 Jan;61(1):117–123. doi: 10.1128/iai.61.1.117-123.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M., Endo J., Hinode D., Nagata A., Maehara R., Sato M., Nakamura R. Isolation and characterization of protease from culture supernatant of Bacteroides gingivalis. J Periodontal Res. 1987 Nov;22(6):491–498. doi: 10.1111/j.1600-0765.1987.tb02060.x. [DOI] [PubMed] [Google Scholar]
- Pike R., McGraw W., Potempa J., Travis J. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J Biol Chem. 1994 Jan 7;269(1):406–411. [PubMed] [Google Scholar]
- Plow E. F., Miles L. A. Plasminogen receptors in the mediation of pericellular proteolysis. Cell Differ Dev. 1990 Dec 2;32(3):293–298. doi: 10.1016/0922-3371(90)90042-u. [DOI] [PubMed] [Google Scholar]
- Saksela O., Rifkin D. B. Cell-associated plasminogen activation: regulation and physiological functions. Annu Rev Cell Biol. 1988;4:93–126. doi: 10.1146/annurev.cb.04.110188.000521. [DOI] [PubMed] [Google Scholar]
- Schenkein H. A. The effect of periodontal proteolytic Bacteroides species on proteins of the human complement system. J Periodontal Res. 1988 May;23(3):187–192. doi: 10.1111/j.1600-0765.1988.tb01356.x. [DOI] [PubMed] [Google Scholar]
- Scott C. F., Whitaker E. J., Hammond B. F., Colman R. W. Purification and characterization of a potent 70-kDa thiol lysyl-proteinase (Lys-gingivain) from Porphyromonas gingivalis that cleaves kininogens and fibrinogen. J Biol Chem. 1993 Apr 15;268(11):7935–7942. [PubMed] [Google Scholar]
- Smalley J. W., Birss A. J., Shuttleworth C. A. The degradation of type I collagen and human plasma fibronectin by the trypsin-like enzyme and extracellular membrane vesicles of Bacteroides gingivalis W50. Arch Oral Biol. 1988;33(5):323–329. doi: 10.1016/0003-9969(88)90065-9. [DOI] [PubMed] [Google Scholar]
- Syed S. A., Loesche W. J. Bacteriology of human experimental gingivitis: effect of plaque age. Infect Immun. 1978 Sep;21(3):821–829. doi: 10.1128/iai.21.3.821-829.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda K., Otsuka M., Ishikawa Y., Sato M., Yamamoto Y., Nakamura R. Thiol-dependent collagenolytic activity in culture media of Bacteroides gingivalis. J Periodontal Res. 1984 Jul;19(4):372–381. doi: 10.1111/j.1600-0765.1984.tb01010.x. [DOI] [PubMed] [Google Scholar]
- Yoshimura F., Nishikata M., Suzuki T., Hoover C. I., Newbrun E. Characterization of a trypsin-like protease from the bacterium Bacteroides gingivalis isolated from human dental plaque. Arch Oral Biol. 1984;29(7):559–564. doi: 10.1016/0003-9969(84)90078-5. [DOI] [PubMed] [Google Scholar]