Abstract
Mitochondrial inheritance is generally assumed to be maternal. However, there is increasing evidence of exceptions to this rule, especially in hybrid crosses. In these cases, mitochondria are also inherited paternally, so “paternal leakage” of mitochondria occurs. It is important to understand these exceptions better, since they potentially complicate or invalidate studies that make use of mitochondrial markers. We surveyed F1 offspring of experimental hybrid crosses of the 17-year periodical cicadas Magicicada septendecim, M. septendecula, and M. cassini for the presence of paternal mitochondrial markers at various times during development (1-day eggs; 3-, 6-, 9-week eggs; 16-month old 1st and 2nd instar nymphs). We found evidence of paternal leakage in both reciprocal hybrid crosses in all of these samples. The relative difficulty of detecting paternal mtDNA in the youngest eggs and ease of detecting leakage in older eggs and in nymphs suggests that paternal mitochondria proliferate as the eggs develop. Our data support recent theoretical predictions that paternal leakage may be more common than previously estimated.
Introduction
Although mitochondrial DNA (mtDNA) exhibits a variety of inheritance patterns in eukaryotes [1], animal mitochondrial DNA is generally assumed to be maternally inherited. In the last 20 years, a few instances have been described in which animal mtDNA is transmitted through patrilines, a phenomenon termed “paternal leakage”. Paternal leakage challenges some of the assumptions involved in using mtDNA as a molecular or forensic marker [2]. Biparental mitochondrial inheritance followed by recombination can complicate phylogenetic reconstruction and molecular dating [3], [4]. Other authors [5], [6] note that divergence time estimates for Drosophila simulans and D. mauritiana differ fourfold, depending on whether an mtDNA polymorphism is ancient or the result of introgression between species.
Table 1 lists animal studies that demonstrate paternal leakage. Much of this work suggests that paternal leakage may be more likely when hybridization is involved, possibly due to the breakdown of mechanisms that normally destroy or exclude paternal mtDNA (for brief review, see [1]). Although reported cases of paternal leakage for animal mtDNA are few, the diversity of the taxa involved and the relative novelty of sensitive PCR-based detection techniques [7] combined with a lack of widespread effort to quantify this phenomenon and the inability of researchers to detect hybridization if maternal and paternal mitochondrial genotypes are identical, raise the possibility that paternal leakage may be more widespread than once thought. Here we present evidence of paternal leakage in hybrid crosses involving three species of 17-year periodical cicada, Magicicada septendecim, M. septendecula and M. cassini.
Table 1. Some examples of paternal leakage in the literature.
| Common Name | Reference | |
| Heterospecific crosses | ||
| Silkmoth | Antheraea pernyi X A. roylei | [49] |
| Fruit fly | Drosophila mauritiana X. D. simulans | [42] |
| Fruit Flies | Drosophila mauritiana X. D. simulans | [44] |
| Tobacco budworm | Heliothis virescens X H. subflexa | [31] |
| Periodical Cicada | Magicicada septendecim X M. cassini | This study |
| Periodical Cicada | Magicicada septendecim X M. septendecula | This study |
| House Mouse | Mus musculus X M. spretus | [50] |
| House Mouse | Mus musculus X M. spretus | [38] |
| House Mouse | Mus musculus X M. spretus | [51] |
| Conspecific crosses | ||
| Honeybee | Apis mellifera carnica X A. mellifera capensis | [33] |
| Cow | Bos taurus | [52] |
| Scorpion | Buthus mardoechi | [53] |
| Frillneck lizard | Chlamydosaurus kingii | [54] |
| Fruit Flies | Drosophila mauritiana X. D. simulans | [44] |
| Anchovy | Engraulis encrasicolus | [55] |
| Human | Homo sapiens | [56] |
| Scorpion | Mesobuthus caucasius | [53] |
| Scorpion | Mesobuthus eupeus | [53] |
| Scorpion | Mesobuthus gibbosus | [53] |
| Sheep | Ovis aries | [57] |
| Eastern tiger swallowtail | Papilio glaucus | [58] |
| Great tit | Parus major major X P. major minor | [59] |
| Flatfish | Platichthys flesus | [60] |
Background
Paternal leakage is of particular interest in the periodical cicadas of North America (Hemiptera: Magicicada spp.) because mitochondrial markers have been central in evolutionary studies of these species. For example, an abrupt mtDNA haplotype (and nuclear color polymorphism) transition has been interpreted as evidence for a lack of gene flow between courtship-song-displaced, synchronic species [8]–[10], and the same haplotype boundary has been interpreted as evidence for sex-biased dispersal [11]–[13]. Either of these interpretations would be complicated by paternal leakage.
The seven currently-recognized 13- and 17-year periodical cicada species (Magicicada septendecim {17}, M. tredecim {13}, M. neotredecim {13}, M. cassini {17}, M. tredecassini {13}, M. septendecula {17}, and M. tredecula {13}), belong to three species groups (-decim, -cassini, and -decula), and each species is most closely related to one with the alternative life cycle (13 or 17 years), suggesting multiple allochronic speciation events. Within each Magicicada species group, mitochondrial genetic differences are slight (0% between 13- and 17-year -decula or -cassini species pairs and 2.6% uncorrected between the M. septendecim/M. neotredecim and M. tredecim). Uncorrected mtDNA distances are 3–4% when comparing -decula to -cassini species and 7–8% between members of either of these species groups and the -decim group [14].
Within a given geographical region, periodical cicadas emerge synchronously in mass numbers, and adults form mixed-species choruses. Different regions are on different emergence schedules with the “brood” year designated by sequential Roman numerals. Although choruses provide opportunities for hybridization, mixed-species matings are rare [15]–[19]. More detailed background information on periodical cicada broods, species, and behavior is available elsewhere [10], [14], [16], [20]–[24].
After emerging from the ground, periodical cicada nymphs undergo ecdysis immediately, after which they spend 5–9 days as relatively inactive, teneral adults [25]–[28]. After the teneral period, adults become more active and mate. Because periodical cicadas are superabundant, unmated teneral adults are relatively easy to obtain. Although these insects are difficult to maintain in the laboratory, they may be maintained and manipulated under semi-natural conditions in outdoor cages containing living, woody vegetation. When males and females are confined in cages and given no choice of mates, they will engage in hybrid matings, and hybrid eggs and nymphs are viable [16], [29].
We developed a PCR-based method that makes use of specific primers and known Magicicada haplotype differences to detect rare mtDNA haplotypes in experimentally-crossed cicadas (our method is similar to that in [30]). We tested this method on mtDNA mixtures made by combining, in different proportions, the DNA of wild-caught individuals of known species, and we then used these primers to investigate the possibility of paternal leakage in reciprocal crosses of M. septendecim with M. cassini and M. septendecula.
Results
Paternal Leakage
We were unable to extract DNA from one pooled sample of 6-week old eggs, and two extractions of 16-month nymphs from M. septendecim homospecific crosses failed to amplify. All other extractions and amplifications were successful. Paternal mtDNA was not found in the 1-day old hybrid eggs, but it was found in all older age groups (4 days, 3 weeks, 6 weeks, 9 weeks, and 16 months; Table 2). The single 4-day old sample was present because a new eggnest from that particular female had to be re-cut because the day-old nest originally cut and dissected was found to be empty. As noted by White [29], heterospecific crosses involving male -decim were fewer in number than other heterospecific crosses because female-cassini and -decula are less likely to mate with the less aggressive -decim males.
Table 2. Number of crosses showing amplification of maternal and paternal haplotypes.* .
| 1 Day* | 3 Weeks | 6 Weeks | 9 Weeks | 16 Months | Total | Total | ||||||
| mat | both | mat | both | mat | both | mat | both | mat | both | mat | both | |
| cassini M×decim F | 10 | 1 | 2 | 12 | 5 | 8 | 0 | 4 | 5 | 10 | 22 | 35 |
| decula M×decim F | 8 | 0 | 9 | 3 | 6 | 3 | - | - | - | - | 23 | 6 |
| decim M×cassini F | 3 | 0 | 0 | 1 | - | - | 4 | 1 | - | - | 7 | 2 |
| decim M×decula F | - | - | 0 | 1 | 0 | 1 | - | - | - | - | 0 | 2 |
| decim×decim | - | - | - | - | - | - | 5 | U | 13 | U | 18 | U |
| cassini×cassini | - | - | - | - | - | - | 3 | U | - | - | 3 | U |
| total | 21 | 1 | 11 | 17 | 11 | 12 | 12 | 5 | 18 | 10 | 73 | 45 |
“mat” tested positive for maternal mtDNA only; “both” tested positive for both maternal and paternal mtDNA; “U” paternal inheritance cannot be determined because the paternal genome is identical to the maternal; “-” no eggs or nymphs were collected for this cross at this time period or samples that were collected failed to extract/amplify (only one pooled sample of 6-week old eggs failed to extract–a -decula M×-decim F; only two extractions failed to amplify and both of these were -decim M×-decim F 16-month old nymphs). Fewer offspring from crosses involving -decim males and heterospecific females were sampled because -decula and -cassini females most often rejected -decim males.
Ruling Out Numts
To ensure that the primers were amplifying mitochondrial COI and not nonfunctional nuclear copies of mitochondrial genes (NUMTs), the amplified paternal and maternal mtDNA from representative sequences of hybrid mixed-haplotype eggnest extractions (8 sequences from -decim×-cassini and 8 from -decim×-decula) were compared to the original COI sequence. All of the sequences exactly corresponded to the original COI sequence of the proper primer set and none of the species-specific primer sets produced sequence that had errors (subpeaks, weak sequence, etc.) that suggested multiple templates. Furthermore, we found no stop codons in any of the sequences, and all species-specific base substitutions were silent (e.g., did not affect amino acid sequence), suggesting that our sequences belong to functional genes.
Ruling Out Contamination
We were careful to avoid contamination that would lead to false positive results (i.e., the detection of mtDNA from heterospecific contamination rather than from paternal leakage). The best evidence against heterospecific contamination was that sample DNA from homospecific crosses amplified with homospecific primers but never with heterospecific primers. Homospecific contamination would be undetectable because contaminant DNA would be identical to sample DNA; however, this would not change our current conclusions about paternal leakage because we cannot detect paternal mtDNA in homospecific crosses for the same reason.
Contaminant DNA is most likely to be amplified when there is little or no target DNA. Every set of PCR reactions (made from a single reaction mix) included a negative control (no maternal or paternal target mtDNA) and none of these control reactions ever showed any sign of amplified DNA.
A final pair of controls included DNA from non-hybrid adults (extracted from a single leg) of both species involved in each cross. This adult DNA was tested with both homo- and hetero-specific primers. These controls were included in every PCR set (made from a single batch of reaction mix) and were visualized on the same gels as the experimental samples (Figs. 1–2). Failure of heterospecific primers and success of the homospecific primers assured us that the correct primers had been added to all reaction mixtures and that contamination was not present.
Figure 1. Example of -decim×-cassini hybrid PCR product in 2% agarose gel stained with Sybrsafe.
First column: primer specific to maternal mtDNA; Lanes 1-5: -decim F×-cassini M; Lane 6: -decim M×-cassini F; Lane 7: Maternal species DNA; Lane 8: 100 bp Ladder; Lane 9: Paternal species DNA; Lane 10: H20 negative control (No DNA); Second column: primer specific to paternal mtDNA. Lanes 1-5: -decim F×-cassini M; Lane 6: -decim M×-cassini F; Lane 7: Paternal species DNA; Lane 8: Ladder; Lane 9: Maternal species DNA; Lane 10: H20 negative control (No DNA). Lanes 4-6 show paternal leakage.
Figure 2. Example of -decim×–decula hybrid PCR product in 2% agarose gel stained with Sybrsafe.
First column: primer specific to maternal mtDNA; Lanes 1-5: -decim F×-decula M; Lane 6: -decim M×-decula F; Lane 7: Maternal species DNA; Lane 8: Ladder; Lane 9: Paternal species DNA; Lane 10: H20 negative control (No DNA); Second column: primer specific to paternal mtDNA. Lanes 1-5: -decim F×-decula M; Lane 6: -decim M×-decula F; Lane 7: Paternal species DNA; Lane 8: Ladder; Lane 9: Maternal species DNA; Lane 10: H20 negative control (No DNA). Lanes 4-6 show paternal leakage.
Discussion
Our results suggest that paternal leakage occurs in hybrid Magicicada. Until recently, the most sensitive techniques for detecting paternal leakage involved backcrossing experiments [31] that could not be used to detect leakage in wild populations or in animals (such as periodical cicadas) with long life cycles. Although PCR-based methods may be susceptible to NUMTs (nonfunctional nuclear copies), whose transmission is biparental, for NUMTs to explain our results each periodical cicada species would need to have an exclusive NUMT not found in the other species, and this exclusive NUMT would need to match the paternal mitochondrial sequence of each cross exactly. We consider this possibility to be highly unlikely. In addition, as explained in the results section, contamination controls argue in favor of paternal leakage.
Our results suggest that paternally-transmitted mitochondria in Magicicada proliferate during development. We detected paternal leakage in all age groups examined except 1-day old eggs. The relative difficulty of detecting leakage in the youngest eggs suggests a scenario in which, as expected, paternal mitochondria are present but extremely rare (and difficult to detect) at first. They then proliferate as the eggs develop (and thus they become more reliably detectable). Our results cannot be explained by ejaculate residue contamination during oviposition; if we were detecting surface contamination rather than leakage, we would expect that paternal haplotypes might be detected in some eggs, but undetectable in hatched nymphs. Given the strong possibility that paternal mitochondria are replicating (if so, then template mtDNA concentrations changed throughout our study), it is not advisable to use the results of these experiments to evaluate the relative frequencies of paternal leakage of the different crosses, since quantitative conclusions from our experiments may be confounded by changing mtDNA template concentrations.
Although we detected paternal mtDNA in hybrid juvenile periodical cicadas, it remains unknown whether paternal mtDNA will persist through development or whether it enters the germ line [32]. In at least one example from holometabolous insects (those with complex metamorphosis), heteroplasmy is not maintained. Meusel [33] found that in honeybees, the paternal contributions disappeared during development. We suspect that the evidence for proliferation of paternal mtDNA in our developing cicadas and the simple metamorphosis of cicadas make it likely that heteroplasmy will persist through to adulthood. We have left some nymphs from this study growing underground and we are continuing to monitor them for evidence of heteroplasmy.
Several factors are thought to contribute to the rarity of paternal leakage. First, in some cases, sperm may not enter oocytes (observed in some tunicates), or sperm may not contain mitochondria (observed in some crayfish species [34]), so paternal leakage is not possible. Even in organisms in which sperm enter oocytes and sperm contain mitochondria, maternal mtDNA outnumber paternal mtDNA by as much as 10,000 fold [35], and the relatively low numbers of paternal relative to maternal mitochondria may have a swamping effect when a zygote is formed [36], [37]. However, one common explanation for the rarity of paternal leakage is that oocytes have mechanisms for actively destroying objects with foreign surface proteins, or, as shown in mammals, that paternal mitochondria are ubiquitinated (either during spermatogenesis or after fertilization) and destroyed by the oocyte [38]–[41].
Some studies of Drosophila [42], [43] suggest that leakage is most likely if the genetic difference between species is approximately 2.5% or greater due to the fact that oocyte enzymes cannot recognize and destroy distantly-related sperm mitochondria, but a more recent study of Drosophila [44] demonstrates paternal leakage between closely related subspecies (<2.5% difference). The Magicicada species used in our experiments exhibit roughly 7–8% sequence divergence (uncorrected) but other Magicicada species pairs are more closely related (3–4% for the -cassini versus -decula siblings, 2.6% for M. septendecim vs. M. tredecim, and close to 0% for M. cassini vs. M. tredecassini and M. sependecula vs. M. tredecula) [14] suggesting the opportunity for further tests of this hypothesis.
Paternal leakage may be more common than previously thought for several reasons. In conspecific crosses paternal mtDNA may be undetectable if mtDNA haplotype variation within populations or among interbreeding populations is slight or absent as in many animal mtDNA studies (e.g., animal species living in previously glaciated areas of North America and Europe [45]), leading to biases against detecting leakage except in cases involving hybridization [46]. Other reasons that paternal leakage may be difficult to detect are that it may occur in some individuals and not others, or with some kinds of crosses and not others. Kondo et al. [42] found that of 331 lines of Drosophila simulans backcrossed with D. mauritiana (backcrossed for ten generations), only four lines showed evidence for paternal leakage of mtDNA. Significantly, in three of these four lines, the maternal mtDNA was completely replaced by the paternal mtDNA while in the fourth, individuals were heteroplasmic. All of these crosses were D. simulans females crossed with D. mauritiana males. In other hybrid crosses, it has been shown that mtDNA from one of the parental species may not survive as well as the other in a hybrid background [5]. Finally, paternal leakage may have gone unnoticed because researchers, expecting it to be virtually non-existent, have not looked for it; evidence for heteroplasmy in mtDNA sequences may have been taken to be artifacts or low-level nuclear copies of mtDNA.
Other evidence that paternal leakage of animal mtDNA might be surprisingly common is presented by Piganeau et al. [4], who found strong evidence of mtDNA recombination (between presumably maternal and paternal mtDNA) based on statistical analysis of 279 animal taxa (156 vertebrates, 57 arthropods, 29 mollusks, 12 nematodes, and 11 echinoderms). Their analyses did not allow them to pinpoint the exact taxa that displayed recombination but they were able to isolate the twenty species that contributed most to the result and were therefore most likely to contain recombinant genotypes. These twenty animal taxa comprised a wide taxonomic sampling (one nematode, one insect, one collembolan, one crustacean, one cephalopod and 13 vertebrates). Two bivalve mollusks were also represented but these species are known to have regular, tissue-specific, double uniparental inheritance of mtDNA. There was no indication that the frequency of recombination varied across taxonomic groups. In two of the twenty strongest cases, recombinant individuals could be recognized and both cases appeared to involve hybridization between subspecies.
Our data add to the growing number of successful interspecific paternal leakage studies. We suggest the need for more surveys of natural populations of hybrid individuals and for more experimental crosses between species and between divergent haplotypes within species to look for paternal leakage. Such studies are important for clarifying potential problems with analyses that rely on exclusively maternal mtDNA inheritance. In addition, such studies might help clarify the reasons why mitochondrial inheritance is ever uniparental.
Materials and Methods
During the emergences of Brood IX (2003) and X (2004) of 17-year periodical cicadas, we collected unmated (newly emerged) cicadas from various locations (Table 3) and performed purebred and cross-species matings by enclosing males and females in small cages (Figures 3– 7; crosses performed and numbers of matings are reported in Table 4). Mating cages contained either males and females of the same species (controls) or males of one species with females of another species (heterospecific crosses) so individuals were not free to choose the species with which they mated. Natural hybridization is rare, partly because females are unresponsive to the songs of heterospecifics [47] (Typical songs from each species group are included in Audio S1, S2, S3, S4). We facilitated hybrid matings by placing heterospecific mating cages near homospecific cages. This arrangement allowed females to hear males of their own species and to signal sexual receptivity, increasing the odds that a heterospecific male in her own cage would mate her. After mating, females were isolated in individually marked cages surrounding live tree branches suitable for oviposition and feeding. The cages were monitored for oviposition, and at four time periods after laying (1 day, 3 weeks, 6 weeks and 9 weeks), approximately 3 eggnests (approximately 60 eggs) were collected from each female's cage. After the eggnests were cut from the branches, the eggs were removed and stored in 100% ethanol. We found that one of the eggnests dissected from the 1-day age group was empty, so we cut and dissected another eggnest from the same female; by the time we did this, the eggnest was 4 days old. All remaining eggnests were clipped from the trees just prior to hatching and the hatching nymphs were allowed to burrow into the ground in marked 1 m2 plots in a second-growth Oak-Hickory forest in Connecticut. After approximately 16 months, cicada nymphs from one control and one hybrid cross (-decim×-decim and -cassini male×-decim female) were excavated and stored in 100% ethanol. All females in this experiment were permitted to mate only once, ruling out mixed paternity among the eggs of an eggnest.
Table 3. Periodical cicada collection sites.
| Brood | Species | Collection Sites |
| XI | M. septendecim | Wilkes County, NC; Pipestem State Park, VA |
| M. cassini | South Gap, VA; Bluestone State Park, VA; Vernick Creek, VA | |
| M. septendecula | Bluestone State Park, VA | |
| X | M. septendecim | Hunterdon County, NJ (Princeton Area) |
| M. cassini | Hunterdon County, NJ (Princeton Area) |
Figure 3. Research organisms and experimental set up.
(A) Magicicada septendecim female (Brood X), (B) Magicicada cassini female (Brood X), (C) Magicicada septendecula male (Brood IX). Photographs in Figs. 3– 7 by C. Simon.
Figure 4. Courtship and mating in periodical cicadas.
(A) A pair of M. septendecim courting inside a screen mesh cage, (B) A pair of M. septendecim mating (Brood IX), (C) A M. septendecim female/M. cassini male mating pair.
Figure 5. M. cassini female ovipositing; note additional eggnest scars on twig.
Figure 6. (A) Three Magicicada eggs removed from an eggnest, (B) A first instar Magicicada nymph.
Figure 7. K. Fontaine inspecting a mating cage for mating pairs.
Table 4. Percentage of caged females that mated for each cross in the 2003 Brood IX and 2004 Brood X experiments.* .
| 2003 Brood IX | -decim male | -cassini male | -decula male |
| -decim female | - | 48.50% | 30.90% |
| -cassini female | 10.90% | - | - |
| -decula female | 5% | - | - |
| 2004 Brood X | |||
| -decim female | 73.30% | 61.50% | |
| -cassini female | 33.30% | 54.60% | |
| -decula female | - | - |
An (-) indicates that that cross was not performed. Better weather may account for the increased mating frequency in Brood X when compared to Brood IX.
DNA was extracted from legs of adult cicadas belonging to the three different Magicicada species groups using the Nucleospin Tissue kit (BD Biosciences Clontech; Palo Alto, CA) following instructions provided by the manufacturer. Extractions were PCR amplified using primers C1-J-2195 and TL2-N-3014 for 30 cycles [48]. PCR product was cleaned and sequenced using BigDye terminator chemistry and an ABI Prism 3100 capillary sequencer. On the basis of these sequences, we developed internal 25-mer COI primers (Table 5) with 3′ ends that anneal to polymorphisms unique to each species group. When annealing temperatures were set to 58° (-decim×–decula crosses) or 60° (-decim×–cassini crosses) these species-specific primers successfully amplified DNA from the appropriate species and failed to amplify heterospecific DNA (Figs. 1–2). To ensure that the primers were amplifying the desired mtDNA segment, the size of the PCR product was estimated with a DNA ladder (exACTGene 100 bp DNA Ladder; Fisher Scientific; Pittsburgh, PA). The PCR product was also sequenced and compared to the COI sequence that was used to develop the primers.
Table 5. Species-specific primer sequences*.
| Primer | Species | Sequence (5′-3′) |
| C1-J-2195 | Universal | TTGATTTTTTGGTCATCCAGAAGT |
| TL2-N-3014 | Universal | TCCAATGCACTAATCTGCCATATTA |
| C1-J-2287 | -decim | GAATCATTTGGATCATTAGGAATGA |
| -cassini | GAATCTTTTGGGTCACTAGGAATAG | |
| C1-N-2712 | -decim | AAAGAAGGTTAAATTTACCCCAAT |
| -cassini | GAAAAAAGTTAAATTTACTCCAAC | |
| C1-N-2787 | -decula | TCTTCTTCCAATAGAAGACATAATA |
| -decim | TCTTCTTCCAATAGAAGATACAATG | |
| C1-J-2607 | -decula | AGGTGCAGTGTTTGCAATCTTGGGG |
| -decim | AGGTGCAGTATTTGCAATTTTAGGA |
differences between the sequences are underlined.
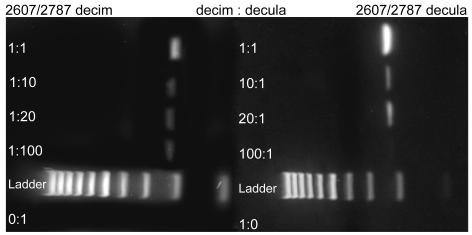
The sensitivity of the diagnostic primers was tested on mixed DNA samples containing DNA from two species in the following ratios: 1∶1, 1∶10, 1∶20, 1∶100, 0∶1, 10∶1, 20∶1 and 100∶1, 1∶0. All mixtures were PCR-tested for the both DNA types with the species-specific primers. Three of the four species-specific primer sets were able to detect the less abundant DNA up to the 1∶100 dilution level (Figs. 8–9). The -decula-specific primers could detect -decula mtDNA up to the 1∶20 dilution level.
Figure 8. Dilution test with different volume ratios of experimentally mixed -decim:-cassini DNA in a 2% agarose gel stained with Sybrsafe.
First column: Amplification with -decim-specific primers 2287/2712; Second column: Amplification with -cassini-specific primers 2287/2712. The less abundant mtDNA type was revealed using species-specific primers. Ladder is a 100 bp ladder with 1000 bp band on left.
Figure 9. Dilution test with different volume ratios of experimentally mixed -decim:-decula in a 2% agarose gel stained with Sybrsafe.
First column: Amplification with -decim-specific primers 2607/2787; Second column: Amplification with -decula-specific primers 2607/2787. The less abundant mtDNA type was revealed using species-specific primers. Ladder is a 100 bp ladder with 1000 bp band on left.
From each eggnest, we pooled 10 fertilized eggs and extracted their DNA; nymphs were extracted singly, and all extractions were performed as above. We probed all collected eggs and nymphs with species-specific primers to detect the presence of both maternal and paternal mtDNA haplotypes. Sample sizes for amplified nymph and pooled-egg samples are listed in Table 2. Each PCR reaction included a negative species control (PCR reaction with heterospecific DNA template from an adult), positive species control (conspecific DNA template from an adult) and negative PCR control (dH20, no DNA but all other reagents).
Supporting Information
Calling song of M. septendecim.
(0.13 MB WAV)
Calling song of M. cassini.
(0.17 MB WAV)
Calling song of M. septendecula.
(0.30 MB WAV)
A Magicicada chorus containing M. septendecim, M. cassini, and M. septendecula.
(4.30 MB WAV)
Acknowledgments
We thank David Rand for helpful discussion, Dan Fontaine, Steve Chiswell, Kathy Hill, and Dave Marshall for assistance in the field, and an anonymous reviewer for helpful comments on earlier drafts of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This material is based upon work partially supported by the National Science Foundation under Grant No. DEB 99-74369, DEB 04-22386, DEB 05-29679, and DEB 06-19012 (REU), and from a University of Connecticut Research Foundation grant to CS. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the NSF or other granting agencies.
References
- 1.Xu J. The inheritance of organelle genes and genomes: patterns and mechanisms. Genome. 2005;48:951–958. doi: 10.1139/g05-082. [DOI] [PubMed] [Google Scholar]
- 2.Budowle B, Allard M-W, Wilson MR, Chakraborty R. Forensics and mitochondrial DNA: Applications, debates, and foundations. Annual Review of Genomics and Human Genetics. 2003;4:119–141. doi: 10.1146/annurev.genom.4.070802.110352. [DOI] [PubMed] [Google Scholar]
- 3.Posada D, Crandall KA. The effect of recombination on the accuracy of phylogeny estimation. Journal Of Molecular Evolution. 2002;54:396–402. doi: 10.1007/s00239-001-0034-9. [DOI] [PubMed] [Google Scholar]
- 4.Piganeau G, Gardner M, Eyre-Walker A. A broad survey of recombination in animal mitochondria. Molecular Biology And Evolution. 2004;21:2319–2325. doi: 10.1093/molbev/msh244. [DOI] [PubMed] [Google Scholar]
- 5.Sackton T, Haney R, Rand D. Cytonuclear coadaptation in Drosophila: Disruption of cytochrome C oxidase activity in backcross genotypes. Evolution. 2003;57:2315–2325. doi: 10.1111/j.0014-3820.2003.tb00243.x. [DOI] [PubMed] [Google Scholar]
- 6.Satta Y, Toyohara N, Ohtaka C, Tatsuno Y, Watanabe TK, et al. Dubious maternal inheritance of mitochondrial DNA in Drosophila simulans and evolution of Drosophila mauritiana. Genetical Research. 1988;52:1–6. [Google Scholar]
- 7.St John JC. The transmission of mitochondrial DNA following assisted reproductive techniques. Theriogenology. 2002;57:109–123. doi: 10.1016/s0093-691x(01)00661-6. [DOI] [PubMed] [Google Scholar]
- 8.Simon C, Tang J, Dalwadi S, Staley G, Deniega J, et al. Genetic Evidence for Assortative Mating between 13-Year Cicadas and Sympatric “17-Year Cicadas with 13-Year Life Cycles” Provides Support for Allochronic Speciation. Evolution. 2000;54:1326–1336. doi: 10.1111/j.0014-3820.2000.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 9.Cooley JR, Simon C, Marshall DC, Slon K, Ehrhardt C. Allochronic speciation, secondary contact, and reproductive character displacement in periodical cicadas (Hemiptera: Magicicada spp.): genetic, morphological, and behavioural evidence. Molecular Ecology. 2001;10:661–671. doi: 10.1046/j.1365-294x.2001.01210.x. [DOI] [PubMed] [Google Scholar]
- 10.Cooley JR, Simon C, Marshall DC. Temporal separation and speciation in periodical cicadas. Bioscience. 2003;53:151–157. [Google Scholar]
- 11.Cox RT, Carlton CE. Evidence of Genetic Dominance of the 13-year Life Cycle in Periodical Cicadas (Homoptera: Cicadidae: Magicicada spp.). American Midland Naturalist. 1991;125:63–74. [Google Scholar]
- 12.Cox RT, Carlton CE. A Commentary on Prime Numbers and Life Cycles of Periodical Cicadas. American Naturalist. 1998;152:162–164. doi: 10.1086/286158. [DOI] [PubMed] [Google Scholar]
- 13.Cox RT, Carlton CE. A comment on gene introgression versus en masse cycle switching in the evolution of 13-year and 17-year life cycles in periodical cicadas. Evolution. 2003;57:428–432. [PubMed] [Google Scholar]
- 14.Williams KS, Simon C. The Ecology, Behavior, And Evolution Of Periodical Cicadas. Annual Review Of Entomology. 1995;40:269–295. [Google Scholar]
- 15.Osborn H. A statistical study of variations in the periodical cicada. Ohio Journal Of Science. 1902;3:323–326. [Google Scholar]
- 16.Alexander RD, Moore TE. The evolutionary relationships of 17-year and 13-year cicadas, and three new species. (Homoptera: Cicadidae, Magicicada). University of Michigan Museum of Zoology Miscellaneous Publication. 1962;121:1–59. [Google Scholar]
- 17.Dybas HS, Lloyd M. Isolation by Habitat in Two Synchronized Species of Periodical Cicadas (Homoptera: Cicadidae: Magicicada). Ecology. 1962;43:444–459. [Google Scholar]
- 18.Dybas HS, Lloyd M. The Habitats of 17-Year Periodical Cicadas (Homoptera: Cicadidae: Magicicada spp.). Ecological Monographs. 1974;44:279–324. [Google Scholar]
- 19.Dunning D, Byers J, Zanger C. Courtship in two species of periodical cicada, Magicicada septendecim and Magicicada cassini. Animal Behaviour. 1979;27:1073–1090. [Google Scholar]
- 20.Marlatt C. United Stated Department of Agriculture, Bureau of Entomology Bulletin 71; 1923. The Periodical Cicada. [Google Scholar]
- 21.Lloyd M, Dybas HS. The Periodical Cicada Problem. I. Population Ecology. Evolution. 1966;20:133–149. doi: 10.1111/j.1558-5646.1966.tb03350.x. [DOI] [PubMed] [Google Scholar]
- 22.Lloyd M, Dybas HS. The Periodical Cicada Problem. II. Evolution. Evolution. 1966;20:466–505. doi: 10.1111/j.1558-5646.1966.tb03381.x. [DOI] [PubMed] [Google Scholar]
- 23.Simon C. Evolution of 13- and 17-year periodical cicadas. Bulletin of the Entomological Society of America. 1988;34:163–176. [Google Scholar]
- 24.Marshall DC, Cooley JR. Reproductive Character Displacement and Speciation in Periodical Cicadas, with Description of a New Species, 13-Year Magicicada neotredecim. Evolution. 2000;54:1313–1325. doi: 10.1111/j.0014-3820.2000.tb00564.x. [DOI] [PubMed] [Google Scholar]
- 25.Karban R. Effects of local density on fecundity and mating speed for periodical cicadas. Oecologia. 1981;51:260–264. doi: 10.1007/BF00540611. [DOI] [PubMed] [Google Scholar]
- 26.Maier C. Observations on the seventeen-year periodical cicada, Magicicada septendecim (Hemiptera: Homoptera: Cicadidae). Annals Of The Entomological Society Of America. 1982;75:14–23. [Google Scholar]
- 27.Young D, Josephson R. Pure-tone songs in cicadas with special reference to the genus Magicicada. Journal of comparative Physiology. 1983;152:197–207. [Google Scholar]
- 28.Cooley JR, Marshall DC. Sexual signaling in periodical cicadas, Magicicada spp. (Hemiptera: Cicadidae). Behaviour. 2001;138:827–855. [Google Scholar]
- 29.White JA. Viable Hybrid Young From Crossmated Periodical Cicadas. Ecology. 1973;54:573–580. [Google Scholar]
- 30.Kondo R, Matsuura ET, Chigusa SI. Further Observation of Paternal Transmission of Drosophila Mitochondrial DNA by PCR Selective Amplification Method. Genetical Research. 1992;59:81–84. doi: 10.1017/s0016672300030287. [DOI] [PubMed] [Google Scholar]
- 31.Lansman RA, Avise JC, Huettel MD. Critical Experimental Test of the Possibility of “Paternal Leakage” of Mitochondrial DNA. PNAS. 1983;80:1969–1971. doi: 10.1073/pnas.80.7.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satta Y, Matsuura ET, Ishiwa H, Takahata N, Chigusa SI. Incomplete Maternal Transmission of Mitochondrial DNA in Drosophila. Genetics. 1990;126:657–664. doi: 10.1093/genetics/126.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meusel MS, Moritz RFA. Transfer of paternal mitochondrial DNA during fertilization of honeybee (Apis mellifera L.) eggs. Current Genetics. 1993;24:539–543. doi: 10.1007/BF00351719. [DOI] [PubMed] [Google Scholar]
- 34.Rokas A, Ladoukakis ED, Zouros E. Trends In Ecology&Evolution 18; 2003. Animal mitochondrial DNA recombination revisited. [Google Scholar]
- 35.Gillham NW. New York: Oxford University Press; 1994. Organelle genes and genomes. [Google Scholar]
- 36.Chesser RK. Heteroplasmy and organelle gene dynamics. Genetics. 1998;150:1309–1327. doi: 10.1093/genetics/150.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birky CW. The inheritance of genes in mitochondria and chloroplasts: Laws, mechanisms, and models. Annual Review of Genetics. 2001;35:125–148. doi: 10.1146/annurev.genet.35.102401.090231. [DOI] [PubMed] [Google Scholar]
- 38.Kaneda H, Hayashi J-I, Takahama S, Taya C, Lindahl KF, et al. Elimination of Paternal Mitochondrial DNA in Intraspecific Crosses During Early Mouse Embryogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:4542–4546. doi: 10.1073/pnas.92.10.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, et al. Ubiquitin tag for sperm mitochondria. Nature. 1999;402:371–372. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- 40.Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, et al. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol Reprod. 2000;63:582–590. doi: 10.1095/biolreprod63.2.582. [DOI] [PubMed] [Google Scholar]
- 41.Sutovsky P, McCauley TC, Sutovsky M, Day BN. Early Degradation of Paternal Mitochondria in Domestic Pig (Sus scrofa) Is Prevented by Selective Proteasomal Inhibitors Lactacystin and MG1321. Biology of Reproduction. 2003;68:1793–1800. doi: 10.1095/biolreprod.102.012799. [DOI] [PubMed] [Google Scholar]
- 42.Kondo R, Satta Y, Matsuura ET, Ishiwa H, Takahata N, et al. Incomplete Maternal Transmission of Mitochondrial DNA in Drosophila. Genetics. 1990;126:657–663. doi: 10.1093/genetics/126.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Molecular Ecology. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 44.Sherengul W, Kondo R, Matsuura ET. Analysis of paternal transmission of mitochondrial DNA in Drosophila. Genes and Genetic Systems. 2006;81:399–404. doi: 10.1266/ggs.81.399. [DOI] [PubMed] [Google Scholar]
- 45.Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal Of The Linnean Society. 1996;58:247–276. [Google Scholar]
- 46.Barr CM, Neiman M, Taylor DR. Inheritance and recombination of mitochondrial genomes in plants, fungi, and animals. New Phytologist. 2005;168:39–50. doi: 10.1111/j.1469-8137.2005.01492.x. [DOI] [PubMed] [Google Scholar]
- 47.Cooley JR. Ann Arbor: The University of Michigan; 1999. Sexual behavior in North American cicadas of the genera Magicicada and Okanagana [Ph.D]. p. 326. [Google Scholar]
- 48.Simon C, Frati F, Beckenbach A, Crespi B, Liu H, et al. Evolution, Weighting, And Phylogenetic Utility Of Mitochondrial Gene-Sequences And A Compilation Of Conserved Polymerase Chain-Reaction Primers. Annals Of The Entomological Society Of America. 1994;87:651–701. [Google Scholar]
- 49.Arunkumar KP, Metta M, Nagaraju J. Molecular phylogeny of silkmoths reveals the origin of domesticated silkmoth, Bombyx mandarina and paternal inheritance of Antheraea proleyi mitochondrial DNA. Molecular Phylogenetics And Evolution. 2006;40:419–427. doi: 10.1016/j.ympev.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 50.Gyllensten U, Wharton D, Joseffson A, Wilson AC. Paternal inheritance of mitochondrial DNA in mice. Nature. 1991;352:255–257. doi: 10.1038/352255a0. [DOI] [PubMed] [Google Scholar]
- 51.Shitara H, Hayashi J-I, Takahama S, Kaneda H. Maternal inheritance of mouse mtDNA in interspecific hybrids: Segregation of the leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics. 1998;148:851–857. doi: 10.1093/genetics/148.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinborn R, Zakhartchenko V, Jelyazkov J, Klein D, Wolf E, et al. Composition of parental mitochondrial DNA in cloned bovine embryos. FEBS Letters. 1998;426:352–356. doi: 10.1016/s0014-5793(98)00350-0. [DOI] [PubMed] [Google Scholar]
- 53.Gantenbein B, Fet V, Gantenbein-Ritter I, Balloux F. Evidence for recombination in scorpioon mitochondrial DNA (Scorpiones: Buthidae). Proceedings Of The Royal Society Of London Series B-Biological Sciences. 2005;272:697–704. doi: 10.1098/rspb.2004.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ujvari B, Dowton M, Madsen T. Mitochondrial DNA recombination in a free-ranging Australian lizard. Biology Letters. 2007;3:189–192. doi: 10.1098/rsbl.2006.0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magoulas A. Restriction-site heteroplasmy in anchovy (Engraulis encrasicholus) indicates incidental biparental inheritance of mitochondrial DNA. Molecular Biology And Evolution. 1993;10:319–325. [Google Scholar]
- 56.Schwartz M, Vissing J. Paternal inheritance of mitochondrial DNA. New England Journal of Medicine. 2002;347:576–580. doi: 10.1056/NEJMoa020350. [DOI] [PubMed] [Google Scholar]
- 57.Zhao X, Li N, Guo X, Hu Z, Gong G, et al. Further evidence for paternal inheritance of mitochondrial DNA in the sheep (Ovis aries). Heredity. 1992;93:399–403. doi: 10.1038/sj.hdy.6800516. [DOI] [PubMed] [Google Scholar]
- 58.Andolfatto P, Scriber JM, Charlesworth B. No association between mitochondrial DNA haplotypes and a female-limited mimicry phenotype in Papilio glaucus. Evolution. 2003;57:305–316. doi: 10.1111/j.0014-3820.2003.tb00265.x. [DOI] [PubMed] [Google Scholar]
- 59.Kvist L, Martens J, Nazarenko A-A, Orell M. Paternal leakage of mitochondrial DNA in the great tit (Parus major). Molecular Biology And Evolution. 2003;20:243–247. doi: 10.1093/molbev/msg025. [DOI] [PubMed] [Google Scholar]
- 60.Hoarau G, Holla S, Lescasse R, Stam WT, Olsen JL. Heteroplasmy and evidence for recombination in thhe mitochondrial control region of the flatfish Platichthys flesus. Molecular Biology And Evolution. 2002;19:2261–2264. doi: 10.1093/oxfordjournals.molbev.a004049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Calling song of M. septendecim.
(0.13 MB WAV)
Calling song of M. cassini.
(0.17 MB WAV)
Calling song of M. septendecula.
(0.30 MB WAV)
A Magicicada chorus containing M. septendecim, M. cassini, and M. septendecula.
(4.30 MB WAV)