Abstract
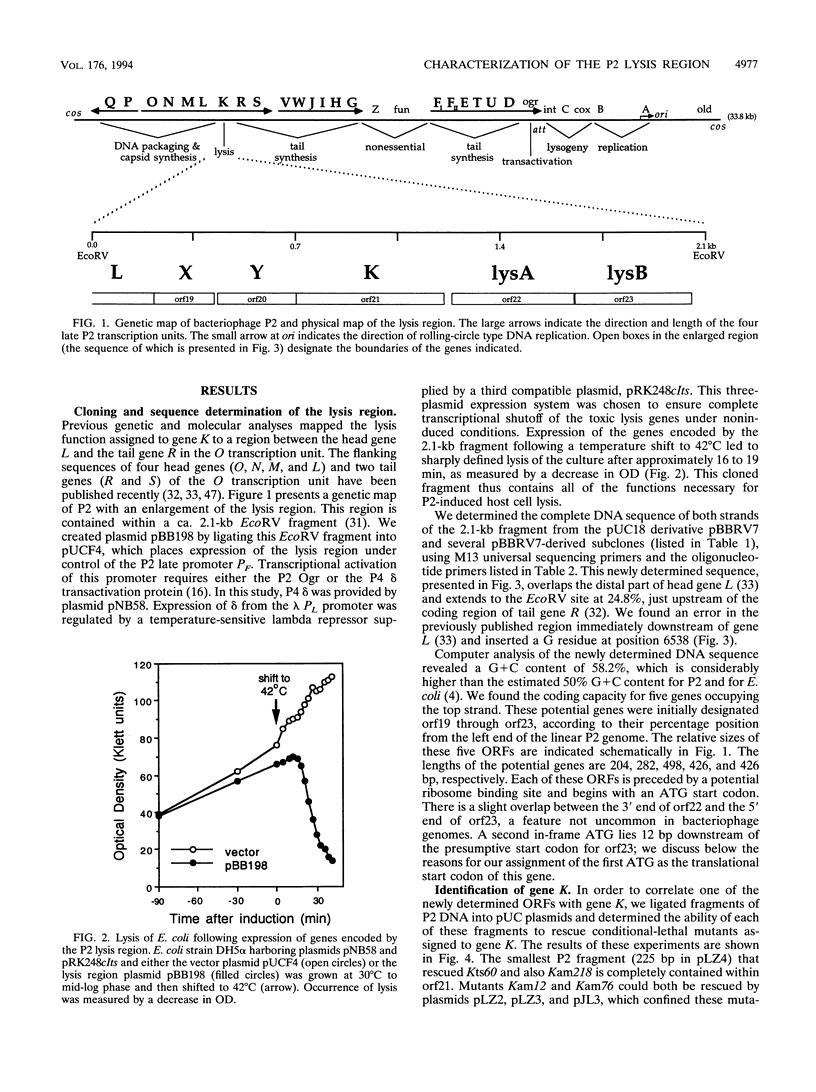
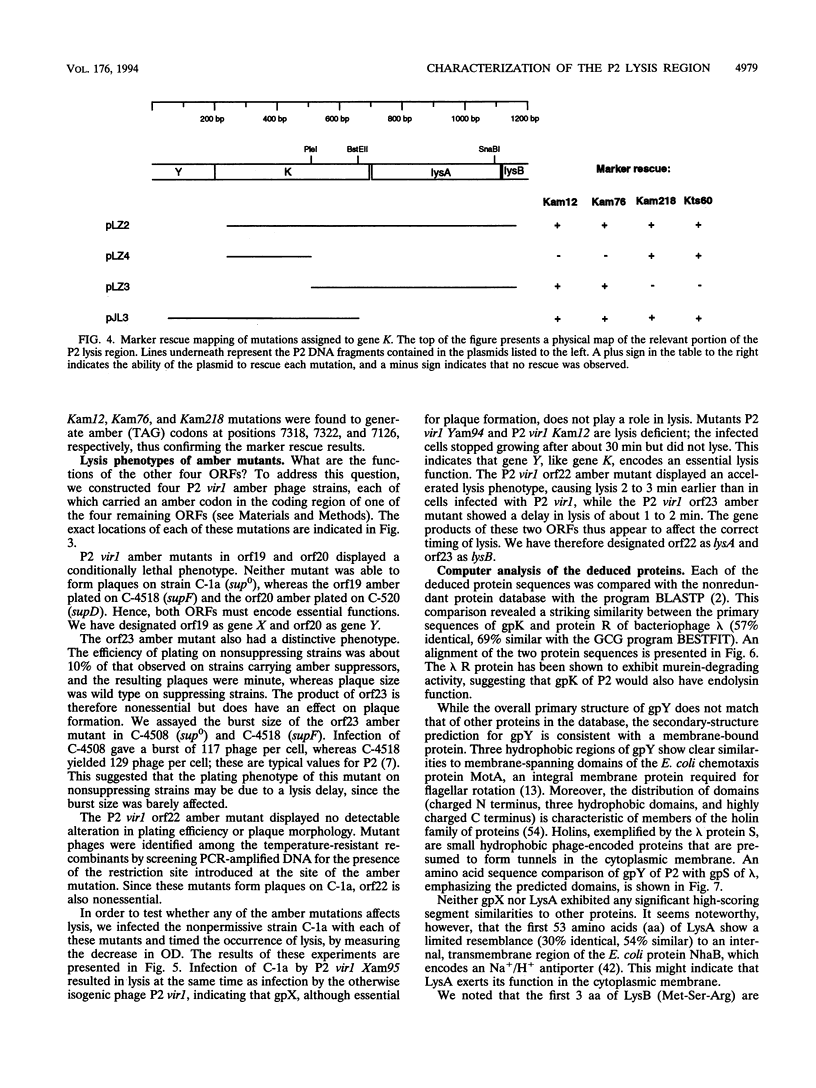
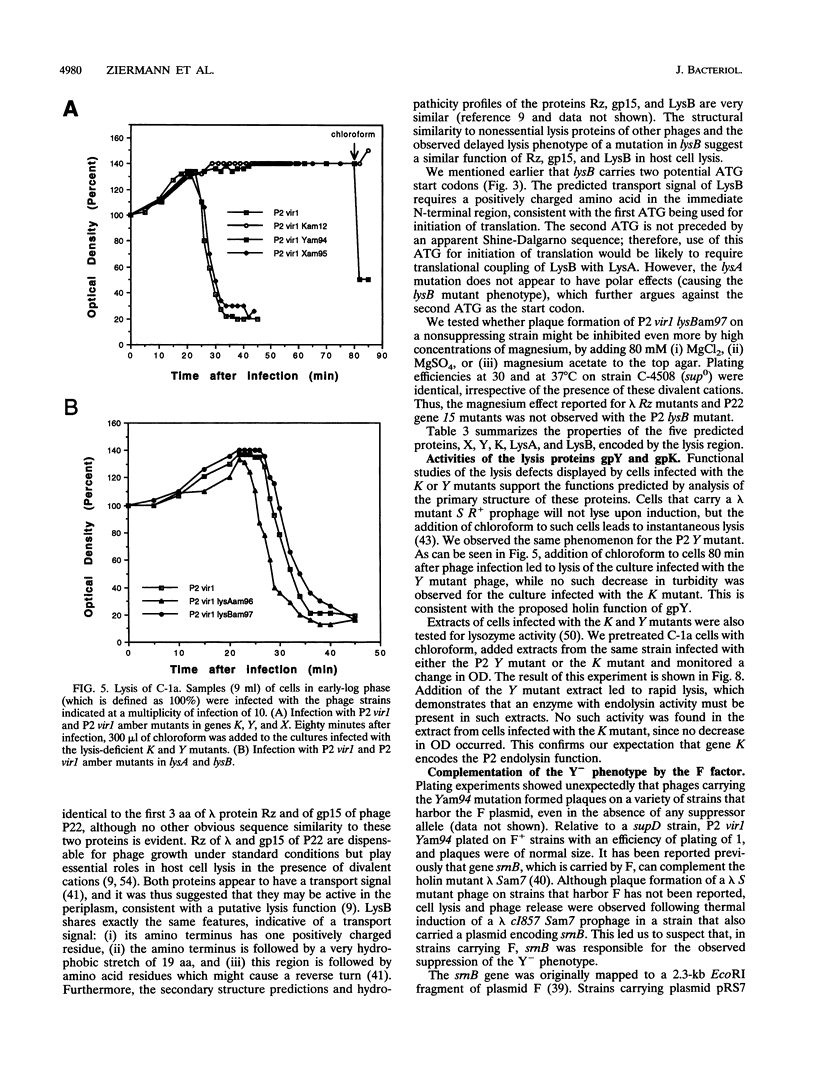
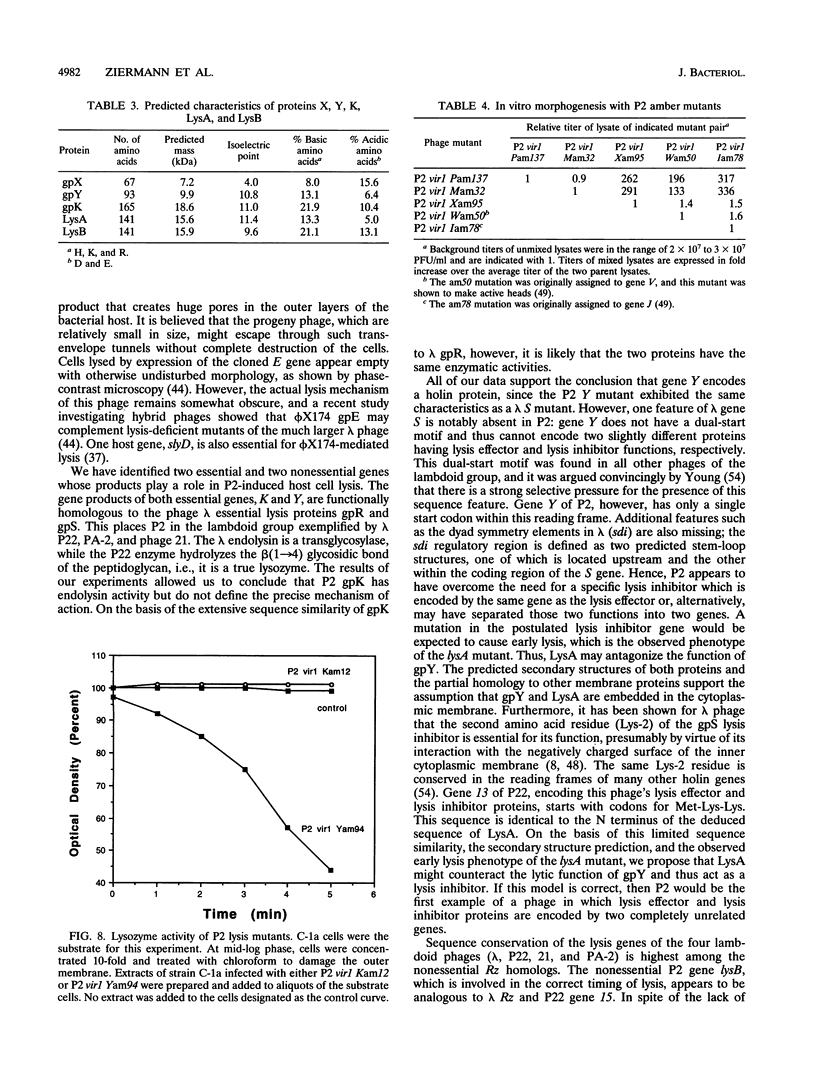
Successful completion of the bacteriophage P2 lytic cycle requires phage-induced lysis of its Escherichia coli host, a process that is poorly understood. Genetic analysis of lysis-deficient mutants defined a single locus, gene K, which lies within the largest late transcription unit of P2 and maps between head gene L and tail gene R. We determined and analyzed the DNA sequence of a ca. 2.1-kb EcoRV fragment that spans the entire region from L to R, thus completing the sequence of this operon. This region contains all of the functions necessary for host cell lysis. Sequence analysis revealed five open reading frames, initially designated orf19 through orf23. All of the existing lysis mutants--ts60, am12, am76, and am218--were located in orf21, which must therefore correspond to gene K. The K gene product has extensive amino acid sequence similarity to the product of gene R of bacteriophage lambda, and its exhibits endolysin function. Site-directed mutagenesis and reverse genetics were used to create P2 amber mutants in each of the four other newly identified open reading frames. Both orf19 (gene X) and orf20 (gene Y) encode essential functions, whereas orf22 (lysA) and orf23 (lysB) are nonessential. Gene Y encodes a polypeptide with striking similarities to the family of holin proteins exemplified by gpS of phage lambda, and the Yam mutant displayed the expected properties of a holin mutant. The gene products of lysA and lysB, although nonessential, appear to play a role in the correct timing of lysis, since a lysA amber mutant caused slightly accelerated lysis and a lysB amber mutant slightly delayed lysis of nonpermissive strains. Gene X must encode a tail protein, since lysates from nonpermissive cells infected with the X amber mutant were complemented in vitro by similar lysates of cells infected with P2 head mutants but not with tail mutants.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- BERTANI G., WEIGLE J. J. Host controlled variation in bacterial viruses. J Bacteriol. 1953 Feb;65(2):113–121. doi: 10.1128/jb.65.2.113-121.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERTANI L. E. The effect of the inhibition of protein synthesis on the establishment of lysogeny. Virology. 1957 Aug;4(1):53–71. doi: 10.1016/0042-6822(57)90043-0. [DOI] [PubMed] [Google Scholar]
- Bernard H. U., Helinski D. R. Use of the lambda phage promoter PL to promote gene expression in hybrid plasmid cloning vehicles. Methods Enzymol. 1979;68:482–492. doi: 10.1016/0076-6879(79)68037-0. [DOI] [PubMed] [Google Scholar]
- Bläsi U., Chang C. Y., Zagotta M. T., Nam K. B., Young R. The lethal lambda S gene encodes its own inhibitor. EMBO J. 1990 Apr;9(4):981–989. doi: 10.1002/j.1460-2075.1990.tb08200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S., Eppler K., Parr R., Poteete A. R. Nucleotide sequence of the bacteriophage P22 gene 19 to 3 region: identification of a new gene required for lysis. Virology. 1989 Aug;171(2):588–598. doi: 10.1016/0042-6822(89)90628-4. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of beta-turns. Biophys J. 1979 Jun;26(3):367–383. doi: 10.1016/S0006-3495(79)85259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie G. E., Calendar R. Interactions between satellite bacteriophage P4 and its helpers. Annu Rev Genet. 1990;24:465–490. doi: 10.1146/annurev.ge.24.120190.002341. [DOI] [PubMed] [Google Scholar]
- Dean G. E., Macnab R. M., Stader J., Matsumura P., Burks C. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984 Sep;159(3):991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Poulsen L. K., Thisted T., Nielsen A. K., Martinussen J., Andreasen P. H. The hok killer gene family in gram-negative bacteria. New Biol. 1990 Nov;2(11):946–956. [PubMed] [Google Scholar]
- Grambow N. J., Birkeland N. K., Anders D. L., Christie G. E. Deletion analysis of a bacteriophage P2 late promoter. Gene. 1990 Oct 30;95(1):9–15. doi: 10.1016/0378-1119(90)90407-i. [DOI] [PubMed] [Google Scholar]
- Halling C., Sunshine M. G., Lane K. B., Six E. W., Calendar R. A mutation of the transactivation gene of satellite bacteriophage P4 that suppresses the rpoA109 mutation of Escherichia coli. J Bacteriol. 1990 Jul;172(7):3541–3548. doi: 10.1128/jb.172.7.3541-3548.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Hocking S. M., Egan J. B. Genetic map of coliphage 186 from a novel use of marker rescue frequencies. Mol Gen Genet. 1982;187(1):87–95. doi: 10.1007/BF00384388. [DOI] [PubMed] [Google Scholar]
- Hocking S. M., Egan J. B. Genetic studies of coliphage 186. II. Genes associated with phage replication and host cell lysis. J Virol. 1982 Dec;44(3):1068–1071. doi: 10.1128/jvi.44.3.1068-1071.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn M. L., Ziermann R., Dehò G., Ow D. W., Sunshine M. G., Calendar R. Bacteriophage P2 and P4. Methods Enzymol. 1991;204:264–280. doi: 10.1016/0076-6879(91)04013-e. [DOI] [PubMed] [Google Scholar]
- King R. A., Anders D. L., Christie G. E. Site-directed mutagenesis of an amino acid residue in the bacteriophage P2 ogr protein implicated in interaction with Escherichia coli RNA polymerase. Mol Microbiol. 1992 Nov;6(22):3313–3320. doi: 10.1111/j.1365-2958.1992.tb02199.x. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lengyel J. A., Goldstein R. N., Marsh M., Calendar R. Structure of the bacteriophage P2 tail. Virology. 1974 Nov;62(1):161–174. doi: 10.1016/0042-6822(74)90312-2. [DOI] [PubMed] [Google Scholar]
- Lindahl G. Characterization of conditional lethal mutants of bacteriophage P2. Mol Gen Genet. 1974 Feb 6;128(3):249–260. doi: 10.1007/BF00267114. [DOI] [PubMed] [Google Scholar]
- Lindahl G. Genetic map of bacteriophage P2. Virology. 1969 Dec;39(4):839–860. doi: 10.1016/0042-6822(69)90021-x. [DOI] [PubMed] [Google Scholar]
- Lindahl G. On the control of transcription in bacteriophage P2. Virology. 1971 Dec;46(3):620–633. doi: 10.1016/0042-6822(71)90065-1. [DOI] [PubMed] [Google Scholar]
- Linderoth N. A., Julien B., Flick K. E., Calendar R., Christie G. E. Molecular cloning and characterization of bacteriophage P2 genes R and S involved in tail completion. Virology. 1994 May 1;200(2):347–359. doi: 10.1006/viro.1994.1199. [DOI] [PubMed] [Google Scholar]
- Linderoth N. A., Ziermann R., Haggård-Ljungquist E., Christie G. E., Calendar R. Nucleotide sequence of the DNA packaging and capsid synthesis genes of bacteriophage P2. Nucleic Acids Res. 1991 Dec;19(25):7207–7214. doi: 10.1093/nar/19.25.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist B. H., Dehò G., Calendar R. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol Rev. 1993 Sep;57(3):683–702. doi: 10.1128/mr.57.3.683-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist B. H. Vegetative DNA of temperate coliphage P2. Mol Gen Genet. 1971;110(2):178–196. doi: 10.1007/BF00332647. [DOI] [PubMed] [Google Scholar]
- Maratea D., Young K., Young R. Deletion and fusion analysis of the phage phi X174 lysis gene E. Gene. 1985;40(1):39–46. doi: 10.1016/0378-1119(85)90022-8. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y., Iguma H., Ono T., Nagaishi H., Clark A. J. Genetic mapping of the F plasmid gene that promotes degradation of stable ribonucleic acid in Escherichia coli. J Bacteriol. 1977 Dec;132(3):784–789. doi: 10.1128/jb.132.3.784-789.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinner E., Padan E., Schuldiner S. Cloning, sequencing, and expression of the nhaB gene, encoding a Na+/H+ antiporter in Escherichia coli. J Biol Chem. 1992 Jun 5;267(16):11064–11068. [PubMed] [Google Scholar]
- Reader R. W., Siminovitch L. Lysis defective mutants of bacteriophage lambda: on the role of the S function in lysis. Virology. 1971 Mar;43(3):623–637. doi: 10.1016/0042-6822(71)90287-x. [DOI] [PubMed] [Google Scholar]
- Roof W. D., Young R. Phi X174 E complements lambda S and R dysfunction for host cell lysis. J Bacteriol. 1993 Jun;175(12):3909–3912. doi: 10.1128/jb.175.12.3909-3912.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki I., Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J Gen Microbiol. 1965 Sep;40(3):365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- Six E. W., Sunshine M. G., Williams J., Haggård-Ljungquist E., Lindqvist B. H. Morphopoietic switch mutations of bacteriophage P2. Virology. 1991 May;182(1):34–46. doi: 10.1016/0042-6822(91)90645-r. [DOI] [PubMed] [Google Scholar]
- Steiner M., Bläsi U. Charged amino-terminal amino acids affect the lethal capacity of Lambda lysis proteins S107 and S105. Mol Microbiol. 1993 May;8(3):525–533. doi: 10.1111/j.1365-2958.1993.tb01597.x. [DOI] [PubMed] [Google Scholar]
- Sunshine M. G., Thorn M., Gibbs W., Calendar R., Kelly B. P2 phage amber mutants: characterization by use of a polarity suppressor. Virology. 1971 Dec;46(3):691–702. doi: 10.1016/0042-6822(71)90071-7. [DOI] [PubMed] [Google Scholar]
- Tsui L., Mark K. The depression of endolysin synthesis in bacteria infected with high multiplicities of phage lambda. Mol Gen Genet. 1976 Feb 2;143(3):269–278. doi: 10.1007/BF00269403. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Witte A., Bläsi U., Halfmann G., Szostak M., Wanner G., Lubitz W. Phi X174 protein E-mediated lysis of Escherichia coli. Biochimie. 1990 Feb-Mar;72(2-3):191–200. doi: 10.1016/0300-9084(90)90145-7. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992 Sep;56(3):430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]



