Abstract
Tn918 mutagenesis of a high-level methicillin-resistant Staphylococcus aureus (methicillin MIC, 800 micrograms/ml) led to the isolation of a low-resistance mutant. The Tn918 insert was transferred back to the parent to produce strain SRM563 (methicillin MIC, 12.5 micrograms/ml), which showed heterogeneous resistance. Twenty-two clinical isolates of methicillin-resistant S. aureus were transformed with DNA of SRM563. In the transformants of most strains, instances of reduced resistance were observed with concomitant increases of autolysis rate induced by Triton X-100 and were generally more profound in high-resistance strains. Two transformants exhibited a decrease of the autolysis rate and little reduction of resistance. In the transformant of methicillin-susceptible strain RN2677, an increase of the autolysis rate and little reduction of resistance were observed. The production of low-affinity penicillin-binding protein (PBP2') did not significantly decrease in the mutants. Insertion of Tn918 occurred within the 3'-terminal region of a novel gene designated llm, which was cloned and sequenced. RNA blot analysis demonstrated that the gene was transcribed. The encoded protein was composed of 351 amino acid residues with a molecular weight of 38,512 and was hydrophobic, suggesting its location on the membrane. The gene was detected by PCR in all S. aureus strains tested but not in the other 26 staphylococcal species. Comparison of the 3'-terminal sequences of the gene among several S. aureus strains showed that, whereas nucleotide substitutions occurred at the third position in seven of eight 3'-terminal codons, only C-terminal amino acid variation of glutamate or aspartate was observed.
Full text
PDF


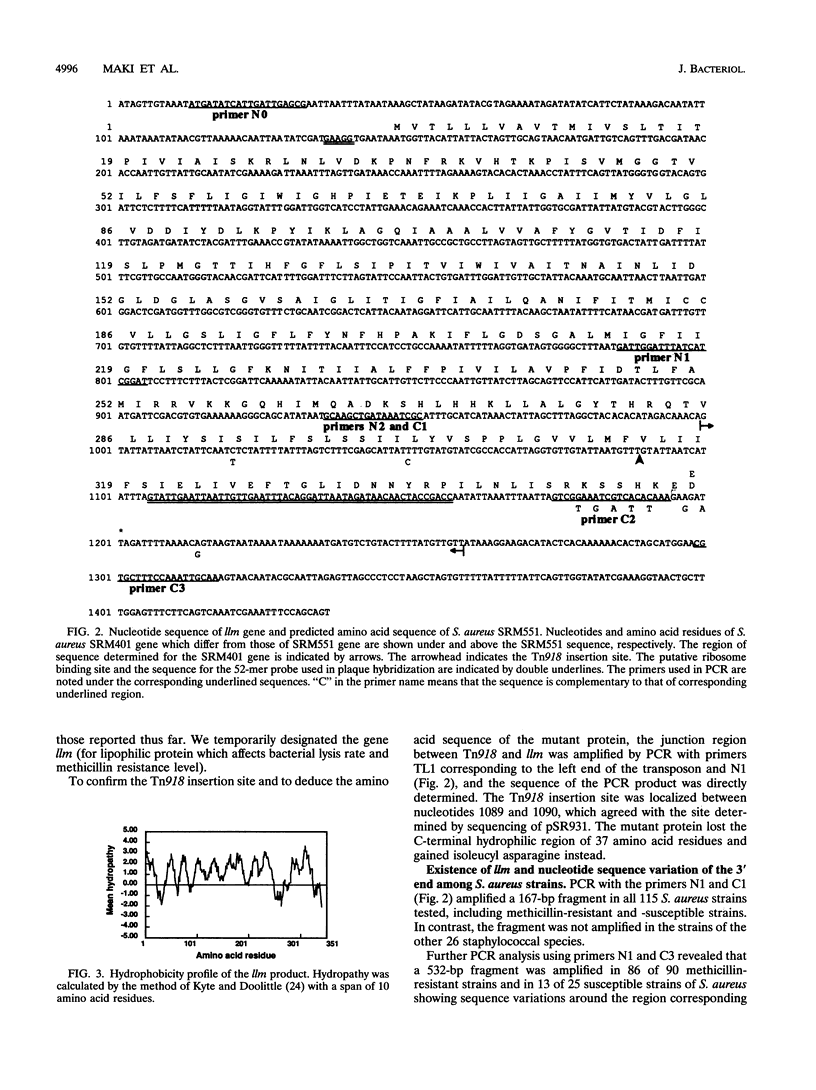


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger-Bächi B., Barberis-Maino L., Strässle A., Kayser F. H. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol Gen Genet. 1989 Oct;219(1-2):263–269. doi: 10.1007/BF00261186. [DOI] [PubMed] [Google Scholar]
- Berger-Bächi B. Insertional inactivation of staphylococcal methicillin resistance by Tn551. J Bacteriol. 1983 Apr;154(1):479–487. doi: 10.1128/jb.154.1.479-487.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B., Strässle A., Gustafson J. E., Kayser F. H. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1992 Jul;36(7):1367–1373. doi: 10.1128/aac.36.7.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B., Strässle A., Kayser F. H. Characterization of an isogenic set of methicillin-resistant and susceptible mutants of Staphylococcus aureus. Eur J Clin Microbiol. 1986 Dec;5(6):697–701. doi: 10.1007/BF02013308. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., An F. Y., White B. A., Gawron-Burke C. Streptococcus faecalis sex pheromone (cAM373) also produced by Staphylococcus aureus and identification of a conjugative transposon (Tn918). J Bacteriol. 1985 Jun;162(3):1212–1220. doi: 10.1128/jb.162.3.1212-1220.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Flannagan S. E., Ike Y., Jones J. M., Gawron-Burke C. Sequence analysis of termini of conjugative transposon Tn916. J Bacteriol. 1988 Jul;170(7):3046–3052. doi: 10.1128/jb.170.7.3046-3052.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Gill J. F., Chakrabarty A. M. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987 Jan;169(1):351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaisford W. C., Reynolds P. E. Methicillin resistance in Staphylococcus epidermidis. Relationship between the additional penicillin-binding protein and an attachment transpeptidase. Eur J Biochem. 1989 Oct 20;185(1):211–218. doi: 10.1111/j.1432-1033.1989.tb15104.x. [DOI] [PubMed] [Google Scholar]
- Gawron-Burke C., Clewell D. B. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J Bacteriol. 1984 Jul;159(1):214–221. doi: 10.1128/jb.159.1.214-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotschlich E. C., Blake M. S., Koomey J. M., Seiff M., Derman A. Cloning of the structural genes of three H8 antigens and of protein III of Neisseria gonorrhoeae. J Exp Med. 1986 Sep 1;164(3):868–881. doi: 10.1084/jem.164.3.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson J. E., Wilkinson B. J. Lower autolytic activity in a homogeneous methicillin-resistant Staphylococcus aureus strain compared to derived heterogeneous-resistant and susceptible strains. FEMS Microbiol Lett. 1989 May;50(1-2):107–111. doi: 10.1016/0378-1097(89)90468-0. [DOI] [PubMed] [Google Scholar]
- Gustafson J., Strässle A., Hächler H., Kayser F. H., Berger-Bächi B. The femC locus of Staphylococcus aureus required for methicillin resistance includes the glutamine synthetase operon. J Bacteriol. 1994 Mar;176(5):1460–1467. doi: 10.1128/jb.176.5.1460-1467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1986 Jan;29(1):85–92. doi: 10.1128/aac.29.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984 May;158(2):513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henze U., Sidow T., Wecke J., Labischinski H., Berger-Bächi B. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J Bacteriol. 1993 Mar;175(6):1612–1620. doi: 10.1128/jb.175.6.1612-1620.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu K., Suzuki E., Takayama H., Katayama Y., Yokota T. Role of penicillinase plasmids in the stability of the mecA gene in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1990 Apr;34(4):600–604. doi: 10.1128/aac.34.4.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblum J. S., Projan S. J., Moghazeh S. L., Novick R. P. A rapid method to quantitate non-labeled RNA species in bacterial cells. Gene. 1988;63(1):75–85. doi: 10.1016/0378-1119(88)90547-1. [DOI] [PubMed] [Google Scholar]
- Kornblum J., Hartman B. J., Novick R. P., Tomasz A. Conversion of a homogeneously methicillin-resistant strain of Staphylococcus aureus to heterogeneous resistance by Tn551-mediated insertional inactivation. Eur J Clin Microbiol. 1986 Dec;5(6):714–718. doi: 10.1007/BF02013311. [DOI] [PubMed] [Google Scholar]
- Kreiswirth B., Kornblum J., Arbeit R. D., Eisner W., Maslow J. N., McGeer A., Low D. E., Novick R. P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993 Jan 8;259(5092):227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- Kuhl S. A., Pattee P. A., Baldwin J. N. Chromosomal map location of the methicillin resistance determinant in Staphylococcus aureus. J Bacteriol. 1978 Aug;135(2):460–465. doi: 10.1128/jb.135.2.460-465.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Maidhof H., Reinicke B., Blümel P., Berger-Bächi B., Labischinski H. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Bacteriol. 1991 Jun;173(11):3507–3513. doi: 10.1128/jb.173.11.3507-3513.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Minamide W., Wada K., Nakamura E., Teraoka H., Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991 Oct;29(10):2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Nomura K., Doi M., Yoshida T. Production of low-affinity penicillin-binding protein by low- and high-resistance groups of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1987 Sep;31(9):1307–1311. doi: 10.1128/aac.31.9.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Tomasz A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J Bacteriol. 1989 Feb;171(2):874–879. doi: 10.1128/jb.171.2.874-879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser J. M., Kapur V. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J Clin Microbiol. 1992 Aug;30(8):2058–2063. doi: 10.1128/jcm.30.8.2058-2063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M. L., Pattee P. A. Confirmation of protoplast fusion-derived linkages in Staphylococcus aureus by transformation with protoplast DNA. J Bacteriol. 1983 Apr;154(1):406–412. doi: 10.1128/jb.154.1.406-412.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson N. E., Pattee P. A. Transformation in Staphylococcus aureus: role of bacteriophage and incidence of competence among strains. J Bacteriol. 1977 Feb;129(2):778–788. doi: 10.1128/jb.129.2.778-788.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsui Y., Yokota T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1985 Sep;28(3):397–403. doi: 10.1128/aac.28.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge B. L., Sidow T., Chang Y. S., Labischinski H., Berger-Bachi B., Gage D. A., Tomasz A. Altered muropeptide composition in Staphylococcus aureus strains with an inactivated femA locus. J Bacteriol. 1993 May;175(9):2779–2782. doi: 10.1128/jb.175.9.2779-2782.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge B. L., de Lencastre H., Tomasz A. Suppression of autolysis and cell wall turnover in heterogeneous Tn551 mutants of a methicillin-resistant Staphylococcus aureus strain. J Bacteriol. 1991 Feb;173(3):1105–1110. doi: 10.1128/jb.173.3.1105-1110.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]