Abstract
PURPOSE
Mutations in the RP1gene account for 6% to 10% of autosomal dominant retinitis pigmentosa (adRP). Previous studies have shown that the RP1gene is expressed specifically in photoreceptor cells. So far, little is known about the RP1 protein or how mutations in RP1lead to photoreceptor cell death. The goal of this study was to identify the RP1 protein and investigate its location in photoreceptor cells.
METHODS
A combination of RT-PCR and rapid amplification of cDNA ends (RACE) was used to isolate the full-length mouse Rp1cDNA. Antibodies against different regions of the predicted mouse Rp1 protein were generated. Western blot analyses were used to identify the RP1/Rp1 proteins. The subcel-lular location of RP1 in human and mouse retinas was determined by immunostaining retinal sections.
RESULTS
The full-length mouse Rp1cDNA is 6944 bp, encoding a predicted protein of 2095 amino acids. Rp1 was found to be a soluble protein of approximately 240 kDa, consistent with predictions based on the cDNA sequence. Immunofluores-cence analyses revealed that both the human RP1 and mouse Rp1 proteins are specifically localized in the connecting cilia of rod and cone photoreceptors.
CONCLUSIONS
The presence of RP1/Rp1 in connecting cilia suggests that it may participate in transport of proteins between the inner and outer segments of photoreceptors or in maintenance of cilial structure. This study forms the basis for further investigation of the function of RP1 in retina and the mechanism by which mutations in RP1lead to photoreceptor cell death.(Invest Ophthalmol Vis Sci. 2002;43:22–2032)
Retinitis pigmentosa (RP) is a group of inherited retinal degeneration disorders characterized by night blindness, progressive loss of peripheral vision, and characteristic pigmentary retinopathy. RP is the most common inherited form of blindness, affecting more than 100,000 people in the United States and 1.5 million people worldwide.1 In addition to variations in clinical phenotype, RP is genetically heterogeneous and can be inherited by autosomal dominant (ad), autosomal recessive (ar), or X-linked transmission as well as a rare digenic mode.1,2 adRP accounts for approximately 15% to 20% of RP cases. Linkage analyses have demonstrated 11 genetic loci for adRP to date.2,3 So far, the genes at four of these loci have been identified.3
The RP1gene was the fourth dominant RP gene to be identified,4–6 after RHO, RDS, and NRL, which encode rhodopsin, peripherin/RDS, and NRL, respectively.7–9 The RP1gene is located on chromosome 8q12 and consists of four exons with an open reading frame of 6468 bp, encoding a predicted protein of 2156 amino acids, mostly by exon 4 (788–6468 bp). The RP1/Rp1gene is expressed only in the photoreceptor cells of the retina, as determined by Northern blot analysis4–6 and in situ hybridization.4 Analysis of homology between human RP1 and other known proteins demonstrates that the N-terminal portion of RP1 is related to dou-blecortin (DCX), which is believed to be involved in directing neuronal migration during development of the central nervous system.10
So far, 20 disease-causing mutations have been identified in the RP1gene.4–6,11–13 These are either nonsense or frame-shift mutations that cluster within a region extending from codons 658-1053 in exon 4. All these mutant alleles would encode truncated proteins without the carboxy 50% to 70% of RP1. Together these mutations account for approximately 6% to 10% of adRP cases in different ethnically diverse populations.4,6,11–13 The most common mutation in RP1, Arg677Ter, is present in approximately 3% of patients with adRP in the United States,4 constituting the third most common adRP mutation, after the Pro23His (9% of cases) and Pro347Leu (4% of cases) mutations in the rhodopsin gene.14 These findings indicate that the RP1 protein plays an important, although as yet unknown, role in photoreceptor function.
To elucidate the function of the RP1 protein and to gain insight into the mechanisms by which mutations in RP1cause retinal degeneration, we cloned and sequenced the full-length mouse Rp1cDNA. Based on the amino acid sequence predicted from Rp1cDNA, we generated antibodies against mouse Rp1 fusion proteins. These antibodies were used to detect the RP1/Rp1 proteins by immunoblotting and to localize the RP1/ Rp1 proteins in human and mouse retinas by immunostaining. Our results show that the RP1/Rp1 protein is located in the connecting cilia of rod and cone photoreceptor cells, making it the second protein specifically localized in this important structure of photoreceptors.
METHODS
Animals and Human Tissues
This research adhered to the tenets of the Declaration of Helsinki, the ARVO Statement on the Use of Animals in Ophthalmic and Vision Research, and the guidelines of the University of Pennsylvania in Animal Care and Use. C57Bl/6J mice and Sprague-Dawley rats were obtained from Jackson Laboratories (Bar Harbor, ME). Frozen cow retinas were purchased from JA Lawson, Inc. (Lincoln, NE). Normal human frozen eyes were provided by the Foundation Fighting Blindness Eye Bank at the Scheie Eye Institute (Philadelphia, PA).
Isolation of Mouse Rp1cDNA and Sequence Analysis
A 1.2-kb fragment of the mouse Rp1cDNA was originally isolated from a mouse retinal cDNA library (GenBank accession number AF141021; hosted by the National Center for Biotechnology Information and available in the public domain at Http://www.ncbi.nlm.nih.gov/genbank).4 A combination of RT-PCR using primers designed from the human RP1cDNA sequence (GenBank, AF155141), 5′ RACE, and 3′ RACE was used to extend this sequence to obtain the full-length Rp1cDNA sequence. RT-PCR of the complete coding region from mouse retinal RNA was performed to confirm the coding sequence. The mouse Rp1 protein sequence was predicted and analyzed using the ExPASy proteomics server (hosted by the Swiss Institute of Bioinfor-matics, Geneva, and available in the public domain at http://www.expasy.org ;Fig. 1).
FIGURE 1.

Predicted amino acid sequence and structural features of RP1/Rp1. The Rp1cDNA encodes a predicted protein of 2095 amino acids. The figure shows the deduced amino acid sequences of both human RP1 and mouse Rp1 proteins, which display 80.1% similarity and 60.4% identity. GenBank accession numbers are AF155141 for human RP1cDNA and AF141021 for mouse Rp1cDNA. The regions of homology with DCX, three nuclear localization signals (NLSs), and the putative nucleotide diphosphate kinase (NDP) motif are boxed.
Construction and Expression of Rp1 Fusion Proteins
Three cDNA fragments corresponding to codons 164-557 (N), 681-1072 (M), and 1671-2095 (C) of predicted Rp1 protein sequence were chosen to make His-tagged proteins using the pET-30a(+) vector (No-vagen, Madison, WI; Fig. 2A). Three smaller fragments corresponding to codons 386-559 (N′), 708-894 (M′), and 1708-1928 (C′) within the above regions were also produced for making glutathione-S-transferase (GST) fusion proteins using the pGEX5.1 vector (Amersham Pharmacia, Arlington Heights, IL; Fig. 2B). To produce these fusion proteins, the indicated regions of Rp1cDNA were amplified by PCR from a full-length Rp1cDNA clone, using primers containing the desired restriction enzyme recognition sites. PCR products were digested with the appropriate restriction enzymes and subcloned into the pET-30a(+) or pGEX-5.1 vectors. BL21 (DE3) Escherichia coli(Novagen) was used to produce fusion proteins according to the manufacturer’s recommendations. All six fusion proteins were found to form inclusion bodies and thus to be insoluble. Two different methods were used for purifying these fusion proteins. For His-tagged fusion proteins, the bacterial lysates were extracted, using a binding buffer (500 mM NaCl, 6 M urea in 20 mM phosphate buffer [pH 7.4]), and affinity-purified using His-Trap columns (Amersham Pharmacia). Purified fusion proteins were eluted with imidazole, dialyzed against PBS with 2 M urea at 4°C, and concentrated. For GST fusion proteins, bacterial protein extracts in SDS sample buffer were separated by SDS-PAGE.15 The gels were transiently stained with a copper staining kit (Bio-Rad, Hercules, CA). The overexpressed fusion proteins of the correct size were cut out of the gel, destained, and eluted in an elution buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.1% SDS, and 0.1 mM EDTA) for 1 hour at room temperature. The eluate was filtered by a syringe and dialyzed against PBS for 24 hours at 4°C. Purified proteins were used to immunize animals.
FIGURE 2.
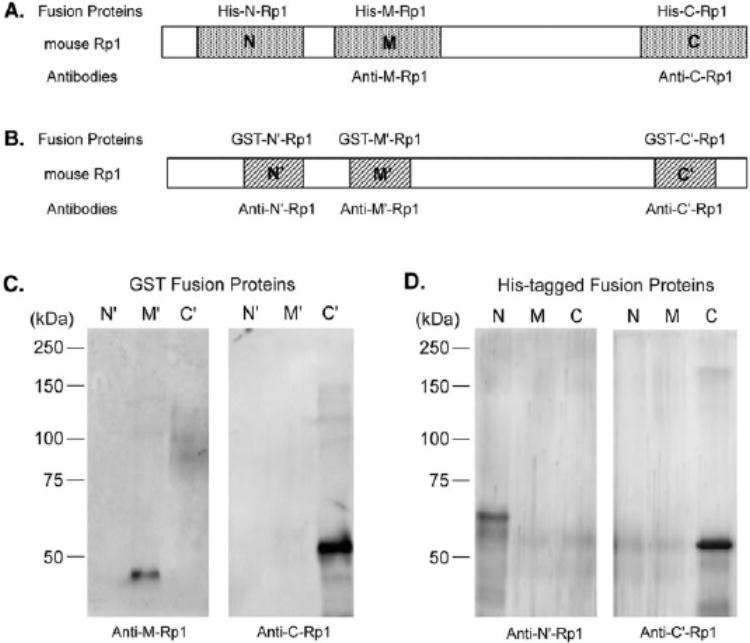
Preparation and specific-ity of anti-Rp1 antibodies. (A)Three different regions of Rp1, each approximately 400 amino acids in length (as indicated by N, M, and C), were chosen to make His-tagged fusion proteins. (B)Three GST fusion proteins containing approximately 200 amino acids of Rp1 (as indicated by N′, M′ and C′) were also made. The Rp1 portions of GST fusion proteins were contained within the corresponding domains of His-tagged fusion proteins. Rabbit polyclonal antibodies anti-M-Rp1 and anti-C-Rp1 were generated against His-tagged fusion protein His-M-Rp1 and His-C-Rp1. Anti-N′-Rp1 and anti-C′-Rp1 were raised against GST fusion protein GST-N′-Rp1 and GST-C′-Rp1 in chickens. Specificity of these antibodies was tested by incubating them with a membrane containing either GST fusion proteins (C)or His-tagged fusion proteins (D).
Preparation of Anti-Rp1 Antibodies
His-tagged fusion proteins His-M-Rp1 and His-C-Rp1 were used to inject rabbits and two polyclonal antisera, anti-M-Rp1 and anti-C-Rp1, were generated (Pocono Rabbit Farm and Laboratory, Canadensis, PA). Spe-cific anti-Rp1 antibodies present in these polyclonal antisera were affinity purified using fusion proteins GST-M′-Rp1 or GST-C′-Rp1 coupled to CNBr-activated Sepharose. Anti-N′-Rp1 and anti-C′-Rp1 antibodies were raised in chicken, using GST-N′-Rp1 and GST-C′-Rp1 fusion proteins as immunogens. Preimmune and immune IgY fractions in egg yolks were purified and stored in PBS (Aves Laboratories, Tigand, OR).
Protein Extracts from Tissues and Western Blot Analysis
Normal adult mouse and rat retinas and additional mouse tissues were dissected and frozen immediately at –80°C. Total protein extracts of each tissue were prepared with SDS sample buffer. Protein samples were boiled and spun at 10,000gfor 10 minutes; the supernatants were then processed for Western blot analysis. One hundred fifty micrograms of each protein sample was separated by SDS-PAGE on 7.5% gels.15 Proteins were then transferred electrophoretically to polyvinyli-dene difluoride (PVDF) membranes.16 The membranes were blocked for 1 hour in TBS-T solution (50 mM Tris-HCl [pH 8.0], 150 mM sodium chloride, 0.1% Tween-20), containing 10% nonfat dry milk, 5% normal goat serum, and 0.05% sodium azide, and incubated either with pre-immune or immune polyclonal antibodies anti-M-Rp1(1 μg/mL) or anti-C′-Rp1 (2.5 μg/mL) in blocking solution for 3 hours. The antibody binding was detected with alkaline phosphatase–conjugated anti-rabbit (1:10000, Vector Laboratories, Burlingame, CA) or anti-chicken secondary antibodies (1:5000, Jackson ImmunoResearch Laboratories, West Grove, PA) and enhanced chemifluorescent (ECF) substrate (Am-ersham Pharmacia). All incubations were done at room temperature. Positive signals were visualized by fluorometry (Storm 860 Imager; Molecular Dynamics, Sunnyvale, CA).
Fractionation of Mouse Retina
Adult mouse retinas were first homogenized in a buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 15 mM EDTA, and 1× protease inhibitor cocktail (Roche Molecular Biochemicals, Indianapolis, IN). The homogenate was centrifuged at 20,000gfor 10 minutes at 4°C, and the supernatant containing soluble proteins was removed. The pellet was resuspended in the same buffer plus 1% Triton X-100. The resultant suspension was separated by centrifugation at 14,000gfor 10 minutes. The residual pellet containing insoluble proteins and cell debris was suspended and extracted with SDS sample buffer.
Preparation of Tissue Sections and Immunostaining Analysis
Eyes of adult mice were dissected after cardiac perfusion with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) and were fixed for 6 hours at 4°C. Fixed eye cups were infiltrated overnight with 30% sucrose in the same buffer, embedded in optimal cutting temperature (OCT) mounting medium, and cryosectioned at 10 μm. Normal adult human retinas were fixed at 6 hours or less after death in 4% parafor-maldehyde and 0.5% glutaraldehyde or in 4% paraformaldehyde alone, all in 0.1 M phosphate buffer. The retinas were processed as for protein extraction and cryosectioned at 12 μm. For immunostaining, human or mouse retinal sections were pretreated with a blocking solution (1% normal horse or goat serum, 1% bovine serum albumin, and 0.05% Triton X-100 in PBS [pH 7.4]) for 1 hour at room temperature and then incubated with primary antibody (two primary antibodies for double staining) diluted in PBS with 0.3% Triton X-100 overnight at 4°C. After they were rinsed with PBS, the sections were treated with one or two secondary antibodies in PBS for 1 hour at room temperature. Slides were then washed in PBS and mounted (Fluoromount-G; Southern Biotechnology Associates, Birmingham, AL). The primary antibodies used were chicken polyclonal anti-N′-Rp1 (5 μg/mL) and anti-C′-Rp1 (5 μg/mL), rabbit affinity purified anti-C-Rp1 (1 μg/mL), mouse monoclonal anti-rhodopsin 4D2 (1:40, from Robert Molday, MD, University of British Columbia, Vancouver, British Columbia, Can-ada),17 and mouse monoclonal human cone-specific antibody 7G6 (1:40, from Peter MacLeish, MD, Morehouse School of Medicine, At-lanta, GA).18 The secondary antibodies were Cy3-conjugated (red) rabbit anti-chicken IgG (1:100), Cy3-conjugated goat anti-rabbit IgG (1:100), and Cy2-conjugated (green) rabbit anti-mouse IgG (1:100) from Jackson ImmunoResearch Laboratories (West Grove, PA). Cell nuclei were counterstained (blue) with 4′, 6′-diamidino-2-phenylindole (DAPI, 1 μg/mL; Molecular Probes, Eugene, OR) added to the mixture of secondary antibodies. Control sections were treated with preim-mune anti-N′-Rp1 or anti-C′-Rp1 at the same dilution. Preabsorption tests were performed by preincubating the diluted affinity purified anti-C'-Rp1 with a 20-fold molar excess of fusion protein GST-C′-Rp1 for 1 hour before the immunostaining procedures. Sections were viewed with a microscope equipped for epifluorescence (Leica, Deerfield, IL) or with a laser scanning confocal microscope (Radiance 2000-MP; Bio-Rad). Confocal image files were processed on computer (Confocal Assistant software; Bio-Rad).
RESULTS
Isolation of Mouse Rp1 cDNA and Sequence Analysis
The full-length mouse Rp1cDNA is 6944 bp, with an open reading frame of 6288 bp, encoding a predicted protein of 2095 amino acids and 234 kDa. Alignment of the predicted human RP1 protein with the mouse Rp1 protein shows 80.1% similarity and 60.4% identity. Several regions show complete identity between the human and mouse proteins, especially in the N-terminal and C-terminal regions. Sequence analysis using publicly available databases demonstrated that the mouse Rp1 protein has homology with DCX, as previously observed for human RP1.4,5 Three potential nuclear localization signal pro-files and a putative nucleoside diphosphate kinase motif were also found to be conserved in both the human and mouse proteins (Fig. 1).
Specificity of Anti-Rp1 Antibodies
Two rabbit polyclonal antibodies, anti-M-Rp1 and anti-C-Rp1, were generated against His-tagged fusion proteins His-M-Rp1 and His-C-Rp1 (Fig. 2A). Two chicken polyclonal antibodies, anti-N′-Rp1 and anti-C′-Rp1, were made against GST fusion proteins GST-N′-Rp1 and GST-C′-Rp1, respectively (Fig. 2B). Before using these antibodies to study Rp1, we first tested their specificity by Western blot analysis. Because the Rp1 portion of the GST fusion proteins were contained within the Rp1 portion of the His-tagged fusion proteins, they were used as the Rp1 antigen providers to test the antibodies raised against His-tagged fusion proteins and vice versa. For example, GST-M′-Rp1 fusion protein was used to test antibody anti-M-Rp1 (made against His-M-Rp1). As shown in Figure 2C, anti-M-Rp1 specifically recognized the corresponding GST fusion protein GST-M′-Rp1, but not GST-N′-Rp1 and GST-C′-Rp1, which were made from different regions of Rp1. Anti-C-Rp1 reacted with GST-C′-Rp1, but not GST-N′-Rp1 or GST-M′-Rp1. In a similar fashion, the His-tagged fusion proteins were also used to test the antibodies raised against GST fusion proteins. Anti-N′-Rp1 detected His-N-Rp1, but not His-M-Rp1 or His-C-Rp1. Anti-C′-Rp1 only detected C-terminal His-tagged fusion protein (Fig. 2D). Therefore, these antibodies were confirmed to have anti-Rp1 specificity and recognized the appropriate domains of the Rp1 protein.
Identification of the Rp1 Protein
Two polyclonal antibodies against distinct regions of predicted mouse Rp1 protein, anti-M-Rp1 and anti-C′-Rp1, were found to specifically detect a protein of approximately 240 kDa on Western blot analysis of mouse retinal extracts, which is in agreement with the predicted size of the Rp1 protein. In addition to the major band at 240 kDa, two weak bands of approximately 220 and 105 kDa were also detected by anti-M-Rp1, and a 90-kDa band was detected by anti-C′-Rp1 (Fig. 3A). These lower molecular weight species may be due to the degradation of Rp1 protein, or nonspecific binding. In the negative control experiments, preimmune antibodies did not detect any specific bands in mouse retinal extracts.
FIGURE 3.
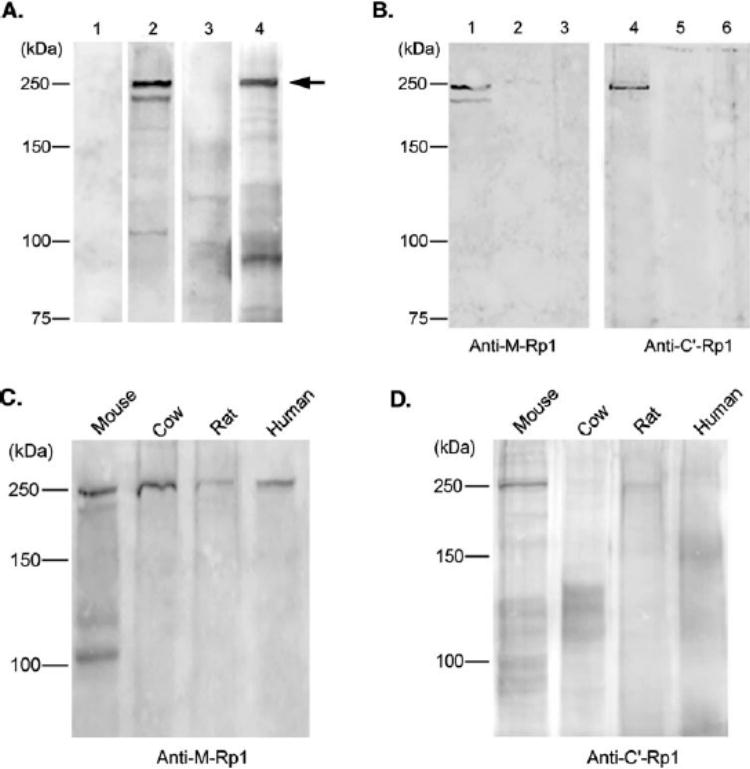
Identification of Rp1 protein by Western blot analysis. (A)Normal mouse retinal protein extracts (˜150 μg) were subjected to immunoblot analysis, using anti-M-Rp1 (lane 2)and anti-C′-Rp1 (lane4). Both antibodies specifically detected a 240-kDa protein (arrow), which is in agreement with the predicted Rp1 size. Negative control experiments, using preimmune anti-M-Rp1 (lane 1)and preimmune anti-C′-Rp1 antibodies are also shown (lane3). (B)Lanes 1and 4: cytosolic soluble fraction; lanes 2and 5: Triton X-100 soluble fraction; lanes 3and 6: insoluble fraction. The 240-kDa Rp1 protein is present exclusively in the cytosol, as detected by anti-M-Rp1 (left)or anti-C′-Rp1 (right). (C)Blots of cow, rat, and human retinal extracts were probed with anti-M-Rp1. (D)Same blot as in (C)was probed with anti-C′-Rp1.
To determine the subcellular distribution of Rp1 protein, mouse retinas were fractionated and analyzed by Western blot analysis (Fig. 3B). The Rp1 protein was found to be concentrated in the soluble fraction, as detected by both anti-M-Rp1 and anti-C′-Rp1 antibodies. Antibodies raised against mouse Rp1 were also tested on retinal extracts from other species, including cow, rat, and human. As shown in Figure 3C, 240-kDa proteins were recognized by anti-M-Rp1 in all four retinal extracts. The lower molecular weight bands detected in mouse retinal extracts were not seen in the retinas of the other species. C-terminal antibody anti-C′-Rp1 also detected the same 240 kDa protein in mouse and rat retinal extracts, but not in the cow or human retinas (Fig. 3D). The human RP1 protein has a predicted molecular weight of 240 kDa, consistent with these findings.
The expression of the mouse Rp1gene was previously shown to be specific to the retina by Northern blot analysis.4,6 To determine the tissue distribution of Rp1 protein, anti-Rp1 antibodies were used to detect Rp1 in total protein extracted from several mouse tissues, including heart, liver, lung, spleen, brain, and skeletal muscle. The 240-kDa band corresponding to the expected Rp1 protein was limited to the retina, using both anti-M-Rp1 (Fig. 4A) and anti-C′-Rp1 (Fig. 4B) antibodies. This observation further confirms that Rp1 is a retina-specific protein. Because the mRNA for Rp1 is not produced in muscle, lung, and spleen, it is likely that the 220-kDa protein detected by the anti-M-Rp1 antibody in these tissues is due to nonspe-cific binding (Fig. 4A).
FIGURE 4.
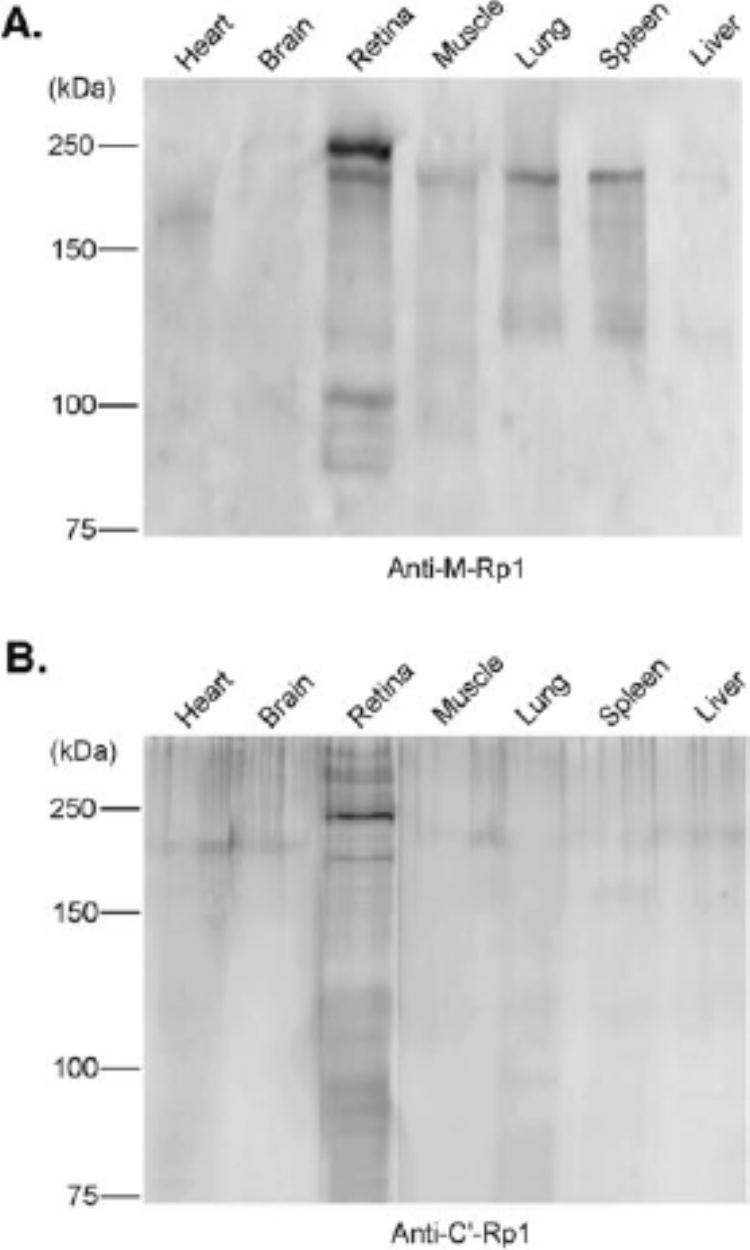
Tissue distribution of mouse Rp1 protein. Equal amounts (˜150 μg) of total protein extracts from indicated mouse tissues were separated by SDS-PAGE on 7.5% gels, followed by Western blot analysis with anti-M-Rp1 (A)and anti-C′-Rp1 (B)antibodies. The 240-kDa band corresponding to Rp1 protein was detected only in retina by both antibodies
Intracellular Location of RP1
We used the anti-Rp1 antibodies to determine the intracellular location of the Rp1 protein in adult mouse retinas by immu-nofluorescence staining. Both anti-N′-Rp1 and anti-C′-Rp1 antibodies produced the same immunostaining pattern, with strong immunostaining in the connecting cilia, between the inner and outer segments of photoreceptors (Figs. 5A, 5C). Fainter immunolabeling was detected in the inner segments; other layers of retina were Rp1 negative (Figs. 5A, 5C). No immunostaining was observed in control sections incubated with preimmune chicken IgY (Figs. 5B, 5D). This localization of Rp1 in the connecting cilia was further confirmed by confocal microscopy at higher magnification (Figs. 5E, 5G). The Rp1 labeling pattern in each connecting cilium matched the location of the axoneme, beginning in the apical inner segment and extending into the proximal outer segment.19–21
FIGURE 5.
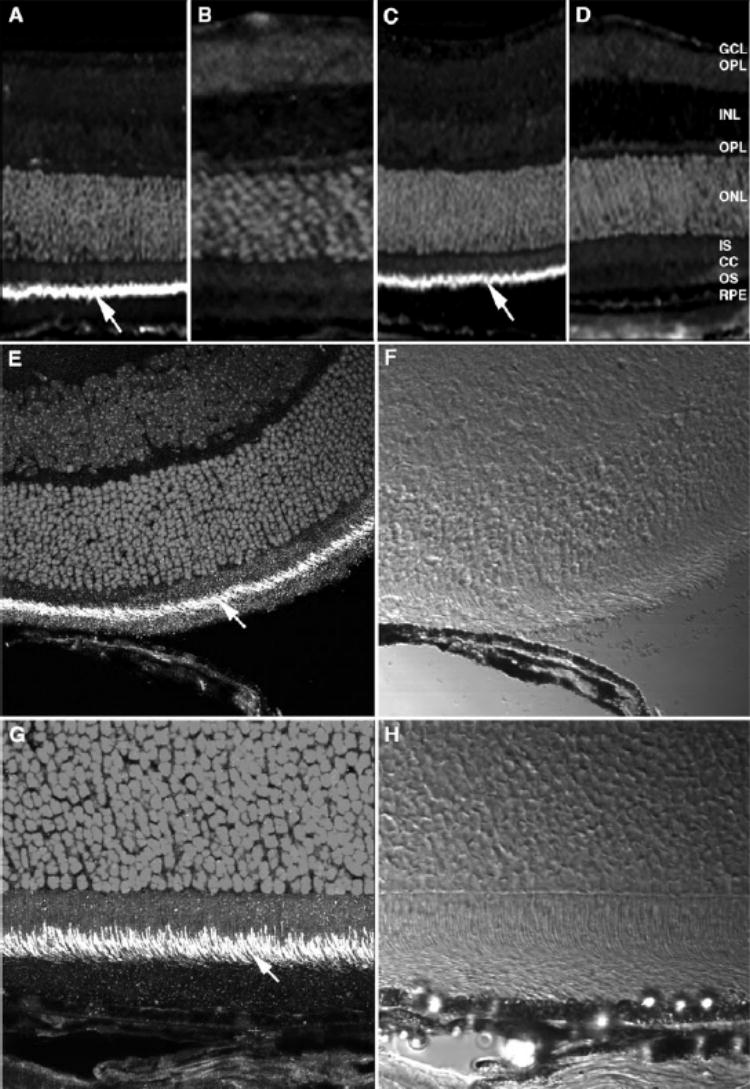
Localization of Rp1 in pho-toreceptor connecting cilia in mouse retina. Frozen sections (10 μm) were prepared from normal adult mouse retina. The retinal sections were incubated with chicken anti-Rp1 polyclonal antibody followed by Cy3-conjugated rabbit anti-chicken secondary antibody. Cell nuclei were stained with DAPI. Specific labeling was detected in the connecting cilia of photoreceptors (arrows). (A, E)Anti-N′-Rp1; (B)preimmune IgY of anti-N′-Rp1; (C, G)anti-C′-Rp1; (D)preimmune IgY of anti-C′-Rp1. (E, G)Confocal microscope images; (F, H)differential interference contrast images of (E)and (G), respectively. Magnification, (A–D)×20; (E, F)×60; (G, H)×100.
We also performed immunostaining on human retinal sections using the anti-Rp1 antibodies. Anti-C-Rp1 antibody labeled the connecting cilia of photoreceptors in human retinas, as found in mouse retinas (Figs. 6A, 6C). The rod and cone inner segments were faintly positive for RP1. Preabsorption with GST-C′-Rp1 fusion protein completely abolished the labeling with the anti-C-Rp1 antibody (Figs. 6B, 6D). The inner plexiform layer and some amacrine cells also were labeled by anti-C-Rp1 in human retinas, but not in adult mouse retinas (data not shown).
FIGURE 6.
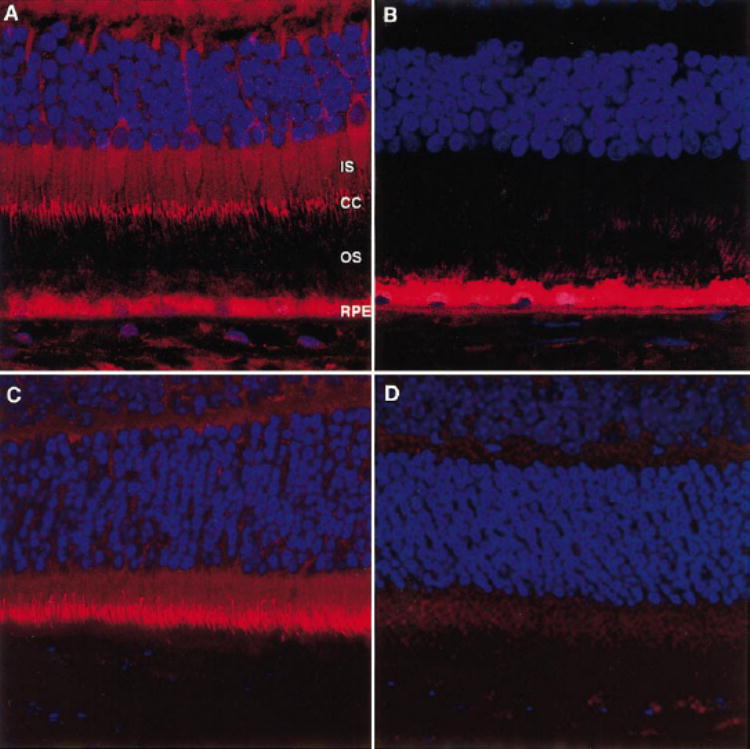
Localization of RP1 in photoreceptor connecting cilia in human retina. Human retinal sections (12 μm) were incubated with rabbit anti-Rp1 antibody, followed by Cy3-conjugated goat anti-rabbit secondary antibody (red). Cell nuclei were stained with DAPI (blue). (A)Human retinal section, anti-C-Rp1. The connecting cilia (CC) of human photoreceptors were labeled. The inner segments (IS) of human photore-ceptors were slightly positive for RP1. The RPE contained autofluores-cent lipofuscin granules. (B)Human retinal section, anti-C-RP1 preab-sorbed with GST-C′-Rp1. (C)Mouse retinal section, anti-C-Rp1; (D)mouse retinal section, anti-C-Rp1 preabsobed with GST-C′-Rp1. Magni-fication, ×60.
To determine whether RP1 immunostaining in the connecting cilia is present in both rods and cones or rods alone, the human cone specific antibody 7G618 was used to perform double labeling with anti-Rp1 antibody on human retinal sections (Fig. 7). All parts of the cones (inner segments, outer segments, somata, and synapses) were strongly labeled by 7G6 (green, Fig. 7A). The same section labeled with the anti-Rp1 antibody showed labeling (red) of the connecting cilia (Fig. 7B). The merged images show yellow staining of the cone connecting cilia (Fig. 7C). Notably, the Rp1 labeling of the cone connecting cilia extended along the sides of the outer segments, consistent with the location of the connecting cilium (Figs. 7B, 7C).19,20
FIGURE 7.
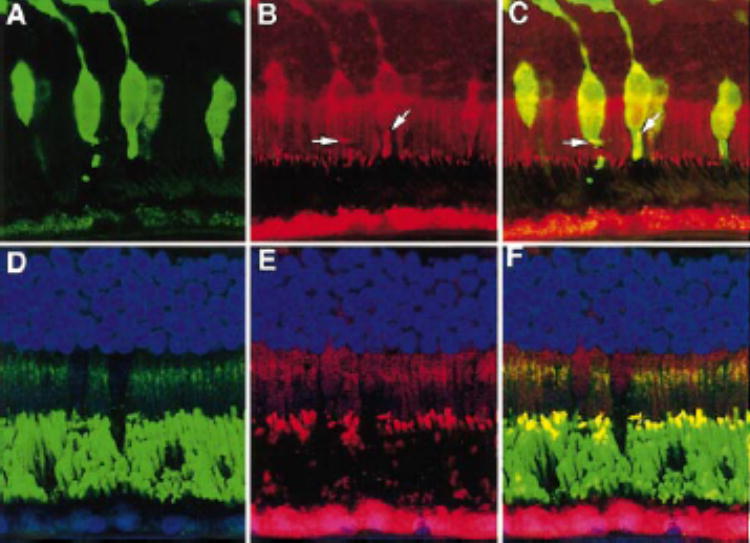
RP1 was present in both rod and cone photoreceptors in human retina. (A)Human retina labeled with human cone-specific antibody 7G6 (green); (B)same section as in (A)labeled with anti-C-Rp1 (red); (C)merged image of (A)and (B). Yellowsignal (arrows)present in the cone-connecting cilia resulted from combination of the redsignal of RP1 and the greensignal of antibody 7G6. (D)Human retina labeled with anti-rhodopsin antibody 4D2. The rod outer segments and the Golgi complexes of the inner segments were labeled. (E)Same section as in (D)labeled with anti-C-Rp1; (F)merged image of (D)and (E). Yellowstaining shows the colocalization of RP1 and rhodopsin in the connecting cilia and bases of the rod outer segments. Magnification, ×60.
Double immunolabeling was also used to verify the location of RP1 in the rods of human retinas. As shown in Figure 7D, a human retina immunolabeled with the anti-rhodopsin antibody 4D2 (green) showed intense staining of the rod outer segments and weaker staining of the Golgi complexes in the inner segments. The same section labeled with anti-C-Rp1 showed labeling of the connecting cilia (red, Fig. 7E). The merged images showed yellow double staining (Fig. 7F), indicating the colo-calization of RP1 and rhodopsin in the connecting cilia and bases of the rod outer segments. Thus, RP1 was found in both rod and cone photoreceptor connecting cilia.
Expression of the Rp1 Protein during Development
Retinas from mice aged 4 to 12 postnatal (P) days (P4–P12) were examined by Western blot analysis (Fig. 8A) and confocal microscopy (Fig. 8B) to determine the pattern of expression of the Rp1 protein during photoreceptor development. For Western blot analysis, total protein from one retina at each time point was used to allow for comparison of the level of Rp1 protein through time course. The Rp1 protein was not detectable at P4. A very low level of Rp1 was detected at P8. The levels of Rp1 protein gradually increased over time, reaching a maximum in adult retinas (6–8 weeks).
FIGURE 8.
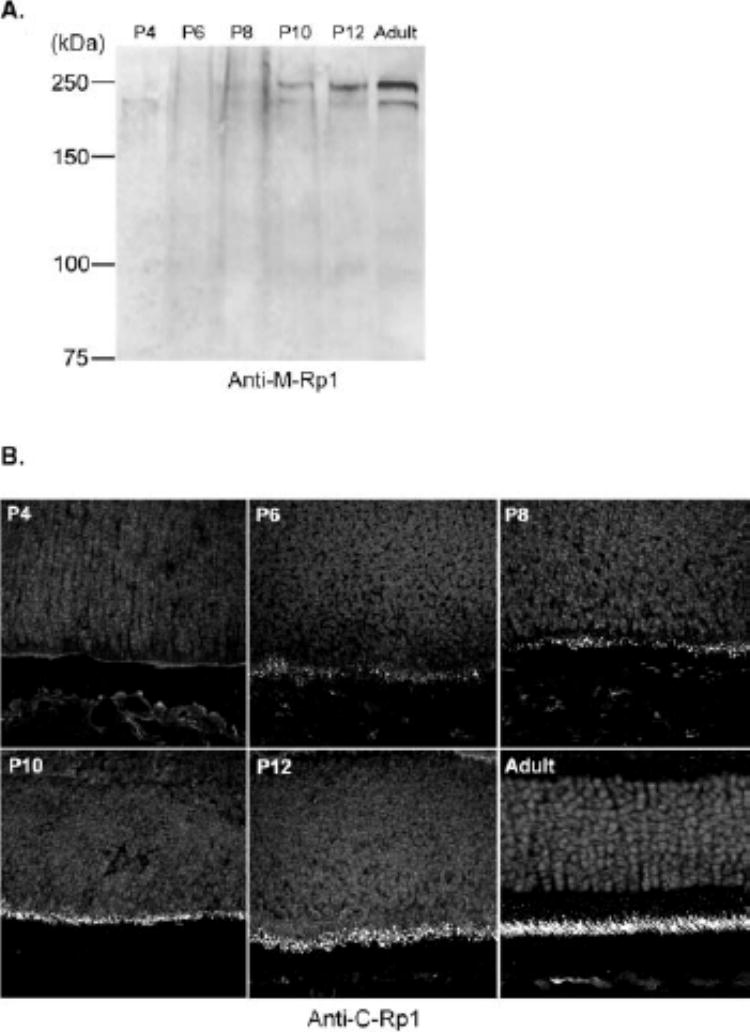
Development of mouse Rp1 protein. (A)Total protein from one retina of P4, P6, P8, P10, and P12 mice was separated on 7.5% SDS-PAGE gels, followed by immunoblot analysis with anti-M-Rp1 antibody. (B)Sections (10 μm) of developing mouse retina were obtained from the same animals as in (A). Anti-C-Rp1 was used to detect the Rp1 protein. At P4, no labeling was detected; at P6, a few positive dots were observed at the outer border of the neuroblastic layer; at P8 and P10, the inner segments had begun to develop and more Rp1 positive structures were present; and at P12, the labeling of Rp1 was localized exclusively to the connecting cilia. Magnification, ×60.
In the sections of developing mouse retinas, no immunore-activity for Rp1 was detected at P4. At P6, a few positive dots were found at the outer border of the neuroblastic layer. In P8 and P10 retinas, the photoreceptor inner segments were Rp1 positive and more Rp1-positive dots were present, corresponding to the short connecting cilia. At P12, the labeling of Rp1 was exclusively localized to the connecting cilia, although the signals in the immature photoreceptors were not as strong as in adult retina (Fig. 8B). The intensity of Rp1 immunolabeling was heavier in the central retina than in the peripheral retina at a given stage, reflecting the central to peripheral gradient of differentiation (data not shown). The inner plexiform layer and amacrine cells of early postnatal mouse retinas were also labeled with anti-C-Rp1 during the period of P4 to P12 (data not shown). This labeling was not observed in the adult mouse retinas. These findings confirm our prior data that the expression of Rp1 begins at the same time that photoreceptor outer segments are forming.4,22
DISCUSSION
Identification of the RP1/Rp1 Protein
In the present study, the RP1/Rp1 protein was found to be specifically localized to the connecting cilia of both rod and cone photoreceptors in human and mouse retinas. To our knowledge, this is the first report of the subcellular localization of RP1/Rp1 protein in photoreceptor cells. These findings are consistent with our previous observation that mouse Rp1mRNA was localized specifically to photoreceptor cell bodies and inner segments by in situ hybridization.4 We also found that the Rp1 protein is soluble and has a molecular weight of 240 kDa, consistent with predictions based on cDNA sequence. The localization of the RP1/Rp1 protein in connecting cilia, despite its solubility, implies that it binds to one or more components of the cilia.
The connecting cilium is a slender structure physically connecting the outer segments and inner segments of retinal photoreceptor cells, with a total length of approximately 200 to 500 nm and a diameter of 170 nm in most mammal photo-receptors.21,23,24 In connecting cilia of photoreceptors, the most obvious cytoskeletal elements are axonemal microtubule doublets (9 × 2 + 0), which arise from the basal body at the distal end of the inner segments, run through the connecting cilia, and extend into the proximal outer segments. The doublets become singlets (9 × 1 + 0) in the outer segments and finally disappear distal to the connecting cilia.21,23,25 The RP1 labeling we observed was longitudinally oriented, extending from the apical inner segment through the connecting cilium into the proximal outer segment. The location of RP1 was consistent with the distribution of the axonemal microtubules in the connecting cilia and outer segments. Accordingly, we hypothesize that the RP1 protein may interact with microtu-bules in the connecting cilia of photoreceptor cells. This hypothesis is further strengthened by the presence of a potential DCX domain at the N-terminal end of predicted RP1 proteins (as will be discussed later).
In addition to the intense labeling of RP1 in the connecting cilia, the inner segments were also faintly positive for RP1 in both human and mouse retinas. This may reflect newly synthesized RP1 in transit to the connecting cilia or may indicate an additional function of RP1 in the inner segments. For example, it is possible that RP1 interacts with cytoplasmic microtubules in the inner segments. The inner plexiform layer and amacrine cells in human retina and neonatal mouse retina were also labeled by anti-C-Rp1, although this pattern was not found in adult mouse retina. Labeling of the inner retinal cells may be due to cross-reactions of the antibody with non-RP1 proteins, in that the expression of Rp1mRNA was detected only in the photoreceptor cells of retina by in situ hybridization.4
Antibodies raised against two distinct regions of mouse Rp1 protein both recognized the same protein of approximately 240 kDa, confirming the predicted size of Rp1. The approximately same size proteins of 240 kDa detected in human, rat, and bovine retinal extracts by anti-Rp1 antibodies indicate that the RP1 proteins in these species also have a similar size. The smaller band of 220 kDa detected by anti-M-Rp1 in various mouse tissues, such as retina, skeletal muscle, lung, and spleen, is likely to be due to nonspecific binding, because this 220-kDa band was not detected by anti-C′-Rp1 antibody and because the mRNA for RP1/Rp1 was detected only in retina by Northern blot analysis.4–6 The two lower molecular weight bands detected by anti-M-Rp1 or anti-C′-Rp1 may result from partial degradation or posttranslational modification of the protein in photoreceptors. The absence of a predicted hydrophobic transmembrane sequence is consistent with our results that the Rp1 is a soluble protein.
The Development of Rp1 Protein
The Rp1 protein is detectable at P6, and the level of Rp1 protein shows a gradual increase in postnatal mouse retinas, consistent with prior data regarding mouse Rp1mRNA.4 The appearance of Rp1 protein is concomitant with the morphologic development of the outer segments, which start forming at approximately day 5 after birth.22 Expression of proteins known to be required for the formation of outer segments, such as rhodopsin and rds/peripherin, also begins at approximately P5.26–28 This implies that, together with other photo-receptor proteins, RP1 may be involved in the formation of outer segments.
Possible Functional Domains of the RP1 Protein
Alignment of the predicted human RP1 and mouse Rp1 protein sequences shows 80.1% similarity and 60.4% identity between the two proteins. Although several regions in the N- and C-terminal regions show complete identity between the human and mouse RP1 proteins, the level of identity between human RP1 and mouse Rp1 is lower than that observed for other photoreceptor proteins. For example, the first two identified adRP genes, rhodopsin and peripherin/RDS, have 99% and 98% identity between human and mouse, respectively.29 This divergence in the RP1 protein sequence may explain why the disease-causing mutations found so far involve large disruptions of this protein and suggests that some amino acid substitutions may not be detrimental to protein function.
Searches for homology between Rp1 and other known protein sequences detected one region of homology at the N-terminal end of Rp1, which is related to DCX, a brain-specific protein implicated in X-linked lissencephaly and double cortex syndrome.10 DCX has strong homology to the human KIAA0369 protein (or DCAMKL1), a central nervous system protein coexpressed in migrating neurons with DCX.30 It has been recently shown that DCX, as well as DCAMKL1, are members of a new family of microtubule-associated proteins (MAPs), based on their colocalization with microtubules, coas-sembly with microtubules, and dramatic effect on microtubule polymerization.30–32 DCX and DCAMKL1 interact with micro-tubules through two tandemly repeated DCX domains (DC).33 It is these two repeated DC domains that represent the homology of RP1 protein with DCX (Fig. 1). Based on the similar distribution of the RP1 protein and axonemal microtubules in the connecting cilia, the DC domains in the RP1 protein sequence provide further evidence that RP1 may interact with microtubules.
Possible Function of the RP1 Protein in Photoreceptor Cells
Photoreceptor cells are highly polarized, and their outer segments have no biosynthetic machinery.34 All components of the outer segment, including the proteins for phototransduc-tion and the lipids of the disc membranes must first be synthesized in the inner segment and transported to the outer segments. Moreover, the outer segment is continually renewed at a rapid rate (˜107 rhodopsin molecules per day per cell) as the distal discs are shed and newly synthesized membrane is added at the base.35 In addition, the concentration of phototransduc-tion proteins changes in response to the light and dark cycle.36,37 An active system(s) in the photoreceptor must exist to transport materials between the inner segments and the outer segments. As the only continuous structural link between the inner segment and outer segment, the connecting cilium is thought to be the critical channel through which the proteins are directionally transported between these compartments. This model for protein transport through the connecting cilia is well supported by experimental evidence.38,39 For example, a recent study demonstrated that rhodopsin was present in and presumably transported through the ciliary membrane.39 However, the exact molecular motors for transport of proteins through the connecting cilia remain to be defined.
Several other proteins also have been localized to the connecting cilia, including a kinesin family member, myosin family members, retinitis pigmentosa guanosine triphosphatase (GT-Pase) regulator (RPGR), and RPGR interacting protein (RP-GRIP).40–44 KIF3A, a component of kinesin-II, has been localized to the connecting cilium and synaptic ribbon in fish and vertebrate photoreceptors.40,45 KIF3A shares a conserved mi-crotubule-based motor domain with kinesin superfamily proteins46 and is required to transport opsin and arrestin from the inner to the outer segment.47
Conventional myosin (myosin II) has been found in the connecting cilium with the same distribution as actin. It has been proposed that the actin-myosin system of the connecting cilium may function to initiate the morphogenesis of a disc membrane.48 Mutations in myosin VIIa were found in patients with Usher syndrome type 1B, an autosomal recessive form of RP and congenital deafness.42 There is a conserved actin-based motor domain in the N-terminal region of myosin VIIa.49 My-osin VIIa has been localized in the retinal pigment epithelium and connecting cilia of photoreceptors.39,50,51 Myosin VIIa is thus a candidate motor for photoreceptor protein transport.
Mutations in the RPGRgene cause RP3, a form of X-linked RP.52 RPGR and its interacting protein, RPGRIP, are colocalized in the outer segment and/or connecting cilia of photoreceptor cells.43,44,53 RPGR is thought to mediate vesicular transport or maintain the polarized protein distribution across the connecting cilium.43,44 RPGRIP is thought to be a structural component of the ciliary axoneme, and one of its functions is to anchor RPGR within the cilium.43
Although localization of a microtubule-associated kinesin family member, an actin-associated myosin family member, and RPGR in the connecting cilia is intriguing, they are all ubiquitously expressed proteins. It remains unknown whether these molecules indeed mediate protein transport between the inner segment and outer segment.43,47 RPGRIP, and now RP1, are the only two proteins found to be uniquely localized in the connecting cilia of photoreceptors.
The location of RP1 in the connecting cilia, and its homology with DCX make RP1 an attractive candidate to participate in transport of newly synthesized outer segment proteins from the inner segments to the site of disc membrane assembly through the connecting cilia. It is possible, for example, that RP1 interacts with microtubules through its N-terminal DC domain, whereas the C-terminal portion of RP1 binds a protein that is headed for the outer segment. Alternative functions for RP1 can also be envisioned, such as regulation of microtubule dynamics through its DC domain, maintenance of the structure and orientation of connecting cilia, or blockage of diffusion between the inner segments and outer segments.
All the disease-causing mutations found in RP1so far cluster in the beginning of exon 4, downstream from the DC domain (exons 2 and 3). These mutations are either nonsense mutations or frame-shift mutations that lead to premature termination of translation. Because these mutations occur after the final intron–exon junction in RP1, it is likely that the mutant RP1mRNAs are not destroyed by nonsense-mediated decay, but that truncated RP1 proteins are produced.54 It is thus possible that mutations in RP1disrupt protein transport through the connecting cilium by separating the N-terminal DC domain from the C-terminal domain(s) of the RP1 protein. Defective protein transport would disrupt outer segment formation and ultimately lead to photoreceptor cell death. Additional studies are needed to test this hypothesis and help elucidate the mechanism by which mutations in RP1cause retinal degeneration.
Acknowledgments
The authors thank Suzanne Pavluk and Wai-Xing Tang for technical assistance.
Supported by National Institutes of Health Grants EY12910 and EY12950; Research to Prevent Blindness; the Foundation Fighting Blindness; the Rosanne Silbermann Foundation, Livingston, New Jersey; the Mackall Foundation Trust, New York, New York; and the March of Dimes Birth Defects Foundation, White Plains, New York.
Footnotes
Commercial relationships policy: N.
References
- 1.Bunker CH, Berson EL, Bromley WC, Hayes RP, Roderick TH. Prevalence of retinitis pigmentosa in Maine. Am J Ophthalmol. 1984;97:357–365. doi: 10.1016/0002-9394(84)90636-6. [DOI] [PubMed] [Google Scholar]
- 2.Dryja TP. Genetic heterogeneity of retinal degenerations and dysfunctions. Les Seminaires Ophtalmologiques d’ISPEN. 1998;10:25–35. [Google Scholar]
- 3.RetNet retinal information network home page. Houston: University of Texas Houston Health Science Center; available at: http://www.sph.uth.tmc.edu/retnet Accessed June 20, 2001.
- 4.Pierce EA, Quinn T, Meehan T, McGee TL, Berson EL, Dryja TP. Mutations in a gene encoding a new oxygen-regulated photore-ceptor protein cause dominant retinitis pigmentosa. Nat Genet. 1999;22:248–254. doi: 10.1038/10305. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan LS, Heckenlively JR, Bowne SJ. Mutations in a novel retina-specific gene cause autosomal dominant retinitis pigmen-tosa. Nat Genet. 1999;22:255–259. doi: 10.1038/10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guillonneau X, Piriev NI, Danciger M. A nonsense mutation in a novel gene is associated with retinitis pigmentosa in a family linked to the RP1 locus. Hum Mol Genet. 1999;8:1541–1546. doi: 10.1093/hmg/8.8.1541. [DOI] [PubMed] [Google Scholar]
- 7.Dryja TP, McGee TL, Hahn LB. Mutations within the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. N Engl J Med. 1990;323:1302–1307. doi: 10.1056/NEJM199011083231903. [DOI] [PubMed] [Google Scholar]
- 8.Kajiwara K, Hahn LB, Mukai S, Travis GH, Berson EL, Dryja TP. Mutations in the human retinal degeneration slow gene in autoso-mal dominant retinitis pigmentosa. Nature. 1991;354:480–483. doi: 10.1038/354480a0. [DOI] [PubMed] [Google Scholar]
- 9.Bessant DA, Payne AM, Mitton KP. A mutation in NRLis associated with autosomal dominant retinitis pigmentosa. Nat Genet. 1999;21:355–356. doi: 10.1038/7678. [DOI] [PubMed] [Google Scholar]
- 10.Gleeson JG, Allen KM, Fox JW. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 11.Bowne SJ, Daiger SP, Hims MM. Mutations in the RP1 gene causing autosomal dominant retinitis pigmentosa. Hum Mol Genet. 1999;8:2121–2128. doi: 10.1093/hmg/8.11.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobson SG, Cideciyan AV, Iannaccone A. Disease expression of RP1 mutations causing autosomal dominant retinitis pig-mentosa. Invest Ophthalmol Vis Sci. 2000;41:1898–1908. [PubMed] [Google Scholar]
- 13.Payne A, Vithana E, Khaliq S. RP1 protein truncating mutations predominate at the RP1 adRP locus. Invest Ophthalmol Vis Sci. 2000;41:4069–4073. [PubMed] [Google Scholar]
- 14.Vaithinathan R, Berson EL, Dryja TP. Further screening of the rhodopsin gene in patients with autosomal dominant retinitis pig-mentosa. Genomics. 1994;21:461–463. doi: 10.1006/geno.1994.1301. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Towbin H, Stahelen T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laird DW, Molday RS. Evidence against the role of rhodopsin in rod outer segment binding to RPE cells. Invest Ophthalmol Vis Sci. 1988;29:419–428. [PubMed] [Google Scholar]
- 18.Chen X, Wikler KC, Macleish PR. Cone properties of retinal margin cells in the monkey (Macaca mulatta) Invest Ophthalmol Vis Sci. 2000;41:2019–2022. [PubMed] [Google Scholar]
- 19.Rohlich P. The sensory cilium of retinal rods is analogous to the transitional zone of motile cilia. Cell Tissue Res. 1975;161:421–430. doi: 10.1007/BF00220009. [DOI] [PubMed] [Google Scholar]
- 20.Peters KR, Palade GE, Schneider BG, Papermaster DS. Fine structure of a periciliary ridge complex of frog retinal rod cells revealed by ultrahigh resolution scanning electron microscopy. J Cell Biol. 1983;96:265–276. doi: 10.1083/jcb.96.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan MW, Iwata RT, Sears RC. Lengths of immunolabeled ciliary microtubules in frog photoreceptor outer segments. Exp Eye Res. 1987;44:623–632. doi: 10.1016/s0014-4835(87)80134-3. [DOI] [PubMed] [Google Scholar]
- 22.Grun G. The Development of the Vertebrate Retina: A Comparative Survey.Berlin. Springer-Verlag. 1982:17–20. [PubMed] [Google Scholar]
- 23.Wen GY, Soifer D, Wisniewski HM. The doublet microtubules of rods of the rabbit retina. Anat Embryol (Berl) 1982;165:315–328. doi: 10.1007/BF00305570. [DOI] [PubMed] [Google Scholar]
- 24.Sung CH, Tai AW. Rhodopsin trafficking and its role in retinal dystrophies. Int Rev Cytol. 2000;195:215–267. doi: 10.1016/s0074-7696(08)62706-0. [DOI] [PubMed] [Google Scholar]
- 25.Roof DJ. Cytoskeletal specializations at the rod photoreceptor distal tip. J Comp Neurol. 1991;305:289–303. doi: 10.1002/cne.903050210. [DOI] [PubMed] [Google Scholar]
- 26.Nir I, Cohen D, Papermaster DS. Immunocytochemical localization of opsin in the cell membrane of developing rat retinal photoreceptors. J Cell Biol. 1984;98:1788–1795. doi: 10.1083/jcb.98.5.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Travis GH. Molecular characterization of the retinal degeneration slow (rds) mutation in mouse. Prog Clin Biol Res. 1991;362:87–114. [PubMed] [Google Scholar]
- 28.Lem J, Krasnoperova NV, Calvert PD. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc Natl Acad Sci USA. 1999;96:736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molday RS. Photoreceptor membrane proteins, phototransduc-tion, and retinal degenerative diseases. Invest Ophthalmol Vis Sci. 1998;39:2491–2513. [PubMed] [Google Scholar]
- 30.Omori Y, Suzuki M, Ozaki K. Expression and chromosomal localization of KIAA0369, a putative kinase structurally related to doublecortin. J Hum Genet. 1998;43:169–177. doi: 10.1007/s100380050063. [DOI] [PubMed] [Google Scholar]
- 31.Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 32.Lin PT, Gleeson JG, Corbo JC, Flanagan L, Walsh CA. DCAMKL1 encodes a protein kinase with homology to doublecortin that regulates microtubule polymerization. J Neurosci. 2000;20:9152–9161. doi: 10.1523/JNEUROSCI.20-24-09152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor KR, Holzer AK, Bazan JF, Walsh CA, Gleeson JG. Patient mutations in doublecortin define a repeated tubulin-binding domain. J Biol Chem. 2000;275:34442–34450. doi: 10.1074/jbc.M007078200. [DOI] [PubMed] [Google Scholar]
- 34.Young RW. Visual cells and the concept of renewal. Invest Oph-thalmol Vis Sci. 1976;15:700–725. [PubMed] [Google Scholar]
- 35.Hollyfield JG, Besharse JC, Rayborn ME. Turnover of rod photore-ceptor outer segments. I: membrane addition and loss in relationship to temperature. J Cell Biol. 1977;75:490–506. doi: 10.1083/jcb.75.2.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangini NJ, Pepperberg DR. Immunolocalization of 48K in rod photoreceptors: light and ATP increase OS labeling. Invest Oph-thalmol Vis Sci. 1988;29:1221–1234. [PubMed] [Google Scholar]
- 37.McGinnis JF, Whelan JP, Donoso LA. Transient, cyclic changes in mouse visual cell gene products during the light-dark cycle. J Neu-rosci Res. 1992;31:584–590. doi: 10.1002/jnr.490310325. [DOI] [PubMed] [Google Scholar]
- 38.Besharse JC, Forestner DM, Defoe DM. Membrane assembly in retinal photoreceptors. III: distinct membrane domains of the connecting cilium of developing rods. J Neurosci. 1985;5:1035–1048. doi: 10.1523/JNEUROSCI.05-04-01035.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfrum U, Schmitt A. Rhodopsin transport in the membrane of the connecting cilium of mammalian photoreceptor cells. Cell Motil Cytoskeleton. 2000;46:95–107. doi: 10.1002/1097-0169(200006)46:2<95::AID-CM2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 40.Beech PL, Pagh-Roehl K, Noda Y, Hirokawa N, Burnside B, Rosen-baum JL. Localization of kinesin superfamily proteins to the connecting cilium of fish photoreceptors. J Cell Sci. 1996;109:889–897. doi: 10.1242/jcs.109.4.889. [DOI] [PubMed] [Google Scholar]
- 41.Williams DS, Hallett MA, Arikawa K. Association of myosin with the connecting cilium of rod photoreceptors. J Cell Sci. 1992;103:183–190. doi: 10.1242/jcs.103.1.183. [DOI] [PubMed] [Google Scholar]
- 42.Weil D, Blanchard S, Kaplan J. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- 43.Hong DH, Yue G, Adamian M, Li T. A retinitis pigmentosa GTPase regulator (RPGR): interacting protein is stably associated with the photoreceptor ciliary axoneme and anchors RPGR to the connecting cilium. J Biol Chem. 2001;276:12091–12099. doi: 10.1074/jbc.M009351200. [DOI] [PubMed] [Google Scholar]
- 44.Hong DH, Pawlyk BS, Shang J, Sandberg MA, Berson EL, Li T. A retinitis pigmentosa GTPase regulator (RPGR)-deficient mouse model for X-linked retinitis pigmentosa (RP3) Proc Natl Acad Sci USA. 2000;97:3649–3654. doi: 10.1073/pnas.060037497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muresan V, Bendala-Tufanisco E, Hollander BA, Besharse JC. Evidence for kinesin-related proteins associated with the axoneme of retinal photoreceptors. Exp Eye Res. 1997;64:895–903. doi: 10.1006/exer.1996.0261. [DOI] [PubMed] [Google Scholar]
- 46.Kondo S, Sato-Yoshitake R, Noda Y. KIF3A is a new micro-tubule-based anterograde motor in the nerve axon. J Cell Biol. 1994;125:1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marszalek JR, Liu X, Roberts EA. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- 48.Williams DS. Actin filaments and photoreceptor membrane turnover. Bioessays. 1991;13:171–178. doi: 10.1002/bies.950130405. [DOI] [PubMed] [Google Scholar]
- 49.Weil D, Kussel P, Blanchard S. The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat Genet. 1997;16:191–193. doi: 10.1038/ng0697-191. [DOI] [PubMed] [Google Scholar]
- 50.el Amraoui A, Sahly I, Picaud S, Sahel J, Abitbol M, Petit C. Human Usher 1B/mouse shaker-1: the retinal phenotype discrepancy explained by the presence/absence of myosin VIIA in the photore-ceptor cells. Hum Mol Genet. 1996;5:1171–1178. doi: 10.1093/hmg/5.8.1171. [DOI] [PubMed] [Google Scholar]
- 51.Liu X, Vansant G, Udovichenko IP, Wolfrum U, Williams DS. Myosin VIIa, the product of the Usher 1B syndrome gene, is concentrated in the connecting cilia of photoreceptor cells. Cell Motil Cytoskeleton. 1997;37:240–252. doi: 10.1002/(SICI)1097-0169(1997)37:3<240::AID-CM6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 52.Meindl A, Dry K, Herrmann K, Manson F. A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3) Nat Genet. 1996;13:35–42. doi: 10.1038/ng0596-35. [DOI] [PubMed] [Google Scholar]
- 53.Roepman R, Bernoud-Hubac N, Schick DE. The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum Mol Genet. 2000;9:2095–2105. doi: 10.1093/hmg/9.14.2095. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE. At least one intron is required for the nonsense-mediated decay of triose-phosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]


