Abstract
The aim of the present investigation was to study the major chromosomal aberrations (CA) like deletion, translocation, inversion and mosaic in prostate cancer patients of Tamilnadu, Southern India. Totally 45 blood samples were collected from various hospitals in Tamilnadu, Southern India. Equal numbers of normal healthy subjects were chosen after signing a consent form. Volunteers provided blood samples (5 ml) to establish leukocyte cultures. Cytogenetic studies were performed by using Giemsa-banding technique and finally the results were ensured by spectral karyotyping (SKY) technique. In the present investigation, major CA like deletion, translocation, inversion and mosaic were identified in experimental subjects. Results showed frequent CA in chromosomes 1, 3, 5, 6, 7, 9, 13, 16, 18 and X. In comparison with experimental subjects, the control subjects exhibited very low levels of major CA (P<0.05). In the present study, the high frequency of centromeric rearrangements indicates a potential role for mitotic irregularities associated with the centromere in prostate cancer tumorigenesis. Identification of chromosome alterations may be helpful in understanding the molecular basis of the disease in better manner.
Keywords: Prostate cancer, Chromosomal aberrations (CA), Giemsa-banding, Spectral karyotyping (SKY)
INTRODUCTION
Prostate cancer is one of the leading cancer types and is the second most common cause of cancer mortality in men in North America (Haas and Sakr, 1997). An influence of diet and environment on prostate cancer risk is suggested by studies in Asian countries and Japanese men who migrated to the US (Cook et al., 1999; Whittemore et al., 1995). In India, the annual mortality in 2000 was 0.7 million, and the annual estimate of cancer for the year 2001 was 0.98 million (Greenlee et al., 2001). It is relatively rare for prostate cancer to be diagnosed in men below 50 years of age, but above this age, the incidence and mortality rates increase exponentially (Haas and Sakr, 1997). The age-specific incidence curve for prostate cancer has a steeper slope than for any other cancer and with the present trend toward an aging population, prostate cancer is a major public health concern (Ross et al., 1979). Nevertheless, the etiology of prostate cancer has not been clearly elucidated, nor is the molecular mechanisms of the disease development and progression well characterized. The most generally accepted model of carcinogenesis postulates that cancer develops through the accumulations of genetic alterations that allow the cells to escape normal growth-regulatory mechanisms (Nowell, 1986).
There has been an ever-growing literature on chromosomal aberrations (CA) in prostate cancer over the past few years. Initial studies suggested that ~75% of prostate cancer tumors had normal male karyotypes. Several candidate chromosomes such as 1, 7, 8, 10, 17, and X are known to play a vital role in the development of prostate cancer (Gibbs et al., 1999; Zucchi et al., 1999). The frequency of chromosome instability in peripheral blood lymphocytes is relevant biomarker for cancer risk in humans, reflecting early biological effects of genotoxic carcinogens and individual cancer susceptibility (Bonassi et al., 2000; Hagmar et al., 1998). An increased frequency of CA in circulating lymphocytes is generally considered indicative of increased cancer risk for those exposed to DNA damaging agents (Bonassi et al., 1995). Although extensive work has been carried out on prostate cancer (Steiner et al., 2002; Verhagen et al., 2002; Wolter et al., 2002), still there are no concrete reports available about the spontaneous background levels of CA in the peripheral blood lymphocytes of prostate cancer patients with respective to the age group. In the present study, the frequency of CA in peripheral blood lymphocytes has been studied in prostate cancer patients (40~80 years) of Tamilnadu and compared with normal healthy subjects of similar age groups to address this issue.
MATERIALS AND METHODS
Subject recruitment and sample collection
A total of 90 subjects aged 40~80 years old including 45 prostate cancer patients and 45 healthy controls, were recruited and subdivided into 4 age groups, namely group I (40~50 years), group II (51~60 years), group III (61~70 years) and group IV (71~80 years). Five millilitres of blood samples were collected from 45 prostate cancer patients (different age groups) in various hospitals of Tamilnadu, who did not undergo any treatment such as hormonal therapy, chemotherapy and radiation therapy. The prostate-specific antigen (PSA) level of collected blood samples was ≤10.0 mg/ml in 39 subjects (86.7%) and ≥10.0 mg/ml in 6 subjects (13.3%). Forty-five healthy controls were selected from the same area as that of the patients. All controls had PSA levels <4.0 mg/ml (90%) and 10% of them had PSA levels <2.0 mg/ml along with confirmation by digital rectal exam. Data on medical and family history of cancer, smoking habits, and occupational history were obtained through an interviewer-administered questionnaire as well as from review of the patient’s hospital records of both control and cancer patients.
Chromosomal aberration assay
All chemical reagents were purchased from Sigma Chemicals (USA), except colcemid that was obtained from Gibco Laboratory (USA). Blood samples were set up to establish leukocyte cultures following standard procedures in our laboratory (Hoyos et al., 1996). Briefly, 0.5 ml blood was added to 4.5 ml RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mmol/L L-glutamine, 1% streptomycin-penicillin, 0.2 ml reagent grade phytohemagglutinin, and was incubated at 37 °C. After 50 h, cultures were treated with 0.1 g/ml colcemid to block cells in mitosis. Lymphocytes were harvested after 52 h by centrifuging cells to remove culture medium (800~1000 r/min), added with hypotonic solution (KCl, 0.075 mol/L) at 37 °C for 20 min to swell the cells, and treated twice with Carnoy’s fixative (3:1 (v/v) ratio of methanol:acetic acid). Slides were carefully dried on a hot plate (56 °C, 2 min), and then stained using the Giemsa-banding (G-banding) technique (Goto et al., 1975). Microscope coordinates of all digitized G-banded preparations were recorded so that the metaphase cells analyzed by G-banding technique could be analyzed concurrently by spectral karyotyping (SKY) methods.
Spectral karyotyping (SKY)
The SKY™ kit probe cocktail from Applied Spectral Imaging (ASI, Carlsbad, CA, USA) was hybridized to metaphase spreads from each prostate cancer slides according to standard protocols (Garini et al., 1996; Schrock et al., 1996; Veldman et al., 1997) and as per the manufacturer’s instructions (ASI, Carlsbad, CA, USA). After destaining the G-banded slides with methanol for 10 min, the slides were rehydrated in a descending ethyl alcohol series (100%, 90%, 70%), and fixed with 1% formaldehyde in 50 mmol/L MgCl2/phosphate buffer solution for 10 min. The slides were then dehydrated using an ascending ethyl alcohol series and denatured for 30~45 s in 70% formamide/2× SSC at 75 °C. The SKY probe was denatured for 7 min at 75 °C, reannealed at 37 °C for 1 h, placed on the slide and covered with a glass coverslip. The coverslip was sealed with rubber cement and the slides placed in a damp container in a 37 °C incubator. After overnight hybridization, the post-hybridization washes were performed as per manufacturer’s instructions (ASI, Carlsbad, CA, USA).
The metaphase images were captured using an SD 200 spectral bio-imaging system attached to a Zeiss microscope and stored on a SKY image-capture workstation. The images were analyzed using the SKY View software version 1.2 which resolves individual fluorochrome spectra by Fourier spectroscopy and distinguishes the spectral signatures for each chromosome to provide a unique pseudocolour for each chromosome (classified image).
Statistical analysis
All statistical analyses were performed using software SPSS for Windows, version 13. To assess the differences between prostate cancer patients and controls, variables expressed by the mean±SE were analyzed using paired t-tests. P<0.05 was used as the criterion for significant difference between the groups.
RESULTS
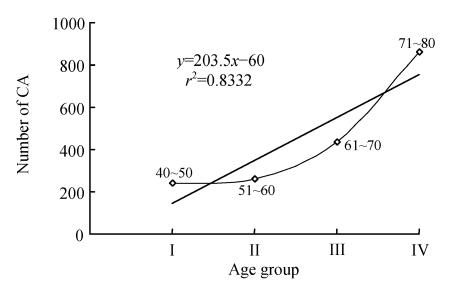
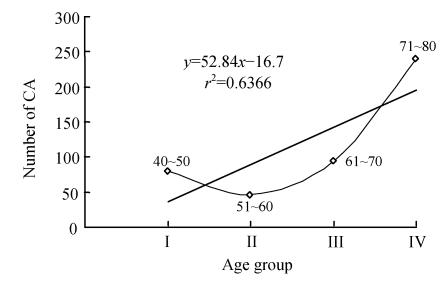
Selected prostate cancer patients with CA are shown in Table 1. Results showed that CA has gradually increased in subjects with respect to their age (Fig.1). The mean±SE was 2.40±0.50 (group I), 2.60±0.21 (group II), 4.35±0.29 (group III), and 8.60±1.56 (group IV). In Table 1, the CA were frequently exhibited in chromosomes 1, 2, 5, 6, 7, 9, 12, 14, 16, 18, 22 and X. Interestingly, controls displayed very low level of major CA (Table 2). In the controls, the mean±SE was 0.80±0.58, 0.46±0.13, 0.95±0.18 and 2.40±0.67 in groups I, II, III and IV, respectively. Statistically significant results were obtained from paired t-test (P<0.05). From the above data, it can be inferred that significant elevation in CA was observed in prostate cancer patients with increase of age when compared with that of control age groups (Fig.2).
Table 1.
Chromosomal abnormalities in prostate cancer patients identified by sequential G-banding and SKY techniques
| No. | Particulars (case No.) | Age (years) | Groups | Chromosome No. | Chromosomal abnormalities identified by G-banding and SKY |
Total | |||
| Deletion | Translocation | Inversion | Mosaic | ||||||
| 1 | PCP001 | 61 | III | 3 | 3 | − | 1 | − | 4 |
| 2 | PCP002 | 48 | I | 13 | 1 | − | 2 | − | 3 |
| 3 | PCP003 | 52 | II | 3, 7 | − | 1 | − | 1 | 2 |
| 4 | PCP004 | 59 | II | 5, 9 | 2 | − | − | 1 | 3 |
| 5 | PCP005 | 60 | II | 7, 9 | 3 | 1 | 1 | − | 5 |
| 6 | PCP006 | 77 | IV | 1, 8, X | 2 | 3 | − | − | 5 |
| 7 | PCP007 | 55 | II | 7 | 1 | 1 | − | − | 2 |
| 8 | PCP008 | 64 | III | 3, 9, 13 | 2 | − | 1 | 1 | 4 |
| 9 | PCP009 | 63 | III | 9, 18 | − | 2 | 1 | − | 3 |
| 10 | PCP010 | 49 | I | 13 | 1 | − | 1 | − | 2 |
| 11 | PCP011 | 52 | II | 7 | 2 | − | − | − | 2 |
| 12 | PCP012 | 54 | II | 9 | − | 1 | 2 | − | 3 |
| 13 | PCP013 | 75 | IV | 3, 5, 9, X | 5 | 1 | − | 1 | 7 |
| 14 | PCP014 | 58 | II | 5 | − | − | − | 2 | 2 |
| 15 | PCP015 | 68 | III | 1, 8 | 3 | 1 | − | − | 4 |
| 16 | PCP016 | 45 | I | 13 | 2 | − | − | − | 2 |
| 17 | PCP017 | 69 | III | 1, 9 | 1 | 2 | − | 3 | 6 |
| 18 | PCP018 | 61 | III | 3, 13 | − | 4 | 1 | − | 5 |
| 19 | PCP019 | 60 | II | 13 | − | − | 2 | − | 2 |
| 20 | PCP020 | 59 | II | 3 | 2 | 1 | − | − | 3 |
| 21 | PCP021 | 63 | III | 9, 18 | − | 2 | − | − | 2 |
| 22 | PCP022 | 68 | III | 7, 9 | 2 | 2 | − | 1 | 5 |
| 23 | PCP023 | 64 | III | 13 | 2 | − | 1 | − | 3 |
| 24 | PCP024 | 58 | II | 9 | − | 1 | 2 | − | 3 |
| 25 | PCP025 | 72 | IV | 3, 7, 9, X | 6 | 3 | − | 1 | 10 |
| 26 | PCP026 | 67 | III | 1, 18 | 2 | 1 | − | 1 | 4 |
| 27 | PCP027 | 41 | I | 7 | − | 1 | − | − | 1 |
| 28 | PCP028 | 54 | II | 3, 5 | − | 2 | − | 1 | 3 |
| 29 | PCP029 | 70 | III | 1, 8 | 3 | 1 | 1 | − | 5 |
| 30 | PCP030 | 49 | I | 7, 13 | 1 | 1 | − | 2 | 4 |
| 31 | PCP031 | 63 | III | 3, 8 | − | 2 | 1 | − | 3 |
| 32 | PCP032 | 66 | III | 13, 18 | 2 | 2 | 1 | − | 5 |
| 33 | PCP033 | 79 | IV | 1, 6, X | 9 | 4 | 1 | − | 14 |
| 34 | PCP034 | 60 | II | 9 | 2 | 1 | − | − | 3 |
| 35 | PCP035 | 68 | III | 3, 9 | 4 | − | 1 | 1 | 6 |
| 36 | PCP036 | 55 | II | 3 | − | − | 2 | − | 2 |
| 37 | PCP037 | 62 | III | 1, 7 | − | 1 | − | 2 | 3 |
| 38 | PCP038 | 68 | III | 8, 13 | 3 | 1 | − | 1 | 5 |
| 39 | PCP039 | 70 | III | 1, 9 | 4 | 2 | − | − | 6 |
| 40 | PCP040 | 58 | II | 7 | 1 | − | − | 1 | 2 |
| 41 | PCP041 | 63 | III | 3, 8 | − | 3 | − | − | 3 |
| 42 | PCP042 | 70 | III | 1, 7 | 1 | 3 | 2 | 1 | 7 |
| 43 | PCP043 | 52 | II | 9 | − | − | 2 | − | 2 |
| 44 | PCP044 | 61 | III | 3, 13 | 1 | − | 3 | − | 4 |
| 45 | PCP045 | 72 | IV | 5, 8, X | 3 | 3 | − | 1 | 7 |
G-banding: Giemsa-banding; SKY: Spectral karyotyping; PCP: Prostate cancer patients
Fig. 1.
Correlation for the total number of chromosomal aberrations (CA) in 100 cells scored per subject prostate cancer. The CA recorded is calculated as per 100 subjects in each group
Table 2.
Chromosomal abnormalities in controls identified by sequential G-banding and SKY techniques
| No. | Particulars (case No.) | Age (years) | Groups | Chromosome No. | Chromosomal abnormalities identified by G-banding and SKY |
Total | |||
| Deletion | Translocation | Inversion | Mosaic | ||||||
| 1 | CS001 | 63 | III | 5 | 1 | − | − | − | 1 |
| 2 | CS002 | 45 | I | 0 | − | − | − | − | 0 |
| 3 | CS003 | 54 | II | 0 | − | − | − | − | 0 |
| 4 | CS004 | 59 | II | 2 | 1 | − | − | − | 1 |
| 5 | CS005 | 59 | II | 7 | − | 1 | − | − | 1 |
| 6 | CS006 | 78 | IV | 2 | 1 | 2 | − | − | 3 |
| 7 | CS007 | 55 | II | 0 | − | − | − | − | 0 |
| 8 | CS008 | 64 | III | 15 | − | 1 | − | 1 | 2 |
| 9 | CS009 | 63 | III | 12 | 1 | − | − | − | 1 |
| 10 | CS010 | 48 | I | 0 | − | − | − | − | 0 |
| 11 | CS011 | 52 | II | 3 | − | 1 | − | − | 1 |
| 12 | CS012 | 54 | II | 0 | − | − | − | − | 0 |
| 13 | CS013 | 74 | IV | 5 | 1 | − | 2 | − | 3 |
| 14 | CS014 | 58 | II | 0 | − | − | − | − | 0 |
| 15 | CS015 | 67 | III | 1 | 1 | 1 | − | − | 2 |
| 16 | CS016 | 45 | I | 0 | − | − | − | − | 0 |
| 17 | CS017 | 67 | III | 7 | 1 | − | − | − | 1 |
| 18 | CS018 | 63 | III | 4 | − | − | 1 | − | 1 |
| 19 | CS019 | 60 | II | 0 | − | − | − | − | 0 |
| 20 | CS020 | 58 | II | 0 | − | − | − | − | 0 |
| 21 | CS021 | 63 | III | 16 | − | 2 | − | − | 2 |
| 22 | CS022 | 66 | III | 0 | − | − | − | − | 0 |
| 23 | CS023 | 65 | III | 2 | 1 | − | 1 | − | 2 |
| 24 | CS024 | 58 | II | 0 | − | − | − | − | 0 |
| 25 | CS025 | 72 | IV | 13 | 2 | − | − | − | 2 |
| 26 | CS026 | 66 | III | 0 | − | − | − | − | 0 |
| 27 | CS027 | 40 | I | 1, 9 | 2 | 1 | − | − | 3 |
| 28 | CS028 | 54 | II | 4 | − | − | − | 1 | 1 |
| 29 | CS029 | 70 | III | 8 | − | 1 | − | − | 1 |
| 30 | CS030 | 47 | I | 5 | − | 1 | − | − | 1 |
| 31 | CS031 | 63 | III | 6 | − | − | 1 | − | 1 |
| 32 | CS032 | 68 | III | 10 | − | 2 | − | − | 2 |
| 33 | CS033 | 79 | IV | 2, 14 | 3 | 1 | − | − | 4 |
| 34 | CS034 | 60 | II | 1 | − | 1 | − | − | 1 |
| 35 | CS035 | 66 | III | 0 | − | − | − | − | 0 |
| 36 | CS036 | 54 | II | 5 | − | − | 1 | − | 1 |
| 37 | CS037 | 61 | III | 0 | − | − | − | − | 0 |
| 38 | CS038 | 67 | III | 0 | − | − | − | − | 0 |
| 39 | CS039 | 70 | III | 8 | − | 1 | − | 1 | 2 |
| 40 | CS040 | 58 | II | 0 | − | − | − | − | 0 |
| 41 | CS041 | 63 | III | 0 | − | − | − | − | 0 |
| 42 | CS042 | 69 | III | 0 | − | − | − | − | 0 |
| 43 | CS043 | 52 | II | 6 | − | 1 | − | − | 1 |
| 44 | CS044 | 61 | III | 9 | 1 | − | − | − | 1 |
| 45 | CS045 | 72 | IV | 0 | − | − | − | − | 0 |
G-banding: Giemsa-banding; SKY: Spectral karyotyping; CS: Control samples
Fig. 2.
Correlation for the total number of chromosomal aberrations (CA) in 100 cells scored per control. The CA recorded is calculated as per 100 controls in each group
DISCUSSION
Cancer is a consequence of genetic or epigenetic alterations in a variety of genes that are fundamental to the process of growth, cell proliferation, differentiation and programmed cell death (Sandberg, 1991). Each alteration whether an initiating or a progression-associated event, may be mediated through gross chromosomal change and hence has the potential to be detected cytogenetically (Solomon et al., 1991). However, a low level of chromosomal instability is detectable in the peripheral blood lymphocytes of patients with skin, breast and bladder cancers and lymphomas (Barrios et al., 1990; Madhavi et al., 1990). Bonassi et al.(1995) reported a significant increase in the mortality ratio for all cancers in subjects who had shown increased levels of CA in their lymphocytes. Essentially, the data from both these studies when pooled indicated that the frequency of chromosome instability in peripheral blood lymphocytes is a relevant biomarker for cancer risks in humans, reflecting early biological effects of genotoxic carcinogens and individual cancer susceptibility (Bonassi et al., 2000; Hagmar et al., 1998).
Although previous studies have shown the presence of chromosome instability in prostate tumors (Verhagen et al., 2002; Wolter et al., 2002), this is the first molecular cytogenetic study to investigate the major CA like deletion, translocation, inversion and mosaics in the peripheral blood lymphocytes of previously untreated prostate cancer patients from Tamilnadu, Southern India among different age groups.
In the present study, chromosome 1 showed the deletion and translocation in prostate cancer patients, groups III and IV showed more number of CA compared to other groups, and group IV particularly had an increase in the number of translocations. The above results of the present study have been supplemented with several reports relating to chromosome 1. Chromosome 1 is involved in prostate cancer whether through the presence of prostate cancer susceptibility genes or through the disruption of common pathways involved in cancer development. Lundgren et al.(1992) reported structural chromosomal changes in prostatic tumor tissues. Carpten et al.(2002) reported recently that mutations on the RNASEL gene (a tumor suppressor gene) on chromosome 1 were responsible for a small fraction of all prostate cancer cases, particularly the most aggressive phenotypes.
In the present study, maximum numbers of CA were deletion, translocation and inversion in the short arm (3p) of chromosome 3 of groups II, III and IV prostate cancer patients. In comparison to groups III and IV, group II showed additional CA in the form of inversions along with deletion and translocation. These results were supported by other studies on chromosome 3. Deletions of the short arm of chromosome 3 (3p24-26 and 3p22-12) were identified in over half of primary prostate cancer cases through LOH (loss of heterozygosity) studies. These deletions were not related to the stage or grade of the tumor (Dahiya et al., 1997). Amplification of chromosome 3q25-27 in primary prostate cancer was identified by CGH (comparative genomic hybridisation), Southern blot and comparative PCR (polymerase chain reaction) (Sattler et al., 2000).
Chromosome 5 showed deletion and mosaic in prostate cancer patients of the present study. Groups II and IV showed the most number of CA compared to control group. Similar studies have reported the loss of alleles of chromosome 5 was detected in a subset of advanced-stage prostate cancer (Cunningham et al., 1996). 5q31-33 was strongly associated with aggressiveness of prostate cancer in a linkage analysis.
In the present study, chromosome 6 showed deletion and translocation in long arm (6q) of prostate cancer patients. This was prominent in group IV patients and controls showed low number of CA in chromosome 6. Other evidences of chromosome 6 involvement in prostate cancer also supported the present study. Almost a third of prostate cancer cases showed LOH as a consequence of deletion of the long arm of chromosome 6, particularly in 6q14-21 (Srikantan et al., 1999). The frequency of 6q deletion in invasive prostate cancer was fivefold higher than in organ-confined prostate cancer (Srikantan et al., 1999). Chromosome translocation involving chromosome 6 was identified in LNCaP cells, where the breakpoint occurred within the TPC gene (homologous to the prokaryotic s10 ribosome protein gene).
Chromosome 7 showed deletion, translocation, inversion and mosaic in prostate cancer patients of the present analysis. Regarding chromosome 7, 7q karyotype was found in all groups and a few of the control group also showed them. Cytogenetic study and FISH studies indicated a gain of chromosome 7 in a substantial number of prostate cancers (Alers et al., 1995; van Dekken et al., 2000), and this gain was associated with higher tumour grade, advanced pathological stage and early prostate cancer death (described as a high Gleason score) (Takahashi et al., 1995). PCR analysis of LOH on prostate cancer indicated a common focus of deletion mapping to 7q31.1-31.2 (Zenklusen et al., 1994).
In chromosome 8, the results identified were similar to chromosome 1 observations. FISH or quantitative PCR showed that allelic imbalance occurred at either the distal terminus of the short arm (8p23) or bordered at 8p22 in almost half of primary prostate cancers (MacGrogan et al., 1994). The 8p22 deletion appeared to accumulate in advanced stages of prostate cancer.
In the present investigation chromosome 9 showed deletion, translocation and inversion in prostate cancer patients of groups II, III, and IV mainly in the long arm (9q). These results were further strengthened by other reports. Loss of chromosome 9 was identified in prostate cancer using FISH. Aneuploidy of chromosome 9 is associated with recurrent disease. Gain of chromosome 9q was found frequently in primary prostate cancer. A high frequency of deletions in 9q21 was also reported in primary prostate cancer (Perinchery et al., 1999).
Chromosome 13 showed deletion and inversion in prostate cancer patients in all groups. Likewise other examples correlating chromosome 13 to prostate cancer include LOH involving the long arm of chromosome 13 in as many as a third of prostate cancers (Cooney et al., 1996; Li et al., 1998). Microsatellite marker analysis indicated that loss of BRCA2 was relatively uncommon in localized prostate cancer but deletion of Rb was more frequent in long arm of chromosome 13 (Li et al., 1998). LOH of 13q was shown to correlate prostate cancer progression (Afonso et al., 1999). Further studies indicated that allelic loss on chromosome 13q14, q21-22 and q33 occurred frequently in metastatic prostate cancer and a subset of prostate cancer (Dong et al., 2001).
The present study also noted deletion and translocation in chromosome 16 in prostate cancer patients in both the long and short arms of groups I, III and IV. An increased rate of LOH was initially identified in chromosome 16q (Kunimi et al., 1991). Structural abnormalities and trisomies of chromosome 16 in prostate cancer were also identified through karyotyping (Miyauchi et al., 1992). FISH analysis mapped the deletions in the long arm of chromosome 16 to 16q22.1-22.3, 16q23.2-24.1 and 16q24.3-ter, encompassing E-cadherin, a putative tumour suppressor gene mapped to 16q22.1.
The present study showed CA like deletion, translocation in the long arm of chromosome 18, in groups III and IV. Deletion of the long arm of chromosome 18 was observed in over 40% of primary prostate cancer (Kunimi et al., 1991; Ueda et al., 1997). LOH was frequently found in chromosome 18q21 (Ueda et al., 1997). Deletion of chromosome 18 occurs more extensively in a metastatic type of prostate cancer.
In chromosome X, deletion and translocation were identified in this study. In group IV, maximum number of the above CA type was observed. Numerical abnormality of the chromosome X was reported in 40% of prostate cancer (Miyoshi et al., 2000). An increased copy number of chromosome X was associated with hormone-refractory prostate cancer, albeit with a high intra tumor heterogeneity (Koivisto et al., 1995).
In the present investigation, increasing age is directly proportional to the increase in CA, and this may also include factors like hormonal inter play, environmental and life style. But age presents a convincing evidence for an increase in the number of aberrations which may be correlated to the onset of this cancer type. In the same way, the study by Sakr et al.(1994) involving the autopsy series of prostate cancer patients revealed small prostatic carcinomas in up to 29% of men aged 30 to 40 years and 64% of men of 60 to 70 years old.
CONCLUSION
This study showed that chromosomal instability in peripheral blood lymphocytes increased in an age wise manner in prostate cancer patients, suggesting the role of age in the development of prostate cancer. CA may hence prove to be a potential biomarker for prostate cancer susceptibility. While the molecular genetic events associated with the initiation and progression of prostate cancer remains poorly understood, it has long been considered that genetic instability plays a pivotal role in the development and progression of human cancer (Loeb, 1991), and as with most types of human cancer, multiple genetic changes are probably necessary for prostate carcinogenesis.
References
- 1.Afonso A, Emmert-Buck MR, Duray PH, Bostwick DG, Linehan WM, Vocke CD. Loss of heterozygosity on chromosome 13 is associated with advanced stage prostate cancer. J Urol. 1999;162(3 Pt 1):922–926. doi: 10.1097/00005392-199909010-00091. [DOI] [PubMed] [Google Scholar]
- 2.Alers JC, Krijtenburg PJ, Vissers KJ, Bosman FT, van der Kwast TH, van Dekken H. Interphase cytogenetics of prostatic adenocarcinoma and precursor lesions: analysis of 25 radical prostatectomies and 17 adjacent prostatic intraepithelial neoplasias. Genes Chromosomes Cancer. 1995;12(4):241–250. doi: 10.1002/gcc.2870120402. [DOI] [PubMed] [Google Scholar]
- 3.Barrios L, Miro R, Caballin MR, Fuster C, Guedea F, Subias A, Egozcue J. Chromosome instability in bladder carcinoma patients. Cancer Genet Cytogenet. 1990;49(1):107–111. doi: 10.1016/0165-4608(90)90170-F. [DOI] [PubMed] [Google Scholar]
- 4.Bonassi S, Abbondandolo A, Camurri L, Dal Prá A, de Ferrari M, Degrassi F, Forni A, Lamberti L, Lando L, Padovani M, et al. Are chromosome aberrations in circulating lymphocytes predicitve of future cancer onset in humans? Prelimiary results of an Italian cohort study. Cancer Genet Cytogenet. 1995;79(2):133–135. doi: 10.1016/0165-4608(94)00131-T. [DOI] [PubMed] [Google Scholar]
- 5.Bonassi S, Hagmar L, Strömberg U, Montagud AH, Tinnerberg H, Forni A, Heikkilä P, Wanders S, Wilhardt P, Hansteen IL, et al. Chromosomal aberrations in lymphocytes predict human cancer independently of exposure to carcinogens. European Study Group on Cytogenetic Biomarkers and Health. Cancer Res. 2000;60(6):1619–1625. [PubMed] [Google Scholar]
- 6.Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, Xu J, Faruque M, Moses T, Ewing C, Gillanders E, et al. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1 . Nat Genet. 2002;30(2):181–184. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- 7.Cook LS, Goldoft M, Schwartz SM, Weiss NS. Incidence of adenocarcinoma of the prostate in Asian immigrants to the United States and their descendants. J Urol. 1999;161(1):152–155. doi: 10.1016/S0022-5347(01)62086-X. [DOI] [PubMed] [Google Scholar]
- 8.Cooney KA, Wetzel JC, Merajver SD, Macoska JA, Singleton TP, Wojno KJ. Distinct regions of allelic loss on 13q in prostate cancer. Cancer Res. 1996;56(5):1142–1145. [PubMed] [Google Scholar]
- 9.Cunningham JM, Shan A, Wick MJ, McDonnell SK, Schaid DJ, Tester DJ, Qian J, Takahashi S, Jenkins RB, Bostwick DG, et al. Allelic imbalance and microsatellite instability in prostatic adenocarcinoma. Cancer Res. 1996;56(19):4475–4482. [PubMed] [Google Scholar]
- 10.Dahiya R, McCarville J, Hu W, Lee C, Chui RM, Kaur G, Deng G. Chromosome 3p24-26 and 3p22-12 loss in human prostatic adenocarcinoma. Int J Cancer. 1997;71(1):20–25. doi: 10.1002/(SICI)1097-0215(19970328)71:1<20::AID-IJC5>3.3.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 11.Dong JT, Boyd JC, Frierson HF. Loss of heterozygosity at 13q14 and 13q21 in high grade, high stage prostate cancer. Prostate. 2001;49(3):166–171. doi: 10.1002/pros.1131. [DOI] [PubMed] [Google Scholar]
- 12.Garini Y, Macville S, du Manoir RA, Buckwald M, Lavi N, Katzir D, Wine I, Bar-Am E, Schröck D, Ried T. Spectral karyotyping. Bioimaging. 1996;4(2):65–72. doi: 10.1002/1361-6374(199606)4:2<65::AID-BIO4>3.3.CO;2-4. [DOI] [Google Scholar]
- 13.Gibbs M, Stanford JL, McIndoe RA, Jarvik GP, Kolb S, Goode EL, Chakrabarti L, Schuster EF, Buckley VA, Miller EL, et al. Evidence for a rare prostate cancer susceptibility locus at chromosome 1p36. Am J Hum Genet. 1999;64(3):776–787. doi: 10.1086/302287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goto K, Akematsu T, Shimazu H, Sugiyama T. Simple differential Giemsa staining of sister chromatids after treatment with photosensitive dyes and exposure to light and the mechanism of staining. Chromosoma. 1975;53(3):223–230. doi: 10.1007/BF00329173. [DOI] [PubMed] [Google Scholar]
- 15.Greenlee RT, Hill-Harmon M, Murry T, Thun M. Cancer statistics. CA Cancer J Clin. 2001;51(1):15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 16.Haas GP, Sakr WA. Epidemiology of prostate cancer. CA Cancer J Clin. 1997;47(5):273–287. doi: 10.3322/canjclin.47.5.273. [DOI] [PubMed] [Google Scholar]
- 17.Hagmar L, Bonassi S, Strömberg U, Brøgger A, Knudsen LE, Norppa H, Reuterwall C European Study Group on Cytogenetic Biomarkers and Health. Chromosomal aberrations in lymphocytes predict human cancer: a report from the European Study Group on Cytogenetic Biomarkers and Health (ESCH) Cancer Res. 1998;58(18):4117–4121. [PubMed] [Google Scholar]
- 18.Hoyos LS, Carvajal S, Solano L, Rodriguez J, Orozco L, López Y, Au WW. Cytogenetic monitoring of farmers exposed to pesticides in Colombia. Environ Health Perspect. 1996;104(Suppl. 3):535–538. doi: 10.2307/3432818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koivisto P, Hyytinen E, Palmberg C, Tammela T, Visakorpi T, Isola J, Kallioniemi OP. Analysis of genetic changes underlying local recurrence of prostate carcinoma during androgen deprivation therapy. Am J Pathol. 1995;147(6):1608–1614. [PMC free article] [PubMed] [Google Scholar]
- 20.Kunimi K, Bergerheim US, Larsson IL, Ekman P, Collins VP. Allelotyping of human prostatic adenocarcinoma. Genomics. 1991;11(3):530–536. doi: 10.1016/0888-7543(91)90059-N. [DOI] [PubMed] [Google Scholar]
- 21.Li C, Larsson C, Futreal A, Lancaster J, Phelan C, Aspenblad U, Sundelin B, Liu Y, Ekman P, Auer G, et al. Identification of two distinct deleted regions on chromosome 13 in prostate cancer. Oncogene. 1998;16(4):481–487. doi: 10.1038/sj.onc.1201554. [DOI] [PubMed] [Google Scholar]
- 22.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51(12):3075–3079. [PubMed] [Google Scholar]
- 23.Lundgren R, Mandahl N, Heim S, Limon J, Henrikson H, Mitelman F. Cytogenetic analysis of 57 primary prostatic adenocarcinomas. Genes Chromosomes Cancer. 1992;4(1):16–24. doi: 10.1002/gcc.2870040103. [DOI] [PubMed] [Google Scholar]
- 24.MacGrogan D, Levy A, Bostwick D, Wagner M, Wells D, Bookstein R. Loss of chromosome arm 8p loci in prostate cancer: mapping by quantitative allelic imbalance. Genes Chromosomes Cancer. 1994;10(3):151–159. doi: 10.1002/gcc.2870100302. [DOI] [PubMed] [Google Scholar]
- 25.Madhavi R, Guntur M, Ghosh R, Ghosh PK. Double minute chromosomes in the leukocytes of a young girl with breast carcinoma. Cancer Genet Cytogenet. 1990;44(2):203–207. doi: 10.1016/0165-4608(90)90048-F. [DOI] [PubMed] [Google Scholar]
- 26.Miyauchi T, Nagayama T, Maruyama K. Chromosomal abnormalities in carcinoma and hyperplasia of the prostate. Nippon Hinyokika Gakkai Zasshi. 1992;83(3):66–74. doi: 10.5980/jpnjurol1989.83.66. [DOI] [PubMed] [Google Scholar]
- 27.Miyoshi Y, Uemura H, Fujinami K, Mikata K, Harada M, Kitamura H, Koizumi Y, Kubota Y. Fluorescence in situ hybridization evaluation of c-myc and androgen receptor gene amplification and chromosomal anomalies in prostate cancer in Japanese patients. Prostate. 2000;43(3):225–232. doi: 10.1002/(SICI)1097-0045(20000515)43:3<225::AID-PROS9>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 28.Nowell PC. Mechanisms of tumor progression. Cancer Res. 1986;46(5):2203–2207. [PubMed] [Google Scholar]
- 29.Perinchery G, Bukurov N, Nakajima K, Chang J, Li LC, Dahiya R. High frequency of deletion on chromosome 9p21 may harbor several tumor-suppressor genes in human prostate cancer. Int J Cancer. 1999;83(5):610–614. doi: 10.1002/(SICI)1097-0215(19991126)83:5<610::AID-IJC7>3.3.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Ross RK, McCurtis JW, Henderson BE, Menck HR, Mack TM, Martin SP. Descriptive epidemiology of testicular and prostatic cancer in Los Angeles. Br J Cancer. 1979;39(3):284–292. doi: 10.1038/bjc.1979.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakr WA, Grignon DJ, Crissman JD, Heilbrun LK, Cassin BJ, Pontes JJ, Haas GP. High grade prostatic intraepithelial neoplasia (HGPIN) and prostatic adenocarcinoma between the ages of 20-69: an autopsy study of 249 cases. In Vivo. 1994;8(3):439–443. [PubMed] [Google Scholar]
- 32.Sandberg AA. Chromosome abnormalities in human cancer and leukemia. Mutat Res. 1991;247(2):231–240. doi: 10.1016/0027-5107(91)90019-k. [DOI] [PubMed] [Google Scholar]
- 33.Sattler HP, Lensch R, Rohde V, Zimmer E, Meese E, Bonkhoff H, Retz M, Zwergel T, Bex A, Stoeckle M, et al. Novel amplification unit at chromosome 3q25-q27 in human prostate cancer. Prostate. 2000;45(3):207–215. doi: 10.1002/1097-0045(20001101)45:3<207::AID-PROS2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 34.Schrock E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273(5274):494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 35.Solomon E, Borrow J, Goodard AD. Chromosome aberrations and cancer. Science. 1991;254(5035):1153–1160. doi: 10.1126/science.1957167. [DOI] [PubMed] [Google Scholar]
- 36.Srikantan V, Sesterhenn IA, Davis L, Hankins G, Avallone F, Livezey JR, Connelly R, Mostofi FK, McLeod DG, Moul JW, et al. Allelic loss on chromosome 6q in primary prostate cancer. Int J Cancer. 1999;84(3):331–335. doi: 10.1002/(SICI)1097-0215(19990621)84:3<331::AID-IJC23>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 37.Steiner T, Junker K, Burkhardt F, Braunsdorf A, Janitzky V, Schubert J. Gain in chromosome 8q correlates with early progression in hormonal treated prostate cancer. Eur Urol. 2002;41(2):167–171. doi: 10.1016/S0302-2838(01)00030-6. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi S, Shan AL, Ritland SR, Delacey KA, Bostwick DG, Lieber MM, Thibodeau SN, Jenkins RB. Frequent loss of heterozygosity at 7q31.1 in primary prostate cancer is associated with tumor aggressiveness and progression. Cancer Res. 1995;55(18):4114–4119. [PubMed] [Google Scholar]
- 39.Ueda T, Komiya A, Emi M, Suzuki H, Shiraishi T, Yatani R, Masai M, Yasuda K, Ito H. Allelic losses on 18q21 are associated with progression and metastasis in human prostate cancer. Genes Chromosomes Cancer. 1997;20(2):140–147. doi: 10.1002/(SICI)1098-2264(199710)20:2<140::AID-GCC4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 40.van Dekken H, Krijtenburg PJ, Alers JC. DNA in situ hybridization (interphase cytogenetics) versus comparative genomic hybridization (CGH) in human cancer: detection of numerical and structural chromosome aberrations. Acta Histochem. 2000;102(1):85–94. doi: 10.1078/0065-1281-00540. [DOI] [PubMed] [Google Scholar]
- 41.Veldman T, Vignon C, Schröck E, Rowley JD, Ried T. Hidden chromosome abnormalities in haematological malignancies detected by multicolour spectral karyotyping. Nat Genet. 1997;15(4):406–410. doi: 10.1038/ng0497-406. [DOI] [PubMed] [Google Scholar]
- 42.Verhagen PC, Hermans KG, Brok MO, van Weerden WM, Tilanus MGJ, de Weger RA, Boon TA, Trapman J. Deletion of chromosomal region 6q14-16 in prostate cancer. Int J Cancer. 2002;102(2):142–147. doi: 10.1002/ijc.10677. [DOI] [PubMed] [Google Scholar]
- 43.Whittemore AS, Kolonel LN, Wu AH, John EM, Gallagher RP, Howe GR, Burch JD, Hankin J, Dreon DM, West DW, et al. Prostate cancer in relation to diet, physical activity, and body size in blacks, whites, and Asians in the United States and Canada. J Natl Cancer Inst. 1995;87(9):652–661. doi: 10.1093/jnci/87.9.652. [DOI] [PubMed] [Google Scholar]
- 44.Wolter H, Trijic D, Gottfried HW, Mattfeldt T. Chromosomal changes in incidental prostatic carcinomas detected by comparative genomic hybridization. Eur Urol. 2002;41(3):328–334. doi: 10.1016/S0302-2838(02)00035-0. [DOI] [PubMed] [Google Scholar]
- 45.Zenklusen JC, Thompson JC, Troncoso P, Kagan J, Conti CJ. Loss of heterozygosity in human primary prostate carcinomas a possible tumor suppressor gene at 7q31.1. Cancer Res. 1994;54:6370–6373. [PubMed] [Google Scholar]
- 46.Zucchi I, Jones J, Affer M, Montagna C, Redolfi E, Susani L, Vezzoni P, Parvari R, Schlessinger D, Whyte MP, et al. Transcription map of Xq27: candidates for several X-linked diseases. Genomics. 1999;57(2):209–218. doi: 10.1006/geno.1999.5768. [DOI] [PubMed] [Google Scholar]