Abstract
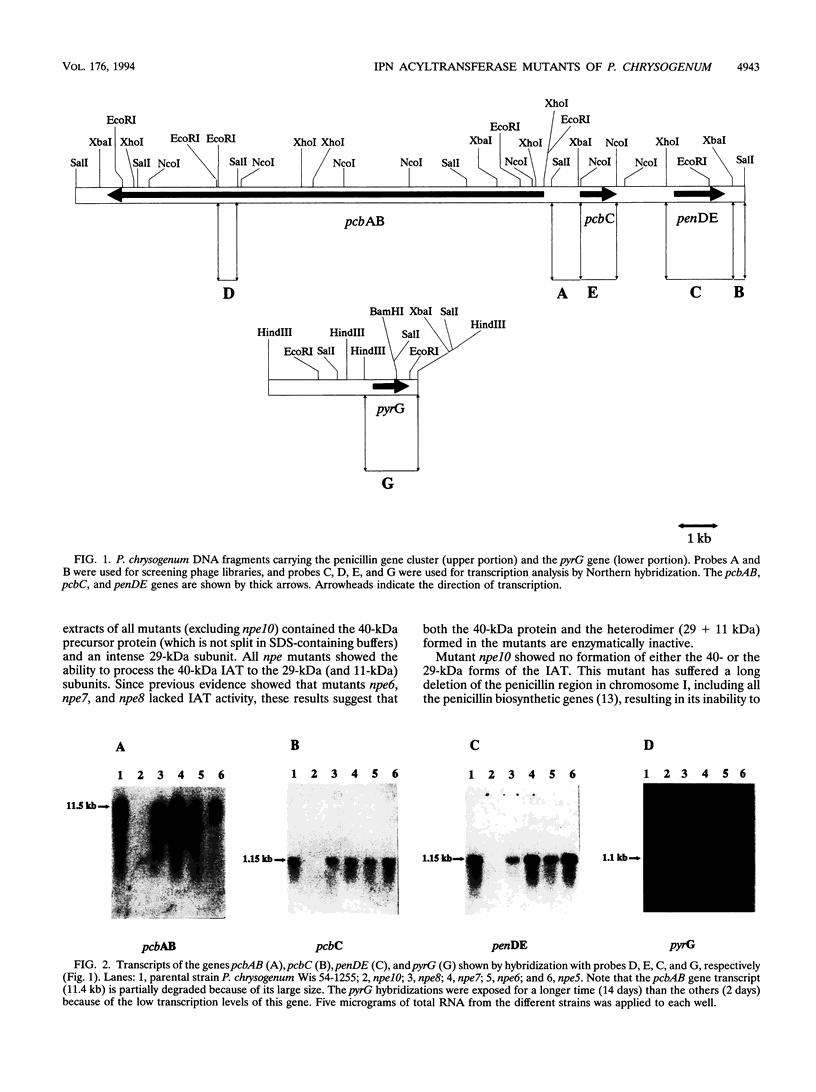
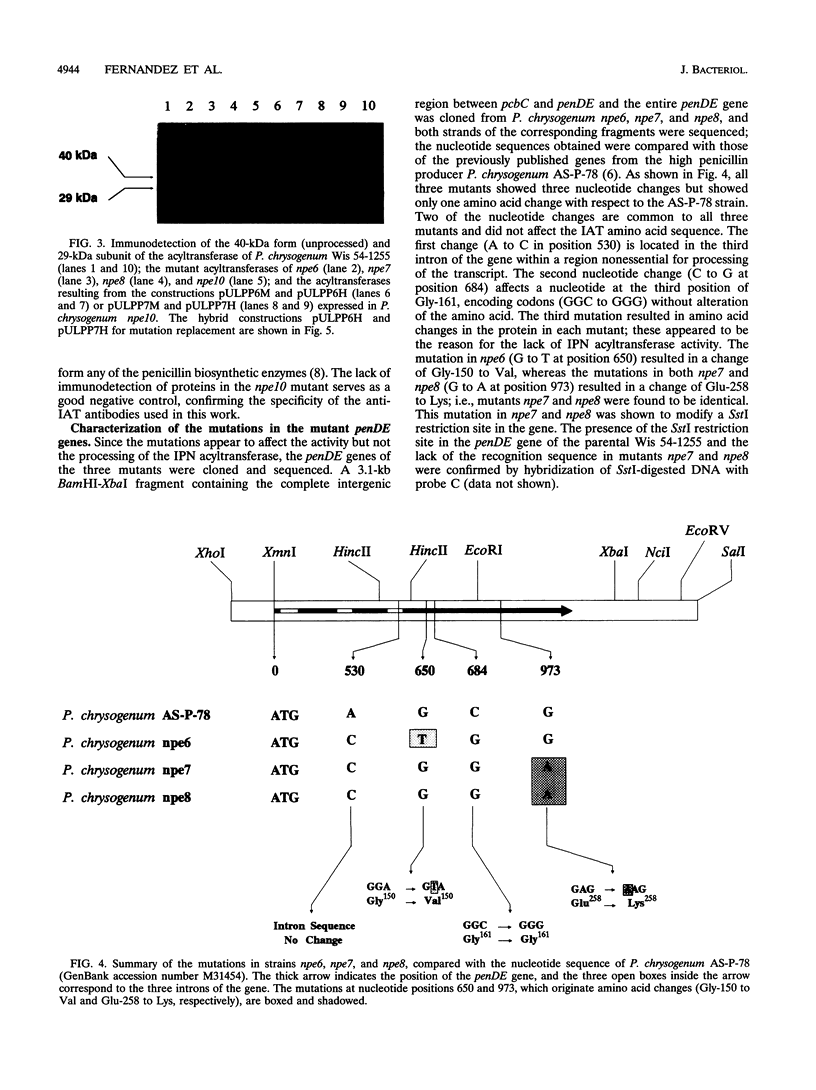
Five mutants of Penicillium chrysogenum blocked in penicillin biosynthesis (npe) which are deficient in isopenicillin N-acyltransferase were isolated previously. Three of these mutants, npe6, npe7, and npe8, have been characterized at the molecular level and compared with npe10, a deletion mutant. Transcripts of normal size (1.15 kb) of the penDE genes, which encode isopenicillin N-acyltransferase, and also of the pcbAB (11.5 kb) and pcbC (1.1 kb) genes were observed in all mutants except for the npe10 mutant. Immunoblotting studies using antibodies against isopenicillin N-acyltransferase showed that all mutants (except npe10) formed the 40-kDa (unprocessed) protein and the 29-kDa subunit of the isopenicillin N-acyltransferase. The 11-kDa subunit could not be observed in the immunoblots. The mutant penDE genes of strains npe6, npe7, and npe8 were cloned and sequenced. These three strains showed a mutation in the penDE genes which results in a single amino acid change in each modified isopenicillin N-acyltransferase. The mutation in npe6 resulted in a change of Gly-150 to Val, whereas the mutation in both npe7 and npe8 introduced a change of Glu-258 to Lys. Replacement of the Val-150 and Lys-258 mutations by constructing hybrid isopenicillin N-acyltransferase molecules led to the recovery of the isopenicillin N-acyltransferase activity. The mutations in npe6, npe7, and npe8 do not affect the ability of the 40-kDa isopenicillin N-acyltransferase to be processed into the component subunits.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alvarez E., Cantoral J. M., Barredo J. L., Díez B., Martín J. F. Purification to homogeneity and characterization of acyl coenzyme A:6-aminopenicillanic acid acyltransferase of Penicillium chrysogenum. Antimicrob Agents Chemother. 1987 Nov;31(11):1675–1682. doi: 10.1128/aac.31.11.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez E., Meesschaert B., Montenegro E., Gutiérrez S., Díez B., Barredo J. L., Martín J. F. The isopenicillin-N acyltransferase of Penicillium chrysogenum has isopenicillin-N amidohydrolase, 6-aminopenicillanic acid acyltransferase and penicillin amidase activities, all of which are encoded by the single penDE gene. Eur J Biochem. 1993 Jul 15;215(2):323–332. doi: 10.1111/j.1432-1033.1993.tb18038.x. [DOI] [PubMed] [Google Scholar]
- Aplin R. T., Baldwin J. E., Cole S. C., Sutherland J. D., Tobin M. B. On the production of alpha, beta-heterodimeric acyl-coenzyme A: isopenicillin N-acyltransferase of Penicillium chrysogenum. Studies using a recombinant source. FEBS Lett. 1993 Mar 15;319(1-2):166–170. doi: 10.1016/0014-5793(93)80060-8. [DOI] [PubMed] [Google Scholar]
- Barredo J. L., Cantoral J. M., Alvarez E., Díez B., Martín J. F. Cloning, sequence analysis and transcriptional study of the isopenicillin N synthase of Penicillium chrysogenum AS-P-78. Mol Gen Genet. 1989 Mar;216(1):91–98. doi: 10.1007/BF00332235. [DOI] [PubMed] [Google Scholar]
- Barredo J. L., van Solingen P., Díez B., Alvarez E., Cantoral J. M., Kattevilder A., Smaal E. B., Groenen M. A., Veenstra A. E., Martín J. F. Cloning and characterization of the acyl-coenzyme A: 6-aminopenicillanic-acid-acyltransferase gene of Penicillium chrysogenum. Gene. 1989 Nov 30;83(2):291–300. doi: 10.1016/0378-1119(89)90115-7. [DOI] [PubMed] [Google Scholar]
- Cantoral J. M., Gutiérrez S., Fierro F., Gil-Espinosa S., van Liempt H., Martín J. F. Biochemical characterization and molecular genetics of nine mutants of Penicillium chrysogenum impaired in penicillin biosynthesis. J Biol Chem. 1993 Jan 5;268(1):737–744. [PubMed] [Google Scholar]
- Díez B., Gutiérrez S., Barredo J. L., van Solingen P., van der Voort L. H., Martín J. F. The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the alpha-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J Biol Chem. 1990 Sep 25;265(27):16358–16365. [PubMed] [Google Scholar]
- Fierro F., Gutiérrez S., Díez B., Martín J. F. Resolution of four large chromosomes in penicillin-producing filamentous fungi: the penicillin gene cluster is located on chromosome II (9.6 Mb) in Penicillium notatum and chromosome I (10.4 Mb) in Penicillium chrysogenum. Mol Gen Genet. 1993 Dec;241(5-6):573–578. doi: 10.1007/BF00279899. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Montenegro E., Barredo J. L., Gutiérrez S., Díez B., Alvarez E., Martín J. F. Cloning, characterization of the acyl-CoA:6-amino penicillanic acid acyltransferase gene of Aspergillus nidulans and linkage to the isopenicillin N synthase gene. Mol Gen Genet. 1990 May;221(3):322–330. doi: 10.1007/BF00259395. [DOI] [PubMed] [Google Scholar]
- Müller W. H., Bovenberg R. A., Groothuis M. H., Kattevilder F., Smaal E. B., Van der Voort L. H., Verkleij A. J. Involvement of microbodies in penicillin biosynthesis. Biochim Biophys Acta. 1992 Apr 22;1116(2):210–213. doi: 10.1016/0304-4165(92)90118-e. [DOI] [PubMed] [Google Scholar]
- Müller W. H., van der Krift T. P., Krouwer A. J., Wösten H. A., van der Voort L. H., Smaal E. B., Verkleij A. J. Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO J. 1991 Feb;10(2):489–495. doi: 10.1002/j.1460-2075.1991.tb07971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queener S. W., Sebek O. K., Vézina C. Mutants blocked in antibiotic synthesis. Annu Rev Microbiol. 1978;32:593–636. doi: 10.1146/annurev.mi.32.100178.003113. [DOI] [PubMed] [Google Scholar]
- Ramsden M., McQuade B. A., Saunders K., Turner M. K., Harford S. Characterization of a loss-of-function mutation in the isopenicillin N synthetase gene of Acremonium chrysogenum. Gene. 1989 Dec 21;85(1):267–273. doi: 10.1016/0378-1119(89)90493-9. [DOI] [PubMed] [Google Scholar]
- Specht C. A., DiRusso C. C., Novotny C. P., Ullrich R. C. A method for extracting high-molecular-weight deoxyribonucleic acid from fungi. Anal Biochem. 1982 Jan 1;119(1):158–163. doi: 10.1016/0003-2697(82)90680-7. [DOI] [PubMed] [Google Scholar]
- Tobin M. B., Baldwin J. E., Cole S. C., Miller J. R., Skatrud P. L., Sutherland J. D. The requirement for subunit interaction in the production of Penicillium chrysogenum acyl-coenzyme A:isopenicillin N acyltransferase in Escherichia coli. Gene. 1993 Oct 15;132(2):199–206. doi: 10.1016/0378-1119(93)90196-a. [DOI] [PubMed] [Google Scholar]
- Tobin M. B., Fleming M. D., Skatrud P. L., Miller J. R. Molecular characterization of the acyl-coenzyme A:isopenicillin N acyltransferase gene (penDE) from Penicillium chrysogenum and Aspergillus nidulans and activity of recombinant enzyme in Escherichia coli. J Bacteriol. 1990 Oct;172(10):5908–5914. doi: 10.1128/jb.172.10.5908-5914.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco J., Gutierrez S., Fernandez F. J., Marcos A. T., Arenos C., Martin J. F. Exogenous methionine increases levels of mRNAs transcribed from pcbAB, pcbC, and cefEF genes, encoding enzymes of the cephalosporin biosynthetic pathway, in Acremonium chrysogenum. J Bacteriol. 1994 Feb;176(4):985–991. doi: 10.1128/jb.176.4.985-991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman P. A., Abraham E. P., Baldwin J. E., Fleming M. D., Schofield C. J., Sutherland J. D., Willis A. C. Acyl coenzyme A: 6-aminopenicillanic acid acyltransferase from Penicillium chrysogenum and Aspergillus nidulans. FEBS Lett. 1990 Mar 26;262(2):342–344. doi: 10.1016/0014-5793(90)80224-7. [DOI] [PubMed] [Google Scholar]