Abstract
Advances in expressed protein ligation (EPL) methods that permit specific introduction of unique modifications into proteins have facilitated protein engineering, structure-function and protein interaction studies. An EPL-generated hybrid exchangeable apolipoprotein has been constructed from recombinant fragments of apolipoprotein E (apoE) and apolipophorin III (apoLp-III). A recombinant fusion protein comprised of human apoE N-terminal residues 1–111, a modified Saccharomyces cerevisiae intein and a chitin binding domain was subjected to 2-mercaptoethanesulfonic acid (MESNA) induced cleavage to generate apoE(1-111)-MESNA. A second fusion protein was comprised of a bacterial pelB leader peptide fused to a variant form of Galleria mellonella apoLp-III residues 1–91. The N-terminal pelB leader sequence directed the newly synthesized fusion protein to the E. coli perisplamic space where endogenous leader peptidase cleavage generated the desired N-terminal cysteine-containing protein fragment. The resulting apoLp-III fragment, which contained no sequence tags or tails, escaped the bacteria and accumulated in the culture medium. When cultured in M9 minimal medium, Asp1Cys apoLp-III(1–91) was produced in high yield and was the sole major protein in the culture supernatant. Ligation reactions with apoE(1–111)-MESNA yielded an engineered hybrid apolipoprotein. The results document the utility of the pelB fusion protein system for generating active N-terminal cysteine containing proteins for EPL applications.
Keywords: Expressed protein ligation, intein, apolipoprotein, apolipophorin, pelB, leader peptidase
Introduction
Native chemical ligation is a useful synthetic method to join independently generated protein fragments via a native peptide bond. Expressed protein ligation (EPL) is a form of native chemical ligation that utilizes intein technology for expression and/or purification of one or more of the fragments to be ligated [1]. EPL has been used to incorporate unnatural amino acids [2,3], biophysical probes [4], post-translational modifications [5] and isotope labels [6,7] in specific locations within a ligated protein product [reviewed in 1,8,9]. These and other EPL strategies have allowed unique problems of protein structure, folding, enzyme mechanism, ion channel function and signaling to be addressed in novel and insightful ways.
EPL involves joining the desired protein fragments via an autocatalytic chemical ligation [8,10]. This reaction, which creates a peptide bond between protein fragments, requires a specific thioester linked leaving group moiety covalently bound to the terminal carboxyl group of one fragment and a cysteine at the amino terminus of the second fragment [8,10]. Adaptations of intein-dependent protein splicing reactions (analogous to intron/exon splicing) originally observed in Saccharomyces cerevisae have made it possible to isolate appropriately modified fragments for subsequent ligation [10–12]. Recombinant production of the protein fragment containing a thioester-linked leaving group moiety normally includes thiol-dependent autocatalytic, intein-mediated cleavage of an engineered fusion protein. Generation of the second protein fragment has relied on three primary approaches including synthetic production by solid-phase peptide synthesis, proteolysis of recombinant proteins by in vitro or in vivo methods [13,14], or thiol and temperature dependent intein mediated fusion protein cleavage [15,16; reviewed in 9,17]. Given limitations on fragment length using solid phase peptide synthesis, the specificity and cost of in vitro protease cleavage and production/yield issues with intein-mediated fusion protein cleavage, the EPL strategy employed requires careful consideration.
Signal peptidases located in the bacterial periplasmic space have been extensively utilized for high-yield production of recombinant proteins [18,19]. The predictable and precise nature of leader peptidase cleavage at the pelB-protein junction combined with high protease activity with a cysteine at position −1 [18], suggests EPL-active protein fragments can be generated by engineering a pelB leader sequence adjacent to the amino-terminal cysteine of the fragment of interest. Furthermore, the pelB sequence directs the newly synthesized protein to the periplasmic space where the membrane-anchored peptidase is localized [20,21]. Studies of bacterially expressed recombinant apolipoproteins have shown that, not only does efficient pelB cleavage occur, the protein product also escapes the periplasm and accumulates in the extracellular culture media [22–25]. This process, for which the mechanism is unknown, facilitates recovery and downstream processing of recombinant proteins from bacterial cultures.
Apolipoprotein E (apoE) is a 299-amino acid glycoprotein that is a well-characterized ligand for the low-density lipoprotein receptor [26]. The X-ray crystal structure of the isolated N-terminal domain revealed a globular bundle of four elongated amphipathic α-helices that is stabilized by interhelical hydrophobic interactions in the absence of lipid [27]. Likewise, insect apolipophorin III (apoLp-III) adopts a helix bundle organization [28,29]. Using recombinant DNA technology, a hybrid apolipoprotein comprised of sequence elements derived from apoE and apoLp-III has been generated [30]. Studies revealed that this engineered hybrid apolipoprotein adopts a folded protein structure that manifests biological activity of the parent proteins. To further pursue hybrid apolipoprotein research, EPL has been employed to generate a protein hybrid comprised of apoE residues 1-111 and Asp1Cys-substituted apoLp-III residues 1-91. To achieve this, the pelB bacterial expression system was employed to generate Asp1Cys apoLp-III(1-91) for use in ligation studies with C-terminal thiol-adducted human apoE(1-111) derived from an intein fusion protein. Optimization studies to determine conditions that promote protein ligation revealed effects of temperature, pH and thiol agent. The approach described expands the strategies available for EPL and provides a means to specifically modify sequence elements within a novel hybrid apolipoprotein.
Materials and Methods
Preparation of apoE(1-111)
Human apoE(1-111) was cloned into the pTYB1 vector (New England Biolabs) and expressed in E. coli ER2566 cells as an S. cerevisiae VMA1 intein and chitin binding domain (CBD) fusion protein. To facilitate optimal intein-mediated fusion protein cleavage [31], valine 111 was mutated to alanine using the QuikChange method (Stratagene) according to the manufacturer’s instructions. Expression and purification procedures for apoE(1-111) followed standardized protocols previously established for generating intein-mediated thioester-adducted proteins [10]. Briefly, saturated overnight cultures were inoculated into 2xYT media containing 50 μg/ml ampicillin, grown to OD600 = 0.6 and induced with 1 mM isopropyl thiogalactopyranoside (IPTG). After 6 h at 30 °C the cells were pelleted by centrifugation (8000 g for 15 min), solubilized with buffer A (20 mM Tris, 150 mM NaCl, 1 mM EDTA, pH=8.0) containing 1% Triton X-100 and stored at −20 °C. Dissolved cell pellets were combined, passed through a microfluidizer, sonicated and centrifuged at 12000 g for 20 min. Isolated clarified cell extract was passed over a chitin bead column pre-equilibrated with buffer A containing 1 % Triton X-100. The column was washed with 10 column volumes of detergent-free buffer A. Fusion protein cleavage was then induced by addition of 2-mercaptoethanesulfonic acid (MESNA) to a final concentration of 60 mM. Flow was arrested for 16–24 h at 22 °C and eluted with 2 bed volumes of buffer A containing 5 mM MESNA. The sample was dialyzed against deionized H20, lyophilized and stored at −20 °C. ApoE(1-111)-MESNA was further purified by semi-preparative C8 reversed-phase high performance liquid chromatography on a Perkin-Elmer Series 200 HPLC.
Preparation of apoLp-III(1-91)
The coding sequence for G. mellonella apoLp-III(1-91) was cloned into the pET22b plasmid (Novagen) directly adjacent to a vector encoded pelB leader sequence. Site directed mutagenesis of aspartate 1 to cysteine was performed using the QuikChange method. Expression and purification of Asp1Cys apoLp-III(1-91) was carried out as previously described for the wild type fragment [25,32]. Briefly, saturated overnight cultures were inoculated into M9 media supplemented with 13.3 mM glucose, 0.1 mM CaCl2, 2 mM MgSO4 and 50 μg/ml ampicillin. At OD600 = 0.6, the culture was induced with 2 mM IPTG. After 6 h at 30 °C, bacteria were pelleted by centrifugation at 8000 g for 15 min and the culture supernatant collected, concentrated by ultrafiltration and chromatographed on a 2.5 × 30 cm column of Sephadex G-75. Fractions containing apoLp-III(1-91) were pooled, dialyzed against deionized H2O, lyophilized and further purified by semi-preparative C8 reversed-phase HPLC.
Analytical methods
Protein purity and/or ligation reaction progress were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using either a 4–20% or fixed 16% acrylamide slab gel. Gels were stained with Amido Black 10B. Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry was performed on a Bruker Daltronics autoflex LRF as described previously [33].
Expressed protein ligation (EPL)
Ligation reactions employed purified apoE(1-111)-MESNA and Asp1Cys apoLp-III(1-91). Unless otherwise stated, fragments were dissolved in 20 mM NaH2PO4, pH=7.2, 150 mM NaCl (PBS) supplemented with 5 % (w/v) MESNA at a final concentration of 5 mg/mL and incubated at 37 °C for 24 h in a final volume of 50 μL. In other experiments, specified reaction parameters were varied as described in the text. Quantification of ligation product yield was performed using ImageJ gel quantification software for Macintosh [34,35]. Percent ligation was calculated from densitometric analysis of stained bands corresponding to apoE(1-111), apoLp-III(1-91) and hybrid apolipoprotein, respectively, as follows: % ligation = [hybrid / (apoE(1-111) + apoLp-III(1-91) + hybrid)] × 100%.
Results and Discussion
Fragment production and characterization
A diagram depicting recombinant apolipoprotein fragment generation and EPL strategy is shown in Figure 1. ApoE(1-111)-MESNA was generated from MESNA induced, intein-mediated cleavage of an apoE·intein·CDB fusion protein. Replacing the commonly used thiol reducing agent dithiothreitol (DTT) with MESNA resulted in a stable adduct that remained covalently bound to the carboxy terminus of alanine 111. Ultimately, the MESNA moiety serves as a leaving group during EPL [8,10,36]. SDS-PAGE analysis verified that the column eluate was highly enriched in apoE(1-111) (Figure 2, panel A) while HPLC analysis gave rise to a single major peak and mass spectrometry yielded a molecular mass = 13,193. The molecular mass increment over that calculated from the amino acid composition of this protein fragment (13,024) is consistent with the presence of a MESNA adduct (MW = 164.2 Da).
Figure 1. Design strategy for EPL-mediated hybrid apolipoprotein production.
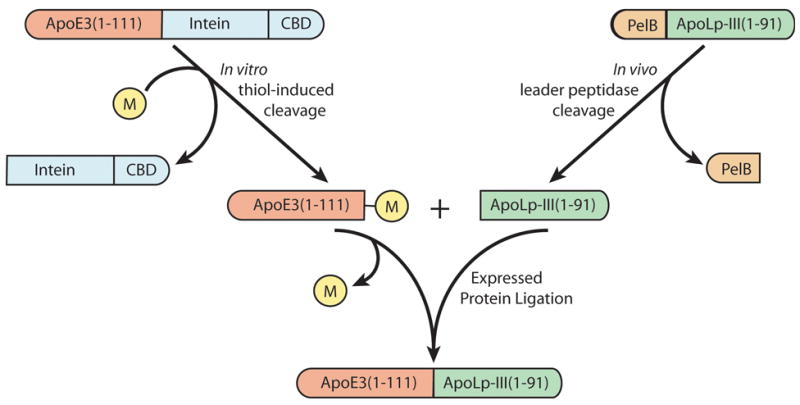
Substrate protein fragments were generated as recombinant fusion proteins, cleaved to generate the desired EPL active fragments and ligated to generate the product hybrid apolipoprotein. M denotes 2-mercaptoethansulfonic acid (MESNA) used to induce intein-mediated cleavage of the apoE · intein · CDB fusion protein.
Figure 2. Characterization of EPL reaction substrate protein fragments.
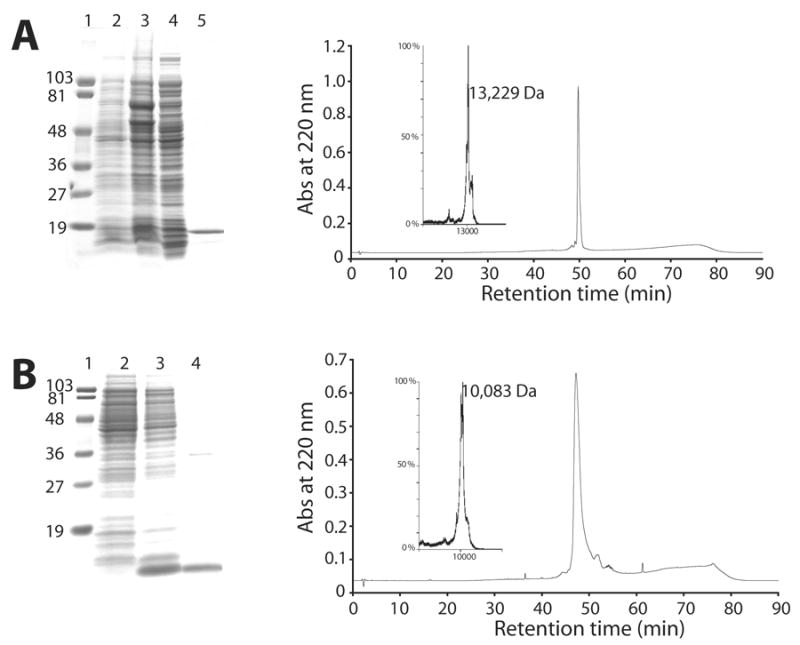
Panel A) apoE(1-111)-MESNA production, purification and analysis. Left; 4–20 % acrylamide gradient SDS-PAGE analysis of fusion protein expression, cleavage and apoE(1-111)-MESNA recovery. Lane 1) molecular weight standards; lane 2) bacterial cell lysate of non-induced ER2566 E. coli cell cultures harboring the apoE-pET22b plasmid; lane 3) lysate of cells induced with 1 mM IPTG; lane 4) chitin column wash flow through following application of an induced cell culture preparation; lane 5) chitin column eluate after exposure to buffer supplemented with 60 mM MESNA. Right; Analytical reversed-phase HPLC of chitin column eluate recovered following exposure to MESNA. Inset: MALDI-TOF analysis of the protein peak with an HPLC retention time = 50 min. Panel B) N-Cys apoLp-III(1-91) production, purification and analysis. Left; 16% acrylamide SDS-PAGE analysis of pelB fusion protein expression, cleavage and Asp1Cys apoLp-III(1-91) recovery. Lane 1) molecular weight standards; lane 2) bacterial cell lysate of non-induced E. coli BL21cells harboring the apoLp-III-pET22b plasmid; lane 3) lysate of cells induced with 2 mM IPTG; lane 4) M9 minimal media cell culture supernatant from induced bacterial cell cultures; Right) Analytical reversed-phase HPLC of induced bacterial cell culture supernatant following Sephadex G-75 column chromatography; Inset: MALDI-TOF analysis of the major HPLC peak (retention time = 48 min).
The EPL substrate fragment containing a reactive N-terminal cysteine (Asp1Cys apoLp-III(1-91)) was expressed in E. coli as a pelB leader peptide fusion protein. The expressed fusion protein was directed to the perisplasmic space where endogenous leader peptidase cleavage generated the desired apoLp-III fragment, which escaped the bacteria and accumulated in the culture medium [25]. ApoLp-III(1-91) was isolated from the culture supernatant by a combination of gel permeation chromatography and reversed phase HPLC (Figure 2, panel B). Mass spectrometry of the isolated fragment yielded a molecular mass of 10,083, in good agreement with the calculated mass of 10,091 for Asp1Cys apoLp-III(1-91), confirming pelB leader peptide cleavage. Final protein yield was ~30–50 mg/L.
Expressed protein ligation
To determine the suitability of apoE(1-111)-MESNA and Asp1Cys apoLp-III(1-91) protein fragments for EPL, incubations were conducted as a function of time (Figure 3). SDS-PAGE analysis revealed a time-dependent accumulation of the expected 23 kDa hybrid apolipoprotein product over the course of 48 h. Appearance of the hybrid apolipoprotein was correlated with a loss of apoE(1-111)-MESNA and apoLp-III(1-91), consistent with a substrate-product relationship. Whereas EPL reactions often require inclusion of chaotropic agents or detergents to maintain substrate fragment solubility [7,37,38], apoE(1-111)-MESNA and apoLp-III(1-91) fragments (as well as the hybrid apolipoprotein product) remain fully soluble in phosphate buffered solution at the millimolar concentrations employed. This finding is in agreement with the known high solubility of intact apoE-N-terminal domain and full-length apoLp-III [29,39]. Based on SDS-PAGE and mass spectrometry analysis, we estimate that, after 48 h, ligation product yield is ~30%. The observed time dependent increase in hybrid apolipoprotein product is in agreement with previous reports that indicate EPL product formation, depending on the size and protein fragment composition, is maximal between 5 and 48 h [31,37,40].
Figure 3. Effect of incubation time on EPL-mediated hybrid apolipoprotein formation.
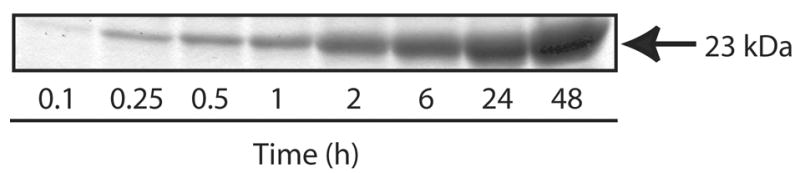
Asp1Cys apoLp-III(1-91) and apoE(1-111)-MESNA were dissolved in PBS supplemented with 5 % (w/v) MESNA (5 mg/mL final concentration) and incubated at 37 °C. Following incubation, hybrid apolipoprotein production was assessed by 16% acrylamide SDS-PAGE analysis. The gel was stained with Amido Black 10B.
In an effort to optimize EPL reaction parameters using this system, further experiments were performed to assess the effect of ligation reaction temperature, pH and thiol agent concentration (Figure 4). While most studies suggest EPL product formation is optimal at 37 °C, isolated reports suggest that room temperature or 4 °C, with longer incubation times, improves product yield [36,40,41]. Whereas hybrid apolipoprotein product formation was low at 4 °C, increasing the temperature to 22 ° C resulted in a dramatic increase in product formation (panel A). Further increases in reaction temperature gave rise to incremental increases in hybrid apolipoprotein product formation.
Figure 4. Effect of incubation parameters on EPL-mediated hybrid apolipoprotein production.
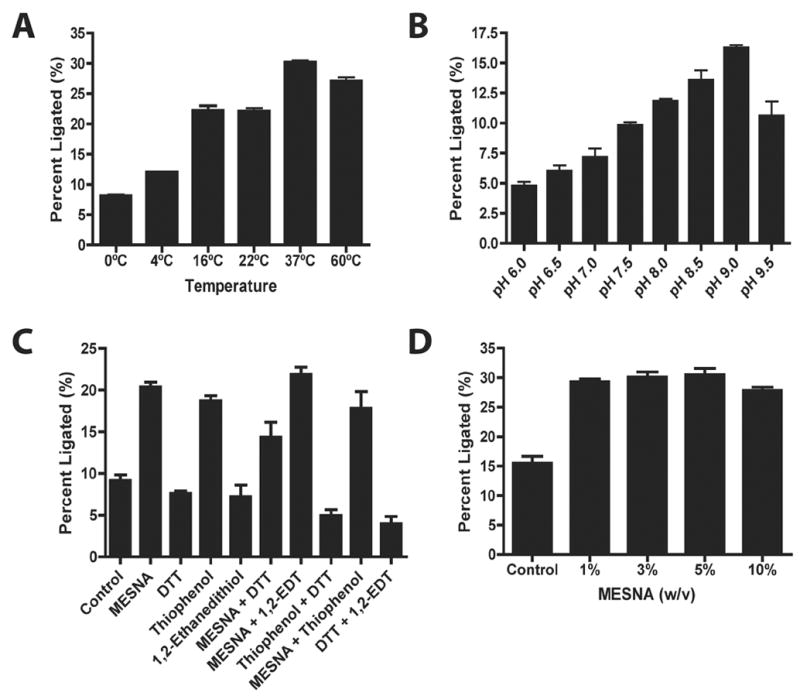
Asp1Cys apoLp-III(1-91) and apoE(1-111)-MESNA were incubated for 48 h at 5 mg/mL final concentration under specified conditions of temperature, buffer pH and thiol agent. Following incubation, aliquots of the reaction mixture were subjected to SDS-PAGE and ligation product formation assessed by densitometry of the stained gel using ImageJ software [30,31]. Panel A) Incubations conducted in PBS supplemented with 5 % (w/v) MESNA at the indicated temperatures; Panel B) incubations conducted at 37 ° in Tris-maleate buffer supplemented with 5 % MESNA and adjusted to the indicated pH; Panel C) incubations as in panel A except for substitution of the specified thiol agent(s) at 10% w/v; Panel D) as in Panel A except for specified MESNA concentration. For panels C and D the control incubation was conducted without added thiol agent. Values reported are the mean ± SEM of three independent determinations.
Using a Tris-maleate buffer system, the effect of solution pH on EPL product formation was examined (panel B). Hybrid apolipoprotein formation was lowest at pH 6.0 and increased steadily with increasing solution pH, reaching a maximum at pH 9.0. The observed decrease in product formation at pH 9.5 suggests a slightly basic pH may be optimal for this EPL reaction. Others have reported that pH, which affects the chemoselectivity of peptide bond formation during ligation, is optimal within a range centered at pH = 8.0 [17].
Studies reporting variable ligation efficiencies using different thiol cofactors stress that choice of thiol agent is critical for ligation reactions [6,7,42], independent of the thiol employed during intein-mediated fusion protein cleavage. Comparison of ligation efficiency in incubations supplemented with different thiol agents including DTT, MESNA, thiophenol and 1,2- ethanedithiol were examined for hybrid apolipoprotein EPL (panel C). As previously documented, DTT resulted in low ligation efficiency, reportedly due to instability of the DTT adduct as a function of time [41,43]. On the other hand, ligation reactions supplemented with MESNA, thiophenol, 1,2-ethanedithiol or a combination of these agents increased hybrid apolipoprotein product formation. The effect of MESNA concentration on product formation was also evaluated (panel D). Compared to the control incubation in which added MESNA was not present, similar product yields were observed between 1 % and 5 % MESNA. The data confirm that MESNA is not only required as a chemical leaving group during EPL but also facilitates ligation reaction progress, most likely by maintaining a reducing environment and preventing unwanted oxidation or hydrolysis.
The effect of substrate concentration on ligation product yield was investigated by varying the amount of either apoE(1-111) or apoLp-III(1-91) in EPL reactions (Figure 5). As expected, in the absence of either substrate protein fragment, no reaction product was detected. When the amount of either apoE(1-111)-MESNA or apoLp-III(1-91) starting substrate was increased relative to the other fragment, a positive correlation with hybrid apolipoprotein product formation was observed as equimolar protein concentrations were approached. However, when the amount of apoE(1-111)-MESNA in the incubation was increased relative to apoLp-III(1-91), maximal product formation was observed below equimolar concentration, indicating that apoE(1-111)-MESNA concentration dependent ligation was saturable (panel A). This suggests that apoE(1-111) is partially limiting with respect to ligation efficiency, perhaps due to the presence of MESNA deficient substrate protein.
Figure 5. Effect of substrate protein fragment concentration on EPL product formation.
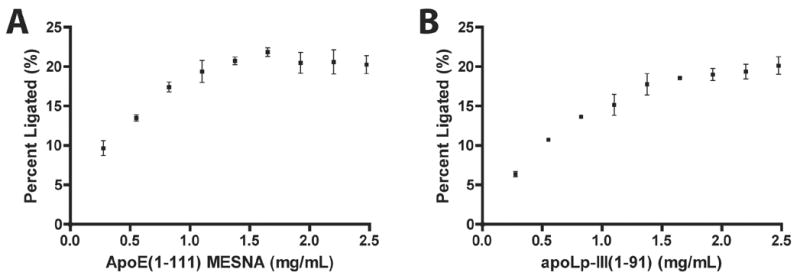
Panel A) Specified concentrations of apoE(1-111)-MESNA and a fixed Asp1Cys apoLp-III(1-91) concentration (2.5 mg/mL final) were incubated in PBS supplemented with 5 % (w/v) MESNA at 37 °C for 24 h (25 μL final volume). Panel B) Specified concentrations of Asp1Cys apoLp-III(1-91) and a fixed apoE(1-111)-MESNA concentration (2.5 mg/mL) were incubated in PBS supplemented with 5 % (w/v) MESNA at 37 °C for 24 h (25 μL final volume). Following incubation, an aliquot of the reaction mixture was subjected to SDS-PAGE and ligation product formation assessed by densitometry of the stained gel using ImageJ software [30,31]. Values reported are the mean ± SEM (n=3).
Summary and Conclusions
The increasing utilization of EPL methodologies for the design of specifically modified proteins has demanded new techniques for manipulating and producing thioester and N-terminal cysteine EPL reaction substrates. The present study employed a novel recombinant system for facile production of an EPL-active, N-Cys protein fragment in high yield. The system described represents an improvement over conventional in vitro protease catalyzed reactions since cleavage occurs in vivo concurrent with protein expression. Using standard protein isolation techniques, the desired apolipoprotein fragment was purified directly from a bacterial culture supernatant without the need to modify or protect the active cysteine. Although other proteins may not escape the bacteria following pelB leader peptide cleavage, localization to the periplasmic space is known to increase protein folding efficiency [44,45]. Additionally, pelB fusion protein expression is readily adaptable to growth in M9 minimal media, permitting stable isotope enrichment for high-resolution structural and biophysical studies [7,38]. Thus, the method described offers a valuable approach for generating EPL active, N-Cys protein fragments for use in ligation reactions, simplifying downstream processing and increasing yield. The establishment of methods to generate hybrid apolipoproteins provides new opportunities for protein engineering and structure function analysis of this and other biologically important protein families.
Acknowledgments
We thank Dr. Vasanthy Narayanaswami for helpful discussions and assistance with intein fusion protein expression and purification and Drs. Vincent Raussens and Darrin A. Lindhout for technical advice and discussions. Additionally, we would like to thank Lydia Krin for assistance with intein fusion protein expression and purification. This research was supported by a grant from the National Institutes of Health (HL-64159).
Abbreviations
- apo
apolipoprotein
- apoLp
apolipophorin
- DTT
dithiothreitol
- EPL
expressed protein ligation
- MESNA
2-mercaptoethanesulfonic acid
- N-cys
amino-terminal cysteine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.David R, Richter MP, Beck-Sickinger AG. Expressed protein ligation. Method and applications. Eur J Biochem. 2004;271:663–677. doi: 10.1111/j.1432-1033.2004.03978.x. [DOI] [PubMed] [Google Scholar]
- 2.Wang D, Cole PA. Protein tyrosine kinase Csk-catalyzed phosphorylation of Src containing unnatural tyrosine analogues. J Am Chem Soc. 2001;123:8883–8886. doi: 10.1021/ja010540b. [DOI] [PubMed] [Google Scholar]
- 3.Yee CS, Chang MC, Ge J, Nocera DG, Stubbe J. 2,3-difluorotyrosine at position 356 of ribonucleotide reductase R2: a probe of long-range proton-coupled electron transfer. J Am Chem Soc. 2003;125:10506–10507. doi: 10.1021/ja036242r. [DOI] [PubMed] [Google Scholar]
- 4.Scheibner KA, Zhang Z, Cole PA. Merging fluorescence resonance energy transfer and expressed protein ligation to analyze protein-protein interactions. Anal Biochem. 2003;317:226–232. doi: 10.1016/s0003-2697(03)00087-3. [DOI] [PubMed] [Google Scholar]
- 5.Zheng W, Zhang Z, Ganguly S, Weller JL, Klein DC, Cole PA. Cellular stabilization of the melatonin rhythm enzyme induced by nonhydrolyzable phosphonate incorporation. Nat Struct Biol. 2003;10:1054–1057. doi: 10.1038/nsb1005. [DOI] [PubMed] [Google Scholar]
- 6.Camarero JA, Shekhtman A, Campbell EA, Chlenov M, Gruber TM, Bryant DA, Darst SA, Cowburn D, Muir TW. Autoregulation of a bacterial sigma factor explored by using segmental isotopic labeling and NMR. Proc Natl Acad Sci USA. 2002;99:8536–8541. doi: 10.1073/pnas.132033899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valiyaveetil FI, MacKinnon R, Muir TW. Semisynthesis and folding of the potassium channel KcsA. J Am Chem Soc. 2002;124:9113–9120. doi: 10.1021/ja0266722. [DOI] [PubMed] [Google Scholar]
- 8.Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 9.Schwarzer D, Cole PA. Protein semisynthesis and expressed protein ligation: chasing a protein’s tail. Curr Opin Chem Biol. 2005;9:561–569. doi: 10.1016/j.cbpa.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 10.Xu MQ, Evans TC., Jr Intein-mediated ligation and cyclization of expressed proteins. Methods. 2001;24:257–277. doi: 10.1006/meth.2001.1187. [DOI] [PubMed] [Google Scholar]
- 11.Kane PM, Yamashiro CT, Wolczyk DF, Neff N, Goebl M, Stevens TH. Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H(+)-adenosine triphosphatase. Science. 1990;250:651–657. doi: 10.1126/science.2146742. [DOI] [PubMed] [Google Scholar]
- 12.Hirata R, Ohsumk Y, Nakano A, Kawasaki H, Suzuki K, Anraku Y. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 1990;265:6726–6733. [PubMed] [Google Scholar]
- 13.Erlanson DA, Chytil M, Verdine GL. The leucine zipper domain controls the orientation of AP-1 in the NFAT.AP-1.DNA complex. Chem Biol. 1996;3:981–991. doi: 10.1016/s1074-5521(96)90165-9. [DOI] [PubMed] [Google Scholar]
- 14.Iwai H, Pluckthun A. Circular beta-lactamase: stability enhancement by cyclizing the backbone. FEBS Lett. 1999;459:166–172. doi: 10.1016/s0014-5793(99)01220-x. [DOI] [PubMed] [Google Scholar]
- 15.Evans TC, Jr, Benner J, Xu MQ. The in vitro ligation of bacterially expressed proteins using an intein from Methanobacterium thermoautotrophicum. J Biol Chem. 1999;274:3923–3926. doi: 10.1074/jbc.274.7.3923. [DOI] [PubMed] [Google Scholar]
- 16.Southworth MW, Amaya K, Evans TC, Xu MQ, Perler FB. Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. BioTechniques. 1999;27:110–4. 116–118, 20. doi: 10.2144/99271st04. [DOI] [PubMed] [Google Scholar]
- 17.Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 18.Dalbey RE, Lively MO, Bron S, van Dijl JM. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paetzel M, Karla A, Strynadka NC, Dalbey RE. Signal peptidases. Chem Rev. 2002;102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 20.Dalbey RE, Wickner W. Leader peptidase catalyzes the release of exported proteins from the outer surface of the Escherichia coli plasma membrane. J Biol Chem. 1985;260:15925–15931. [PubMed] [Google Scholar]
- 21.Paetzel M, Dalbey RE, Strynadka NC. The structure and mechanism of bacterial type I signal peptidases. A novel antibiotic target. Pharmacol Ther. 2000;87:27–49. doi: 10.1016/s0163-7258(00)00064-4. [DOI] [PubMed] [Google Scholar]
- 22.Ryan RO, Schieve D, Wientzek M, Narayanaswami V, Oikawa K, Kay CM, Agellon LB. Bacterial expression and site-directed mutagenesis of a functional recombinant apolipoprotein. J Lipid Res. 1995;36:1066–1072. [PubMed] [Google Scholar]
- 23.Fisher CA, Wang J, Francis GA, Sykes BD, Kay CM, Ryan RO. Bacterial overexpression, isotope enrichment, and NMR analysis of the N-terminal domain of human apolipoprotein E. Biochem Cell Biol. 1997;75:45–53. [PubMed] [Google Scholar]
- 24.Weers PM, Wang J, Van der Horst DJ, Kay CM, Sykes BD, Ryan RO. Recombinant locust apolipophorin III: characterization and NMR spectroscopy. Biochim Biophys Acta. 1998;1393:99–107. doi: 10.1016/s0005-2760(98)00063-0. [DOI] [PubMed] [Google Scholar]
- 25.Dettloff M, Weers PM, Niere M, Kay CM, Ryan RO, Wiesner A. An N-terminal three-helix fragment of the exchangeable insect apolipoprotein apolipophorin III conserves the lipid binding properties of wild-type protein. Biochemistry. 2001;40:3150–3157. doi: 10.1021/bi0013804. [DOI] [PubMed] [Google Scholar]
- 26.Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 27.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 28.Breiter DR, Kanost MR, Benning MM, Wesenberg G, Law JH, Wells MA, Rayment I, Holden HM. Molecular structure of an apolipoprotein determined at 2.5-A resolution. Biochemistry. 1991;30:603–608. doi: 10.1021/bi00217a002. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Sykes BD, Ryan RO. Structural basis for the conformational adaptability of apolipophorin III, a helix-bundle exchangeable apolipoprotein. Proc Natl Acad Sci USA. 2002;99:1188–1193. doi: 10.1073/pnas.032565999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiss RS, Weers PM, Narayanaswami V, Cohen J, Kay CM, Ryan RO. Structure-guided protein engineering modulates helix bundle exchangeable apolipoprotein properties. J Biol Chem. 2003;278:21952–21959. doi: 10.1074/jbc.M302676200. [DOI] [PubMed] [Google Scholar]
- 31.Hackeng TM, Griffin JH, Dawson PE. Protein synthesis by native chemical ligation: expanded scope by using straightforward methodology. Proc Natl Acad Sci USA. 1999;96:10068–10073. doi: 10.1073/pnas.96.18.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dettloff M, Niere M, Ryan RO, Luty R, Kay CM, Wiesner A, Weers PM. Differential lipid binding of truncation mutants of Galleria mellonella apolipophorin III. Biochemistry. 2002;41:9688–9695. doi: 10.1021/bi0200108. [DOI] [PubMed] [Google Scholar]
- 33.Beckstead JA, Block BL, Bielicki JK, Kay CM, Oda MN, Ryan RO. Combined N- and C-terminal truncation of human apolipoprotein A-I yields a folded, functional central domain. Biochemistry. 2005;44:4591–4599. doi: 10.1021/bi0477135. [DOI] [PubMed] [Google Scholar]
- 34.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics Intl. 2004;11:36–42. [Google Scholar]
- 35.Rasband WS. Image J. U.S. National Institutes of Health; Bethesda, Maryland, USA: http://rsb.info.nih.gov/ij/1997–2006. [Google Scholar]
- 36.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 37.Romanelli A, Shekhtman A, Cowburn D, Muir TW. Semisynthesis of a segmental isotopically labeled protein splicing precursor: NMR evidence for an unusual peptide bond at the N-extein-intein junction. Proc Natl Acad Sci USA. 2004;101:6397–6402. doi: 10.1073/pnas.0306616101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muralidharan V, Cho J, Trester-Zedlitz M, Kowalik L, Chait BT, Raleigh DP, Muir TW. Domain-specific incorporation of noninvasive optical probes into recombinant proteins. J Am Chem Soc. 2004;126:14004–14012. doi: 10.1021/ja0466199. [DOI] [PubMed] [Google Scholar]
- 39.Wetterau JR, Aggerbeck LP, Rall SC, Jr, Weisgraber KH. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J Biol Chem. 1988;263:6240–6248. [PubMed] [Google Scholar]
- 40.Xu R, Ayers B, Cowburn D, Muir TW. Chemical ligation of folded recombinant proteins: segmental isotopic labeling of domains for NMR studies. Proc Natl Acad Sci USA. 1999;96:388–393. doi: 10.1073/pnas.96.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans TC, Jr, Benner J, Xu MQ. Semisynthesis of cytotoxic proteins using a modified protein splicing element. Protein Sci. 1998;7:2256–2264. doi: 10.1002/pro.5560071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson EC, Kent SB. Insights into the mechanism and catalysis of the native chemical ligation reaction. J Am Chem Soc. 2006;128:6640–6646. doi: 10.1021/ja058344i. [DOI] [PubMed] [Google Scholar]
- 43.Chong S, Mersha FB, Comb DG, Scott ME, Landry D, Vence LM, Perler FB, Benner J, Kucera RB, Hirvonen CA, Pelletier JJ, Paulus H, Xu MQ. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 44.Missiakas D, Raina S. Protein folding in the bacterial periplasm. J Bacteriol. 1997;179:2465–2471. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sone M, Kishigami S, Yoshihisa T, Ito K. Roles of disulfide bonds in bacterial alkaline phosphatase. J Biol Chem. 1997;272:6174–6178. doi: 10.1074/jbc.272.10.6174. [DOI] [PubMed] [Google Scholar]


