Abstract
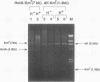
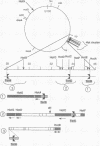
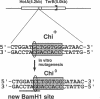
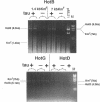
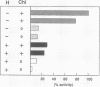
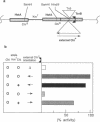
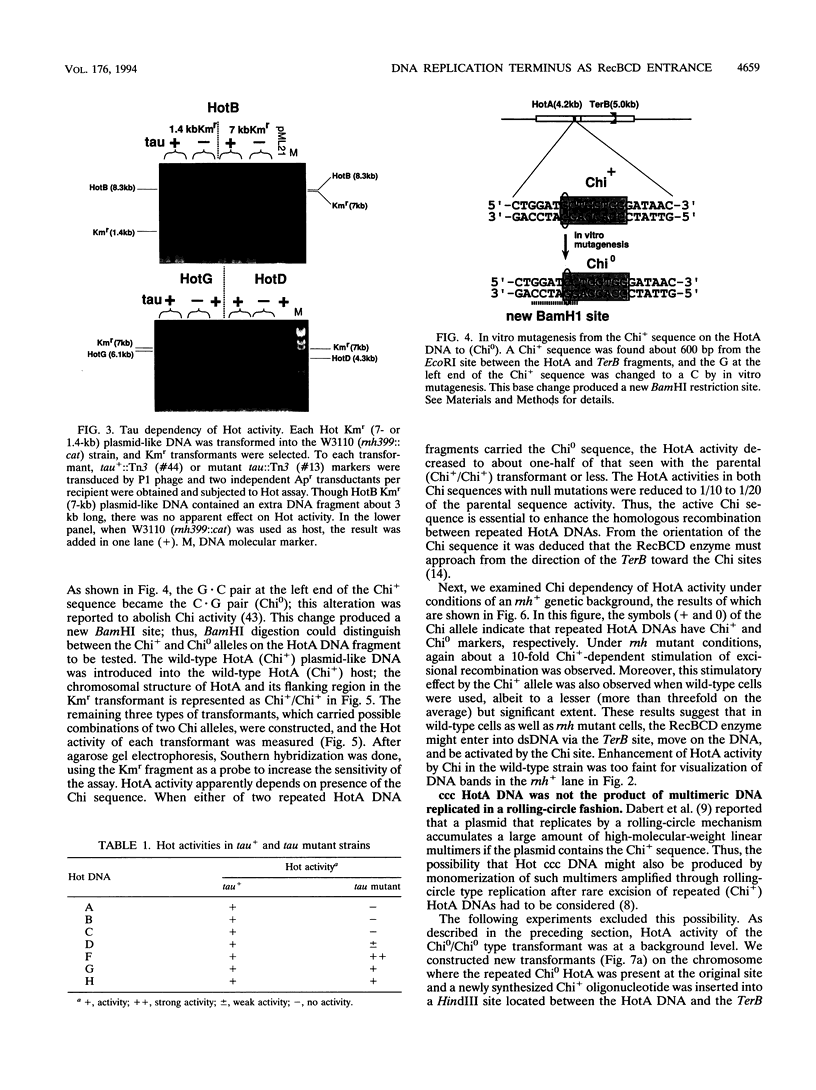
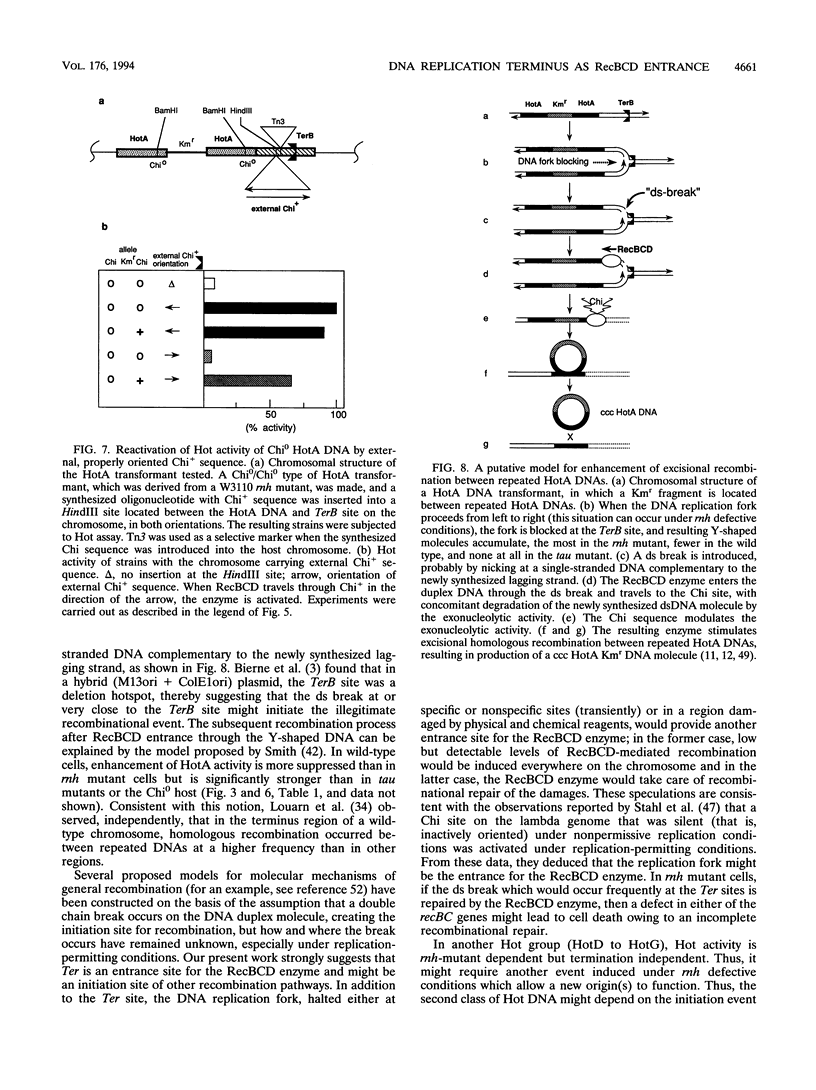
In Escherichia coli, eight kinds of chromosome-derived DNA fragments (named Hot DNA) were found to exhibit homologous recombinational hotspot activity, with the following properties. (i) The Hot activities of all Hot DNAs were enhanced extensively under RNase H-defective (rnh) conditions. (ii) Seven Hot DNAs were clustered at the DNA replication terminus region on the E. coli chromosome and had Chi activities (H. Nishitani, M. Hidaka, and T. Horiuchi, Mol. Gen. Genet. 240:307-314, 1993). Hot activities of HotA, -B, and -C, the locations of which were close to three DNA replication terminus sites, the TerB, -A, and -C sites, respectively, disappeared when terminus-binding (Tau or Tus) protein was defective, thereby suggesting that their Hot activities are termination event dependent. Other Hot groups showed termination-independent Hot activities. In addition, at least HotA activity proved to be dependent on a Chi sequence, because mutational destruction of the Chi sequence on the HotA DNA fragment resulted in disappearance of the HotA activity. The HotA activity which had disappeared was reactivated by insertion of a new, properly oriented Chi sequence at the position between the HotA DNA and the TerB site. On the basis of these observations and positional and orientational relationships between the Chi and the Ter sequences, we propose a model in which the DNA replication fork blocked at the Ter site provides an entrance for the RecBCD enzyme into duplex DNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H., Ehrlich S. D., Michel B. The replication termination signal terB of the Escherichia coli chromosome is a deletion hot spot. EMBO J. 1991 Sep;10(9):2699–2705. doi: 10.1002/j.1460-2075.1991.tb07814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely G., Colloms S., May G., Burke M., Sherratt D. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 1991 Aug;3(8):789–798. [PubMed] [Google Scholar]
- Bouché J. P. Physical map of a 470 x 10(3) base-pair region flanking the terminus of DNA replication in the Escherichia coli K12 genome. J Mol Biol. 1982 Jan 5;154(1):1–20. doi: 10.1016/0022-2836(82)90413-2. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Lockshon D., Fangman W. L. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992 Oct 16;71(2):267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- Burland V., Plunkett G., 3rd, Daniels D. L., Blattner F. R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics. 1993 Jun;16(3):551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- Clerget M. Site-specific recombination promoted by a short DNA segment of plasmid R1 and by a homologous segment in the terminus region of the Escherichia coli chromosome. New Biol. 1991 Aug;3(8):780–788. [PubMed] [Google Scholar]
- Dabert P., Ehrlich S. D., Gruss A. Chi sequence protects against RecBCD degradation of DNA in vivo. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):12073–12077. doi: 10.1073/pnas.89.24.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D. A., Kowalczykowski S. C. Homologous pairing in vitro stimulated by the recombination hotspot, Chi. Cell. 1991 Jul 26;66(2):361–371. doi: 10.1016/0092-8674(91)90625-9. [DOI] [PubMed] [Google Scholar]
- Dixon D. A., Kowalczykowski S. C. The recombination hotspot chi is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell. 1993 Apr 9;73(1):87–96. doi: 10.1016/0092-8674(93)90162-j. [DOI] [PubMed] [Google Scholar]
- Dower N. A., Stahl F. W. Chi activity during transduction-associated recombination. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7033–7037. doi: 10.1073/pnas.78.11.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulds D., Dower N., Stahl M. M., Stahl F. W. Orientation-dependent recombination hotspot activity in bacteriophage lambda. J Mol Biol. 1979 Jul 15;131(4):681–695. doi: 10.1016/0022-2836(79)90197-9. [DOI] [PubMed] [Google Scholar]
- François V., Louarn J., Louarn J. M. The terminus of the Escherichia coli chromosome is flanked by several polar replication pause sites. Mol Microbiol. 1989 Aug;3(8):995–1002. doi: 10.1111/j.1365-2958.1989.tb00250.x. [DOI] [PubMed] [Google Scholar]
- Henderson D., Weil J. Recombination-deficient deletions in bacteriophage lambda and their interaction with chi mutations. Genetics. 1975 Feb;79(2):143–174. doi: 10.1093/genetics/79.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka M., Akiyama M., Horiuchi T. A consensus sequence of three DNA replication terminus sites on the E. coli chromosome is highly homologous to the terR sites of the R6K plasmid. Cell. 1988 Nov 4;55(3):467–475. doi: 10.1016/0092-8674(88)90033-5. [DOI] [PubMed] [Google Scholar]
- Hidaka M., Kobayashi T., Horiuchi T. A newly identified DNA replication terminus site, TerE, on the Escherichia coli chromosome. J Bacteriol. 1991 Jan;173(1):391–393. doi: 10.1128/jb.173.1.391-393.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka M., Kobayashi T., Takenaka S., Takeya H., Horiuchi T. Purification of a DNA replication terminus (ter) site-binding protein in Escherichia coli and identification of the structural gene. J Biol Chem. 1989 Dec 15;264(35):21031–21037. [PubMed] [Google Scholar]
- Hill T. M., Pelletier A. J., Tecklenburg M. L., Kuempel P. L. Identification of the DNA sequence from the E. coli terminus region that halts replication forks. Cell. 1988 Nov 4;55(3):459–466. doi: 10.1016/0092-8674(88)90032-3. [DOI] [PubMed] [Google Scholar]
- Hill T. M., Tecklenburg M. L., Pelletier A. J., Kuempel P. L. tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1593–1597. doi: 10.1073/pnas.86.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T., Hidaka M. Core sequence of two separable terminus sites of the R6K plasmid that exhibit polar inhibition of replication is a 20 bp inverted repeat. Cell. 1988 Aug 12;54(4):515–523. doi: 10.1016/0092-8674(88)90073-6. [DOI] [PubMed] [Google Scholar]
- Horiuchi T., Maki H., Sekiguchi M. RNase H-defective mutants of Escherichia coli: a possible discriminatory role of RNase H in initiation of DNA replication. Mol Gen Genet. 1984;195(1-2):17–22. doi: 10.1007/BF00332717. [DOI] [PubMed] [Google Scholar]
- Itaya M., Crouch R. J. A combination of RNase H (rnh) and recBCD or sbcB mutations in Escherichia coli K12 adversely affects growth. Mol Gen Genet. 1991 Jul;227(3):424–432. doi: 10.1007/BF00273933. [DOI] [PubMed] [Google Scholar]
- Keil R. L., Roeder G. S. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984 Dec;39(2 Pt 1):377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- Khatri G. S., MacAllister T., Sista P. R., Bastia D. The replication terminator protein of E. coli is a DNA sequence-specific contra-helicase. Cell. 1989 Nov 17;59(4):667–674. doi: 10.1016/0092-8674(89)90012-3. [DOI] [PubMed] [Google Scholar]
- Kobayashi I., Stahl M. M., Stahl F. W. The mechanism of the chi-cos interaction in RecA-RecBC-mediated recombination in phage lambda. Cold Spring Harb Symp Quant Biol. 1984;49:497–506. doi: 10.1101/sqb.1984.049.01.056. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Hidaka M., Horiuchi T. Evidence of a ter specific binding protein essential for the termination reaction of DNA replication in Escherichia coli. EMBO J. 1989 Aug;8(8):2435–2441. doi: 10.1002/j.1460-2075.1989.tb08374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Hidaka M., Nishizawa M., Horiuchi T. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol Gen Genet. 1992 Jun;233(3):355–362. doi: 10.1007/BF00265431. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kuempel P. L., Henson J. M., Dircks L., Tecklenburg M., Lim D. F. dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol. 1991 Aug;3(8):799–811. [PubMed] [Google Scholar]
- Lam S. T., Stahl M. M., McMilin K. D., Stahl F. W. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics. 1974 Jul;77(3):425–433. doi: 10.1093/genetics/77.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. H., Kornberg A., Hidaka M., Kobayashi T., Horiuchi T. Escherichia coli replication termination protein impedes the action of helicases. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9104–9108. doi: 10.1073/pnas.86.23.9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louarn J. M., Louarn J., François V., Patte J. Analysis and possible role of hyperrecombination in the termination region of the Escherichia coli chromosome. J Bacteriol. 1991 Aug;173(16):5097–5104. doi: 10.1128/jb.173.16.5097-5104.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone R. E., Chattoraj D. K., Faulds D. H., Stahl M. M., Stahl F. W. Hotspots for generalized recombination in the Escherichia coli chromosome. J Mol Biol. 1978 Jun 5;121(4):473–491. doi: 10.1016/0022-2836(78)90395-9. [DOI] [PubMed] [Google Scholar]
- Nishitani H., Hidaka M., Horiuchi T. Specific chromosomal sites enhancing homologous recombination in Escherichia coli mutants defective in RNase H. Mol Gen Genet. 1993 Sep;240(3):307–314. doi: 10.1007/BF00280380. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Pickett G. G., Kogoma T., Kornberg A. RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1040–1044. doi: 10.1073/pnas.81.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli A. S., Schultz D. W., Taylor A. F., Smith G. R. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell. 1985 May;41(1):145–151. doi: 10.1016/0092-8674(85)90069-8. [DOI] [PubMed] [Google Scholar]
- Sharma B., Hill T. M. TerF, the sixth identified replication arrest site in Escherichia coli, is located within the rcsC gene. J Bacteriol. 1992 Dec;174(23):7854–7858. doi: 10.1128/jb.174.23.7854-7858.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R., Amundsen S. K., Chaudhury A. M., Cheng K. C., Ponticelli A. S., Roberts C. M., Schultz D. W., Taylor A. F. Roles of RecBC enzyme and chi sites in homologous recombination. Cold Spring Harb Symp Quant Biol. 1984;49:485–495. doi: 10.1101/sqb.1984.049.01.055. [DOI] [PubMed] [Google Scholar]
- Smith G. R., Kunes S. M., Schultz D. W., Taylor A., Triman K. L. Structure of chi hotspots of generalized recombination. Cell. 1981 May;24(2):429–436. doi: 10.1016/0092-8674(81)90333-0. [DOI] [PubMed] [Google Scholar]
- Smith G. R. Mechanism and control of homologous recombination in Escherichia coli. Annu Rev Genet. 1987;21:179–201. doi: 10.1146/annurev.ge.21.120187.001143. [DOI] [PubMed] [Google Scholar]
- Smith G. R., Schultz D. W., Crasemann J. M. Generalized recombination: nucleotide sequence homology between Chi recombinational hotspots. Cell. 1980 Mar;19(3):785–793. doi: 10.1016/s0092-8674(80)80054-7. [DOI] [PubMed] [Google Scholar]
- Stahl F. W., Crasemann J. M., Stahl M. M. Rec-mediated recombinational hot spot activity in bacteriophage lambda. III. Chi mutations are site-mutations stimulating rec-mediated recombination. J Mol Biol. 1975 May 15;94(2):203–212. doi: 10.1016/0022-2836(75)90078-9. [DOI] [PubMed] [Google Scholar]
- Stahl F. W., Thomason L. C., Siddiqi I., Stahl M. M. Further tests of a recombination model in which chi removes the RecD subunit from the RecBCD enzyme of Escherichia coli. Genetics. 1990 Nov;126(3):519–533. doi: 10.1093/genetics/126.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M. M., Kobayashi I., Stahl F. W., Huntington S. K. Activation of Chi, a recombinator, by the action of an endonuclease at a distant site. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2310–2313. doi: 10.1073/pnas.80.8.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61(1):63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- Thaler D. S., Sampson E., Siddiqi I., Rosenberg S. M., Thomason L. C., Stahl F. W., Stahl M. M. Recombination of bacteriophage lambda in recD mutants of Escherichia coli. Genome. 1989;31(1):53–67. doi: 10.1139/g89-013. [DOI] [PubMed] [Google Scholar]
- Thaler D. S., Stahl F. W. DNA double-chain breaks in recombination of phage lambda and of yeast. Annu Rev Genet. 1988;22:169–197. doi: 10.1146/annurev.ge.22.120188.001125. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Keil R. L., Roeder G. S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987 Mar 27;48(6):1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- Yagil E., Dower N. A., Chattoraj D., Stahl M., Pierson C., Stahl F. W. Chi mutation in a transposon and the orientation-dependence of Chi phenotype. Genetics. 1980 Sep;96(1):43–57. doi: 10.1093/genetics/96.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap W. Y., Kreuzer K. N. Recombination hotspots in bacteriophage T4 are dependent on replication origins. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6043–6047. doi: 10.1073/pnas.88.14.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., Fayet O., Kogoma T. Multiple origin usage for DNA replication in sdrA(rnh) mutants of Escherichia coli K-12. Initiation in the absence of oriC. J Mol Biol. 1984 Sep 15;178(2):227–236. doi: 10.1016/0022-2836(84)90141-4. [DOI] [PubMed] [Google Scholar]