Abstract
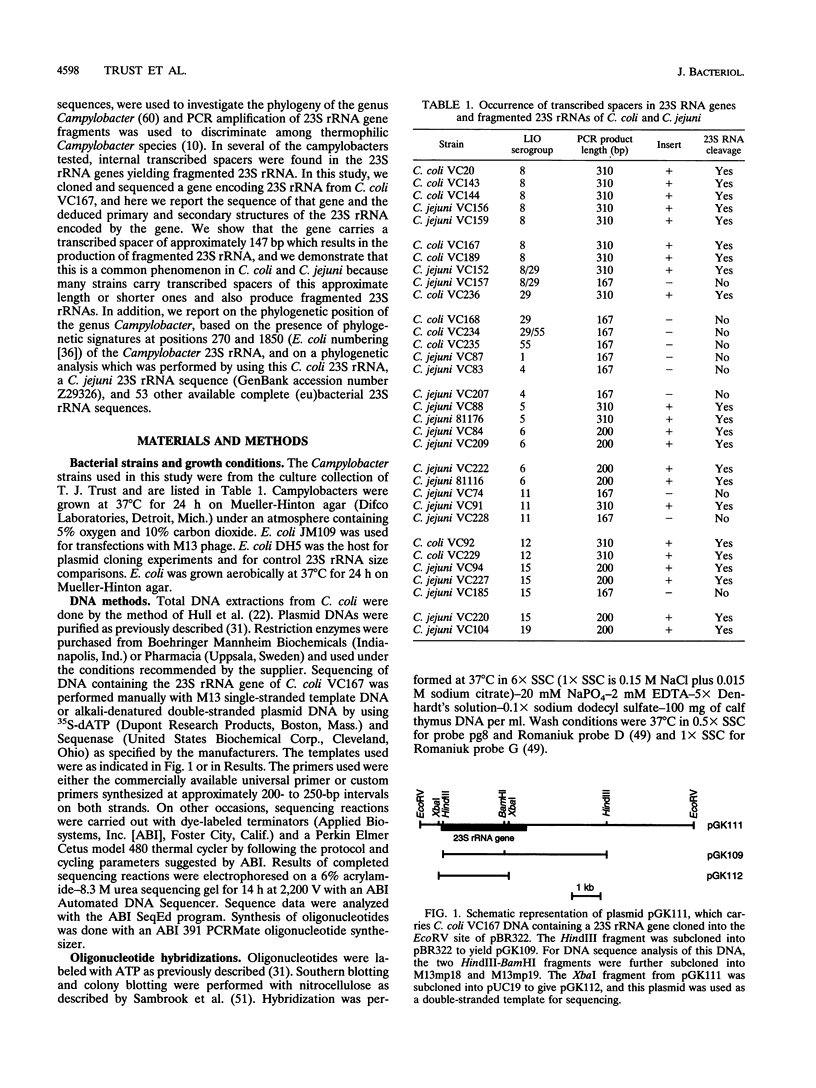
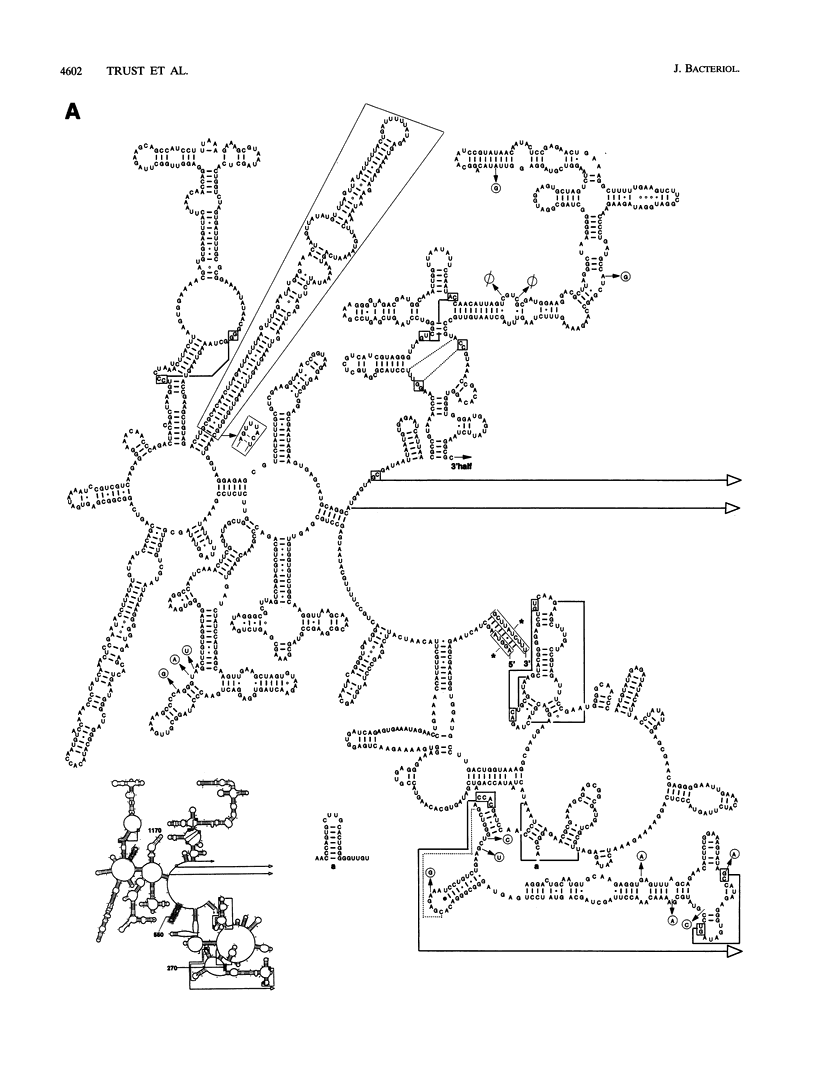
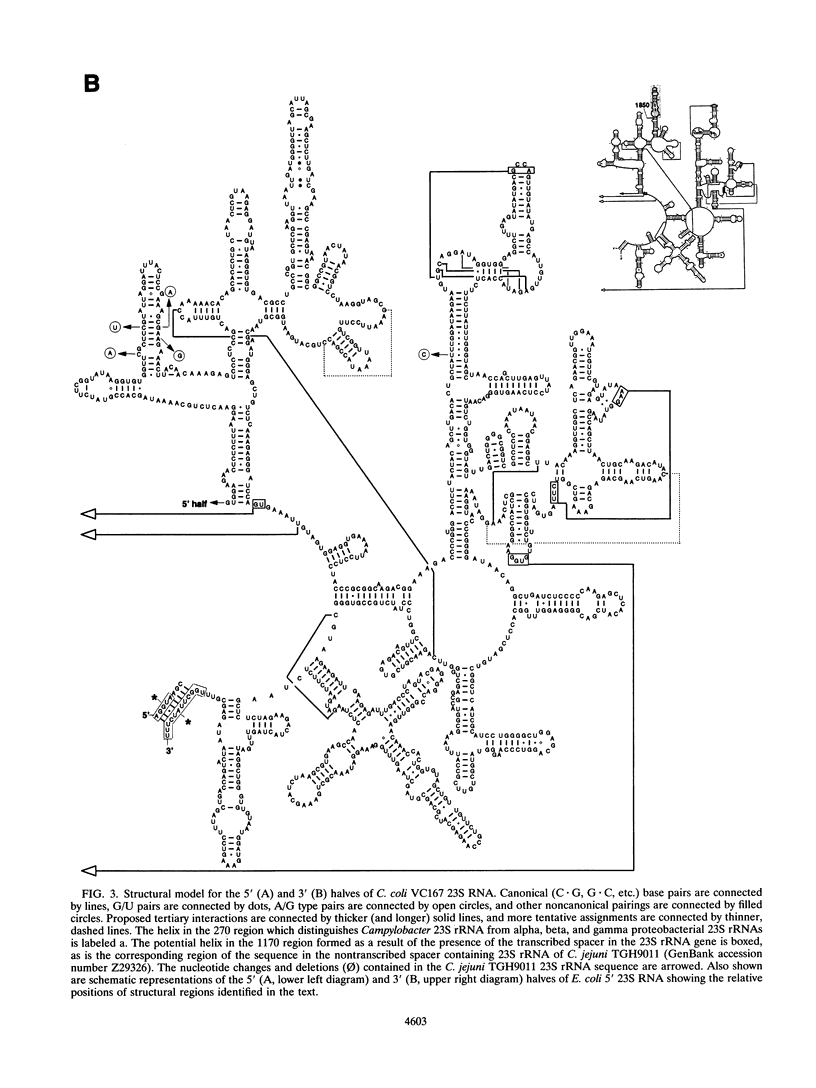
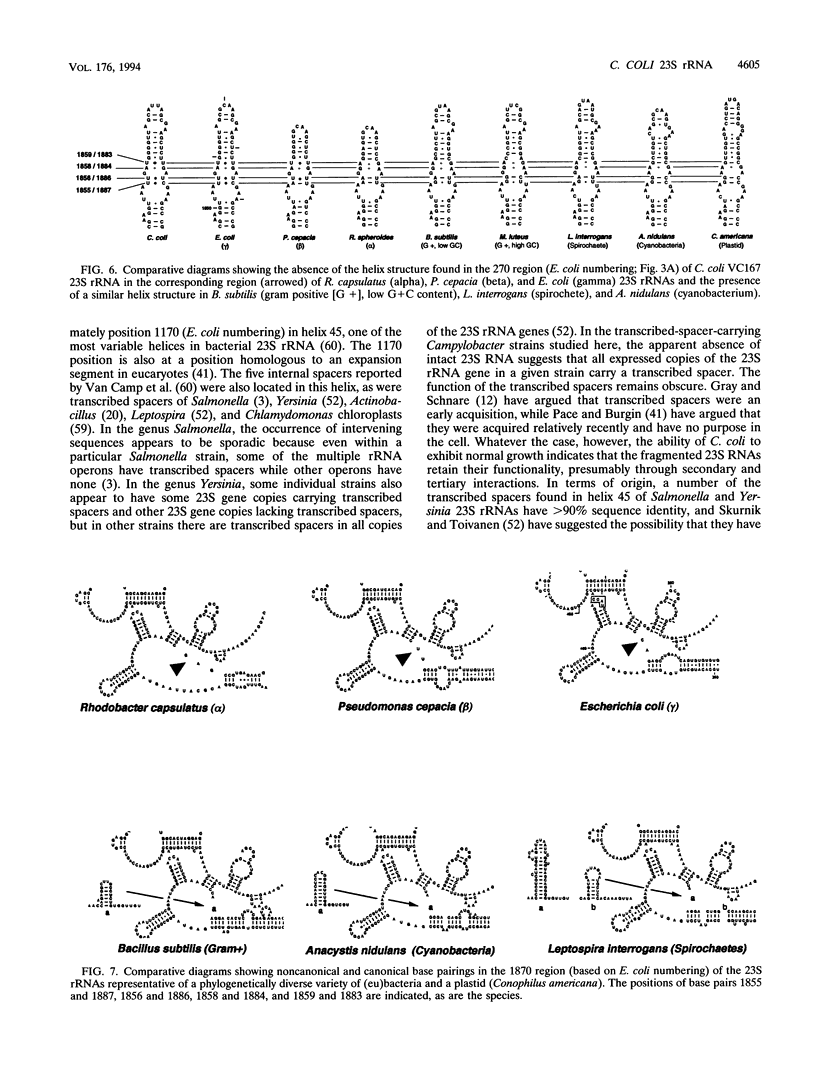
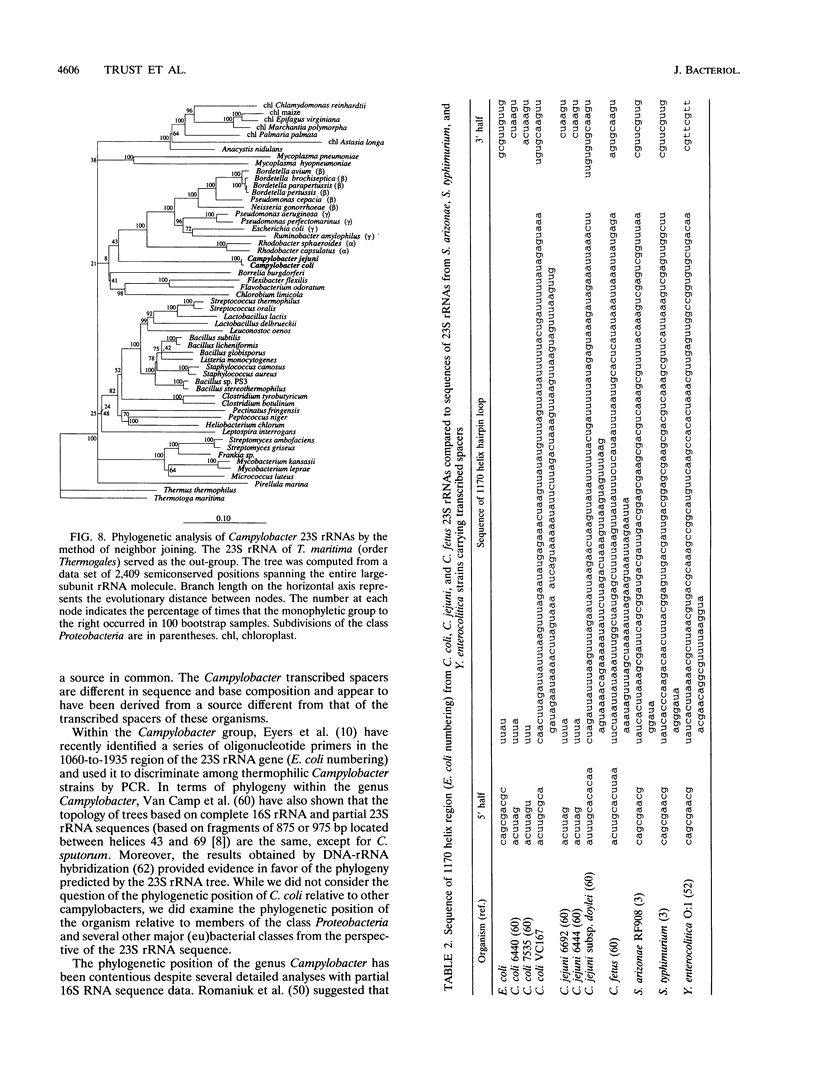
The nucleotide sequence of a 23S rRNA gene of Campylobacter coli VC167 was determined. The primary sequence of the C. coli 23S rRNA was deduced, and a secondary-structure model was constructed. Comparison with Escherichia coli 23S rRNA showed a major difference in the C. coli rRNA at approximately position 1170 (E. coli numbering) in the form of an extra sequence block approximately 147 bp long. PCR analysis of 31 other strains of C. coli and C. jejuni showed that 69% carried a transcribed spacer of either ca. 147 or ca. 37 bp. Comparison of all sequenced Campylobacter transcribed spacers showed that the Campylobacter inserts were related in sequence and percent G+C content. All Campylobacter strains carrying transcribed spacers in their 23S rRNA genes produced fragmented 23S rRNAs. Other strains which produced unfragmented 23S rRNAs did not appear to carry transcribed spacers at this position in their 23S rRNA genes. At the 1850 region (E. coli numbering), Campylobacter 23S rRNA displayed a base pairing signature most like that of the beta and gamma subdivisions of the class Proteobacteria, but in the 270 region, Campylobacter 23S rRNA displayed a helix signature which distinguished it from the alpha, beta, and gamma subdivisions. Phylogenetic analysis comparing C. coli VC167 23S rRNA and a C. jejuni TGH9011 (ATCC 43431) 23S rRNA with 53 other completely sequenced (eu)bacterial 23S rRNAs showed that the two campylobacters form a sister group to the alpha, beta, and gamma proteobacterial 23S rRNAs, a positioning consistent with the idea that the genus Campylobacter belongs to the epsilon subdivision of the class Proteobacteria.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belland R. J., Trust T. J. Deoxyribonucleic acid sequence relatedness between thermophilic members of the genus Campylobacter. J Gen Microbiol. 1982 Nov;128(11):2515–2522. doi: 10.1099/00221287-128-11-2515. [DOI] [PubMed] [Google Scholar]
- Burgin A. B., Parodos K., Lane D. J., Pace N. R. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell. 1990 Feb 9;60(3):405–414. doi: 10.1016/0092-8674(90)90592-3. [DOI] [PubMed] [Google Scholar]
- Butler P. D., Moxon E. R. A physical map of the genome of Haemophilus influenzae type b. J Gen Microbiol. 1990 Dec;136(12):2333–2342. doi: 10.1099/00221287-136-12-2333. [DOI] [PubMed] [Google Scholar]
- De Rijk P., Neefs J. M., Van de Peer Y., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1992 May 11;20 (Suppl):2075–2089. doi: 10.1093/nar/20.suppl.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBry R. W. The consistency of several phylogeny-inference methods under varying evolutionary rates. Mol Biol Evol. 1992 May;9(3):537–551. doi: 10.1093/oxfordjournals.molbev.a040740. [DOI] [PubMed] [Google Scholar]
- Eaton K. A., Dewhirst F. E., Radin M. J., Fox J. G., Paster B. J., Krakowka S., Morgan D. R. Helicobacter acinonyx sp. nov., isolated from cheetahs with gastritis. Int J Syst Bacteriol. 1993 Jan;43(1):99–106. doi: 10.1099/00207713-43-1-99. [DOI] [PubMed] [Google Scholar]
- Etoh Y., Dewhirst F. E., Paster B. J., Yamamoto A., Goto N. Campylobacter showae sp. nov., isolated from the human oral cavity. Int J Syst Bacteriol. 1993 Oct;43(4):631–639. doi: 10.1099/00207713-43-4-631. [DOI] [PubMed] [Google Scholar]
- Eyers M., Chapelle S., Van Camp G., Goossens H., De Wachter R. Discrimination among thermophilic Campylobacter species by polymerase chain reaction amplification of 23S rRNA gene fragments. J Clin Microbiol. 1993 Dec;31(12):3340–3343. doi: 10.1128/jcm.31.12.3340-3343.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., Logan S. M., Trust T. J. Genomic rearrangements associated with antigenic variation in Campylobacter coli. J Bacteriol. 1988 Jan;170(1):316–319. doi: 10.1128/jb.170.1.316-319.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson C. E., Thomas C. J., Trust T. J. Detection of Aeromonas salmonicida from fish by using polymerase chain reaction amplification of the virulence surface array protein gene. Appl Environ Microbiol. 1992 Dec;58(12):3816–3825. doi: 10.1128/aem.58.12.3816-3825.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Gray M. W., Schnare M. N. A compilation of large subunit (23S and 23S-like) ribosomal RNA structures: 1993. Nucleic Acids Res. 1993 Jul 1;21(13):3055–3074. doi: 10.1093/nar/21.13.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Larsen N., Woese C. R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994 Mar;58(1):10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Power A., Hertz G. Z., Putz E. J., Stormo G. D. Identifying constraints on the higher-order structure of RNA: continued development and application of comparative sequence analysis methods. Nucleic Acids Res. 1992 Nov 11;20(21):5785–5795. doi: 10.1093/nar/20.21.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Woese C. R. Higher order structural elements in ribosomal RNAs: pseudo-knots and the use of noncanonical pairs. Proc Natl Acad Sci U S A. 1990 Jan;87(2):663–667. doi: 10.1073/pnas.87.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraszthy V. I., Sunday G. J., Bobek L. A., Motley T. S., Preus H., Zambon J. J. Identification and analysis of the gap region in the 23S ribosomal RNA from Actinobacillus actinomycetemcomitans. J Dent Res. 1992 Sep;71(9):1561–1568. doi: 10.1177/00220345920710090401. [DOI] [PubMed] [Google Scholar]
- Hull R. A., Gill R. E., Hsu P., Minshew B. H., Falkow S. Construction and expression of recombinant plasmids encoding type 1 or D-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect Immun. 1981 Sep;33(3):933–938. doi: 10.1128/iai.33.3.933-938.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. W., Bingham H., Khawaja R., Louie H., Hani E., Neote K., Chan V. L. Physical map of Campylobacter jejuni TGH9011 and localization of 10 genetic markers by use of pulsed-field gel electrophoresis. J Bacteriol. 1992 Jun;174(11):3494–3498. doi: 10.1128/jb.174.11.3494-3498.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. W., Lombardi R., Bingham H., Hani E., Louie H., Ng D., Chan V. L. Fine mapping of the three rRNA operons on the updated genomic map of Campylobacter jejuni TGH9011 (ATCC 43431). J Bacteriol. 1993 Nov;175(22):7468–7470. doi: 10.1128/jb.175.22.7468-7470.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawiec S., Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990 Dec;54(4):502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labigne-Roussel A., Courcoux P., Tompkins L. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J Bacteriol. 1988 Apr;170(4):1704–1708. doi: 10.1128/jb.170.4.1704-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J., Harrison A. P., Jr, Stahl D., Pace B., Giovannoni S. J., Olsen G. J., Pace N. R. Evolutionary relationships among sulfur- and iron-oxidizing eubacteria. J Bacteriol. 1992 Jan;174(1):269–278. doi: 10.1128/jb.174.1.269-278.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Olsen G. J., Maidak B. L., McCaughey M. J., Overbeek R., Macke T. J., Marsh T. L., Woese C. R. The ribosomal database project. Nucleic Acids Res. 1993 Jul 1;21(13):3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P. P., DeBrunner-Vossbrinck B., Dunn B., Miotto K., MacDonnell M. T., Rollins D. M., Pillidge C. J., Hespell R. B., Colwell R. R., Sogin M. L. Phylogenetic diversity and position of the genus Campylobacter. Syst Appl Microbiol. 1987;9:231–238. doi: 10.1016/s0723-2020(87)80027-9. [DOI] [PubMed] [Google Scholar]
- Le Bourgeois P., Mata M., Ritzenthaler P. Genome comparison of Lactococcus strains by pulsed-field gel electrophoresis. FEMS Microbiol Lett. 1989 May;50(1-2):65–69. doi: 10.1016/0378-1097(89)90460-6. [DOI] [PubMed] [Google Scholar]
- Logan S. M., Trust T. J., Guerry P. Evidence for posttranslational modification and gene duplication of Campylobacter flagellin. J Bacteriol. 1989 Jun;171(6):3031–3038. doi: 10.1128/jb.171.6.3031-3038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs B., Kaplan S. 23 s precursor ribosomal RNA of Rhodopseudomonas spheroides. J Mol Biol. 1970 Apr 28;49(2):297–317. doi: 10.1016/0022-2836(70)90247-0. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Kop J., Wheaton V., Brosius J., Gutell R. R., Kopylov A. M., Dohme F., Herr W., Stahl D. A., Gupta R. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981 Nov 25;9(22):6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Nuijten P. J., Bartels C., Bleumink-Pluym N. M., Gaastra W., van der Zeijst B. A. Size and physical map of the Campylobacter jejuni chromosome. Nucleic Acids Res. 1990 Nov 11;18(21):6211–6214. doi: 10.1093/nar/18.21.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Matsuda H., Hagstrom R., Overbeek R. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994 Feb;10(1):41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Woese C. R., Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994 Jan;176(1):1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner J. L. The genus Campylobacter: a decade of progress. Clin Microbiol Rev. 1988 Apr;1(2):157–172. doi: 10.1128/cmr.1.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph D., McClelland M. Intervening sequence with conserved open reading frame in eubacterial 23S rRNA genes. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6864–6868. doi: 10.1073/pnas.90.14.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A., Sykes J. A study of the atypical ribosomal RNA components of Rhodopseudomonas spheroides. Biochim Biophys Acta. 1971 Apr 29;238(1):99–115. doi: 10.1016/0005-2787(71)90014-1. [DOI] [PubMed] [Google Scholar]
- Roller C., Ludwig W., Schleifer K. H. Gram-positive bacteria with a high DNA G+C content are characterized by a common insertion within their 23S rRNA genes. J Gen Microbiol. 1992 Jun;138(6):1167–1175. doi: 10.1099/00221287-138-6-1167. [DOI] [PubMed] [Google Scholar]
- Romaniuk P. J., Trust T. J. Rapid identification of Campylobacter species using oligonucleotide probes to 16S ribosomal RNA. Mol Cell Probes. 1989 Jun;3(2):133–142. doi: 10.1016/0890-8508(89)90024-8. [DOI] [PubMed] [Google Scholar]
- Romaniuk P. J., Zoltowska B., Trust T. J., Lane D. J., Olsen G. J., Pace N. R., Stahl D. A. Campylobacter pylori, the spiral bacterium associated with human gastritis, is not a true Campylobacter sp. J Bacteriol. 1987 May;169(5):2137–2141. doi: 10.1128/jb.169.5.2137-2141.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurnik M., Toivanen P. Intervening sequences (IVSs) in the 23S ribosomal RNA genes of pathogenic Yersinia enterocolitica strains. The IVSs in Y. enterocolitica and Salmonella typhimurium have a common origin. Mol Microbiol. 1991 Mar;5(3):585–593. doi: 10.1111/j.1365-2958.1991.tb00729.x. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Stanley J., Linton D., Burnens A. P., Dewhirst F. E., Owen R. J., Porter A., On S. L., Costas M. Helicobacter canis sp. nov., a new species from dogs: an integrated study of phenotype and genotype. J Gen Microbiol. 1993 Oct;139(10):2495–2504. doi: 10.1099/00221287-139-10-2495. [DOI] [PubMed] [Google Scholar]
- Taylor D. E., Eaton M., Yan W., Chang N. Genome maps of Campylobacter jejuni and Campylobacter coli. J Bacteriol. 1992 Apr;174(7):2332–2337. doi: 10.1128/jb.174.7.2332-2337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. E. Genetics of Campylobacter and Helicobacter. Annu Rev Microbiol. 1992;46:35–64. doi: 10.1146/annurev.mi.46.100192.000343. [DOI] [PubMed] [Google Scholar]
- Turmel M., Gutell R. R., Mercier J. P., Otis C., Lemieux C. Analysis of the chloroplast large subunit ribosomal RNA gene from 17 Chlamydomonas taxa. Three internal transcribed spacers and 12 group I intron insertion sites. J Mol Biol. 1993 Jul 20;232(2):446–467. doi: 10.1006/jmbi.1993.1402. [DOI] [PubMed] [Google Scholar]
- Vandamme P., Falsen E., Rossau R., Hoste B., Segers P., Tytgat R., De Ley J. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int J Syst Bacteriol. 1991 Jan;41(1):88–103. doi: 10.1099/00207713-41-1-88. [DOI] [PubMed] [Google Scholar]
- Wesley I. V., Wesley R. D., Cardella M., Dewhirst F. E., Paster B. J. Oligodeoxynucleotide probes for Campylobacter fetus and Campylobacter hyointestinalis based on 16S rRNA sequences. J Clin Microbiol. 1991 Sep;29(9):1812–1817. doi: 10.1128/jcm.29.9.1812-1817.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]




