Abstract
Recent efforts have defined a cis-acting DNA regulatory element in plants, the C-repeat/dehydration responsive element (DRE), that stimulates transcription in response to low temperature and water deficit. Here we report the isolation of an Arabidopsis thaliana cDNA that encodes a C-repeat/DRE binding factor, CBF1 (C-repeat/DRE Binding Factor 1). Analysis of the deduced CBF1 amino acid sequence indicates that the protein has a molecular mass of 24 kDa, a potential nuclear localization sequence, and a possible acidic activation domain. CBF1 also has an AP2 domain, which is a DNA-binding motif of about 60 aa present in the Arabidopsis proteins APETALA2, AINTEGUMENTA, and TINY; the tobacco ethylene response element binding proteins; and numerous other plant proteins of unknown function. The transcript levels for CBF1, which appears to be a single or low copy number gene, did not change appreciably in plants exposed to low temperature or in detached leaves subjected to water deficit. Binding of CBF1 to the C-repeat/DRE was demonstrated in gel shift assays using recombinant CBF1 protein expressed in Escherichia coli. Moreover, expression of CBF1 in yeast was found to activate transcription of reporter genes containing the C-repeat/DRE as an upstream activator sequence but not mutant versions of the DNA element. We conclude that CBF1 can function as a transcriptional activator that binds to the C-repeat/DRE DNA regulatory element and, thus, is likely to have a role in cold- and dehydration-regulated gene expression in Arabidopsis.
Keywords: cold acclimation, COR genes, drought stress, signal transduction, yeast
Freezing temperatures and drought are two adverse environmental conditions that limit the geographical distribution of plants and account for significant reductions in the yields of agriculturally important crops. Consequently, considerable effort has been directed at determining the nature of freezing and drought injury and the protection mechanisms plants have evolved that increase tolerance for these environmental “stresses.” Significantly, these studies have revealed that freezing injury is largely a consequence of freeze-induced dehydration (1). Thus, freezing and drought tolerance might have certain protective mechanisms in common. Indeed, it has been observed that some plants increase in freezing tolerance in response to dehydration stress (2, 3). The molecular basis of this cross-protection is not fully understood but appears to include the expression of common genes. For instance, certain COR (cold-regulated) genes of Arabidopsis thaliana that encode novel hydrophilic polypeptides, such as COR15a and COR78, are induced during cold acclimation, the process whereby plants increase in freezing tolerance in response to low nonfreezing temperatures, and are up-regulated in response to water deficit (4). It has been hypothesized that these and other COR genes might help plant cells tolerate the potentially damaging effects of dehydration associated with freezing and drought (4). Indeed, recent studies indicate that constitutive expression of COR15a, which encodes a chloroplast-targeted polypeptide, enhances the freezing tolerance of chloroplasts and has multiple effects on the freezing tolerance of plasma membranes (5).
A major goal now is to determine how plants sense low temperature and water deficit and process this information to alter gene expression. An important advance in this regard was the recent identification of the C-repeat/dehydration responsive element (DRE) cis-acting DNA regulatory element (6–8). The element, which has a 5-bp core sequence of CCGAC, designated the C-repeat (7), is present in one to multiple copies in the promoters of many cold-regulated plant genes, including the Arabidopsis genes COR15a (7) and COR78/RD29A (ref. 6; COR78 and RD29A are alternative designations for the same gene) and the Brassica napus gene BN115 (8). Deletion analysis of the Arabidopsis COR15a promoter indicated that the C-repeat might be part of a cis-acting cold-regulatory DNA element (7). This was first demonstrated by Yamaguchi-Shinozaki and Shinozaki (6), who showed that two 9-bp DNA elements containing the C-repeat, present in the promoter of COR78/RD29A, induce cold-regulated gene expression when fused to a reporter gene. Moreover, they found that the two DNA elements also stimulated transcription in response to dehydration stress. These C-repeat-containing elements were, therefore, given the designation DRE.
Many of the changes in gene expression that occur in response to low temperature and drought require the action of abscisic acid (ABA; refs. 9 and 10). However, studies employing the ABA-insensitive (abi) mutants of Arabidopsis indicate that there are also ABA-independent signal transduction pathways for cold- and dehydration-regulated gene expression (11, 12). Indeed, the results of Yamaguchi-Shinozaki and Shinozaki (6) indicate that the C-repeat/DRE is not responsive to ABA levels and, thus, appears to impart cold- and dehydration-regulated gene expression through an ABA-independent pathway.
How does the C-repeat/DRE impart cold- and dehydration-regulated gene expression? A critical step toward answering this key question is determining the nature of the protein factors that interact with the DNA regulatory element. Here we describe the isolation and characterization of an Arabidopsis cDNA encoding the first identified C-repeat/DRE binding factor, CBF1 (C-repeat/DRE Binding Factor 1), and demonstrate that the protein functions as a transcriptional activator in yeast.
MATERIALS AND METHODS
Plant Material.
A. thaliana (L.) Heyn. ecotype RLD plants were grown in pots in controlled environment chambers at 22°C under constant illumination with cool-white fluorescent lamps (≈100 μmol·m−2·s−1) as described (13). The effect of low temperature on CBF1 expression was determined by placing pots of plants in a cold room at 2.5°C under constant illumination with cool-white fluorescent lamps (≈25 μmol·m−2·s−1) for various times. CBF1 expression in response to water deficit was determined by placing detached leaves on dry filter paper in an uncovered Petri dish for 1.5 h at room temperature on a laboratory bench, at which time the tissues had lost approximately 20% of their fresh weight. The Petri dish was covered, placed in a sealed plastic bag, and “incubated” on a laboratory bench for an additional 5.5 h. For the control, leaves were placed on water-saturated filter paper in a covered Petri dish that was then sealed in a plastic bag and incubated on a laboratory bench for 7 h.
Yeast Reporter Strains.
Oligonucleotides encoding either wild-type or mutant versions of the C-repeat/DRE (Table 1), synthesized at the Michigan State University Macromolecular Structure Facility, were ligated into the BglII site of the lacZ reporter vector pBgl-lacZ (kindly provided by J. Li; ref. 15). The resulting reporter constructs were integrated into the the ura3 locus of Saccharomyces cerevisiae strain GGY1 (MATα Δgal4 Δgal80 ura3 leu2 his3 ade2 tyr; provided by J. Li; ref. 15) by transformation and selection for uracil prototrophy.
Table 1.
Oligonucleotides encoding wild-type and mutant versions of the C-repeat/DRE
| Oligonucleotide | C-repeat/DRE* | Sequence† |
|---|---|---|
| MT50 | COR15a | gatcATTTCATGGCCGACCTGCTTTTT |
| MT52 | M1COR15a | CACAATTTCAaGaattcaCTGCTTTTTT |
| MT80 | M2COR15a | gatcATTTCATGGtatgtCTGCTTTTT |
| MT125 | M3COR15a | gatcATTTCATGGaatcaCTGCTTTTT |
| MT68 | COR15b | gatcACTTGATGGCCGACCTCTTTTTT |
| MT66 | COR78-1 | gatcAATATACTACCGACATGAGTTCT |
| MT86 | COR78-2 | ACTACCGACATGAGTTCCAAAAAGC |
The C-repeat/DRE sequences tested are either wild-type versions found in the promoters of COR15a (7), COR15b (ref. 14; K. Wilhelm & M.F.T., unpublished data), or COR78/RD29A (6) or are mutant versions of the COR15a C-repeat/DRE (M1COR15a, M2COR15a, and M3COR15a) in which the entire C-repeat, CCGAC, had been altered.
Uppercase letters designate bases in wild-type C-repeat/DRE sequences. The core C-repeats are indicated in bold type. Lowercase letters at the beginning of a sequence indicate bases added to facilitate cloning. The lowercase letters that are underlined indicate the mutations in the C-repeat/DRE sequence of COR15a.
Screen of Arabidopsis cDNA Library.
The Arabidopsis pACT cDNA expression library donated by John Walker (deposited in the Arabidopsis Biological Resource Center as stock #CD4-10; funded by National Science Foundation/Department of Energy/U.S. Department of Agriculture Collaborative Research in Plant Biology Program Grant USDA 92–37105-7675) was screened for clones encoding C-repeat/DRE binding domains. The cDNA library harbored in Escherichia coli BNN132 was amplified, and plasmid DNA was isolated and transformed into the yeast GGY1 reporter strains selecting for leucine prototrophy. Yeast transformants that had been grown for 2 or 3 days at 30°C were overlaid with either a nitrocellulose membrane filter (Schleicher & Schuell) or Whatman #50 filter paper and incubated overnight at 30°C. The yeast-impregnated filters were lifted from the plates and treated with X-Gal (5-bromo-4-chloro-3-indolyl β-d-galactoside) to assay colonies for β-galactosidase activity (15). Plasmid DNA from transformants that formed blue colonies on X-Gal-treated filters was recovered (16), propagated in E. coli DH5α, and transformed back into the yeast reporter strains to confirm activity.
Yeast Transformation and Quantitative β-Galactosidase Assays.
Yeast were transformed by either electroporation (17) or the lithium acetate/carrier DNA method (18). Quantitative in vitro β-galactosidase assays were performed as described by Rose and Botstein (19).
Expression of CBF1 Protein in E. coli and Yeast.
CBF1 was expressed in E. coli using the pET-28a(+) vector (Novagen). The BglII–BclI restriction fragment of pACT-11 (pACT-11 is described in Results) encoding CBF1 was ligated into the BamHI site of the vector bringing CBF1 under control of the T7 phage promoter. The construct resulted in a histidine tag, a thrombin recognition sequence, and a T7 epitope tag being fused to the N terminus of CBF1. The construct was transformed into E. coli BL21 (DE3), and the recombinant CBF1 protein was expressed as recommended by the supplier (Novagen). Expression of CBF1 in yeast was accomplished by ligating restriction fragments encoding CBF1 (the BclI–BglII and BglII–BglII fragments from pACT-11) into the BglII site of pDB20.1 (kindly provided by S. Triezenberg; ref. 20), bringing CBF1 under control of the yeast ADC1 promoter.
Gel Shift Assays.
Total soluble E. coli protein (40 ng) was incubated at room temperature in 10 μl of 1× binding buffer (15 mM Hepes, pH 7.9/1 mM EDTA/30 mM KCl/5% glycerol/1 mM DTT) plus 50 ng of poly(dI-dC)·poly (dI-dC) (Pharmacia) with or without 100 ng of competitor DNA. After 10 min, a double-stranded DNA probe (1 ng) that was 32P-labeled by end-filling (21) was added, and the mixture was incubated for an additional 10 min. Samples were loaded onto polyacrylamide gels (4% wt/vol) and fractionated by electrophoresis at 150 V for 2 h (21). Probes and competitor DNAs were prepared from oligonucleotide inserts ligated into the BamHI site of pUC118 (21). Orientation and concatenation number of the inserts were determined by dideoxy DNA sequence analysis (21). Inserts were recovered after restriction digestion with EcoRI and HindIII and fractionation on polyacrylamide gels (12% wt/vol; ref. 21).
DNA and RNA Hybridization.
Total RNA was isolated from Arabidopsis plants (13) and Northern transfers prepared and hybridized as described (22), except that high-stringency wash conditions were at 50°C in 0.1× SSPE (1× SSPE is 3.6 M NaCl/20 mM EDTA/0.2 M Na2HPO4, pH 7.7)/0.5% SDS. Total Arabidopsis DNA was isolated (23) and transferred to nylon membranes (Micron Separations, Westboro, MA) as described (21). High-stringency hybridization and wash conditions were done according to Walling et al. (24). Low-stringency hybridization was in 6× SSPE/0.5% SDS/0.25% low-fat dried milk at 60°C. Low-stringency washes were in 1× SSPE/0.5% SDS at 50°C. Hybridization probes used for the entire CBF1 coding sequence and the 3′ end of CBF1 were the BclI–BglII and EcoRV–BglII restriction fragments from pACT-11, respectively, that had been gel-purified (21). DNA probes were radiolabeled with 32P-nucleotides by either random priming or by directed priming with internal oligonucleotides (21). Autoradiography was performed using Hyperfilm-MP (Amersham).
RESULTS
Screen for Arabidopsis cDNAs Encoding a C-Repeat/DRE Binding Domain.
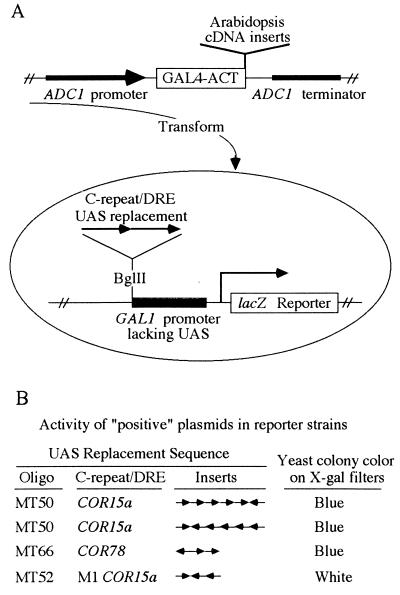
The “one-hybrid” strategy (15) was used to screen for Arabidopsis cDNAs encoding a C-repeat/DRE binding domain. In particular, yeast strains were constructed that contained a lacZ reporter gene with either wild-type or mutant verions of C-repeat/DRE sequences from the promoters of COR15a or COR78 (Table 1) in place of the normal upstream activator sequence of the GAL1 promoter (Fig. 1A). Yeast strains carrying these reporter constructs produced low levels of β-galactosidase and formed white colonies on filters containing X-Gal. The reporter strains carrying the wild-type C-repeat/DRE sequences were transformed with a cDNA expression library that contained random Arabidopsis cDNA inserts fused to the acidic activator domain of the yeast GAL4 transcription factor GAL4-ACT (Fig. 1A). The notion was that some recombinant plasmids in the expression library might contain a cDNA insert encoding a C-repeat/DRE binding domain fused to GAL4-ACT and that such a hybrid protein might bind upstream of the lacZ reporter genes carrying the C-repeat/DRE sequence, activate transcription of the lacZ gene, and result in yeast forming blue colonies on X-Gal-treated filters.
Figure 1.
Approach used to isolate cDNA inserts encoding a C-repeat/DRE binding domain. (A) Screening strategy (see text for details). (B) Activity of the “positive” plasmids in yeast reporter strains carrying the indicated upstream activator sequences. The oligonucleotides (oligo) used to make the upstream activator sequence elements (Table 1), the number of inserts, and their direction of insertion are indicated.
Upon screening about 2 × 106 yeast transformants, three positive plasmids were isolated that caused yeast strains carrying lacZ reporters fused to wild-type C-repeat/DRE inserts to form blue colonies on X-Gal-treated filters (Fig. 1B). The three plasmids did not cause a yeast strain carrying a mutant C-repeat/DRE fused to lacZ to turn blue (Fig. 1B). Thus, activation of the reporter genes by the plasmids appeared to depend on the C-repeat/DRE sequence. Restriction enzyme analysis and DNA sequencing indicated that the three plasmids had an identical 1.8-kb cDNA insert (Fig. 2A). One of the plasmids, designated pACT-11, was chosen for further study.
Figure 2.
Analysis of the pACT-11 cDNA clone. (A) Schematic drawing of the pACT-11 cDNA insert. (B) DNA and amino acid sequence of the 24-kDa polypeptide. The AP2 domain is indicated by double underlines. The basic amino acids that potentially act as a nuclear localization signal are indicated with asterisks. The BclI site immediately upstream of the 24-kDa polypeptide used in subcloning the 24-kDa polypeptide and the EcoRV site used in subcloning the 3′ end of CBF1 are indicated by single underlines. (C) Schematic drawing showing locations of the potential nuclear localization signal (NLS), the AP2 domain, and the acidic region of the 24-kDa polypeptide. Numbers indicate amino acid residues. (D) Comparison of the AP2 domains from the 24-kDa polypeptide and the tobacco DNA binding protein EREBP2 (25). Identical amino acids are indicated with single vertical lines; similar amino acids are indicated by colons; amino acids that are invariant in AP2 domains (26) are indicated with asterisks; and the histidine residue present in CBF1 and TINY (27) that is a tyrosine residue in all other described AP2 domains is indicated with a caret. A single amino acid gap in the CBF1 sequence is indicated by a dot.
pACT-11 Encodes a 24-kDa Polypeptide with an AP2 Domain.
Our expectation was that the cDNA insert in pACT-11 would have a C-repeat/DRE binding domain fused to the yeast GAL4-ACT sequence. However, DNA sequence analysis indicated that an ORF of only 9 aa had been added to the C terminus of GAL4-ACT. It seemed highly unlikely that this short amino acid sequence encoded a DNA binding domain. Also surprising was the fact that about half of the cDNA insert in pACT-11 corresponded to 25S rRNA sequences (Fig. 2A). Further analysis, however, indicated that the insert had an ORF, in opposite orientation to the GAL4-ACT sequence, deduced to encode a 24-kDa polypeptide (Fig. 2 A–C; presumably, the sequences for the 24-kDa polypeptide and the 25S RNA were ligated together during the cloning process). The polypeptide had a basic region that could potentially serve as a nuclear localization signal (28) and an acidic C-terminal half (pI of 3.6) that might act as an acidic transcription activator domain (29). Searches of the GenBank nonredundant nucleotide, peptide, and EST databases (Aug. 25, 1995, and Mar. 24, 1996) indicated that there was no previously described homolog of the 24-kDa polypeptide. However, the polypeptide did have an AP2 domain (ref. 30; Fig. 2 B and D), a DNA binding motif of about 60 aa present in numerous plant proteins, including the APETALA2 (31) protein of Arabidopsis and the EREBPs (ethylene response element binding proteins) of tobacco (25).
The 24-kDa Polypeptide Binds to the C-Repeat/DRE and Activates Transcription in Yeast.
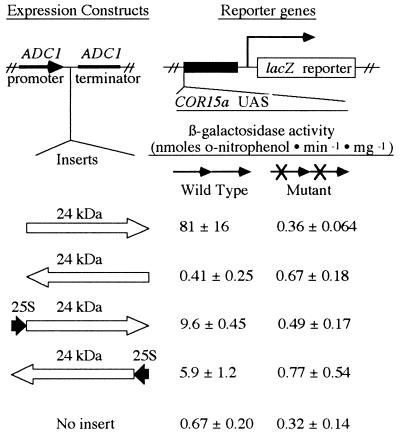
We hypothesized that the 24-kDa polypeptide was responsible for activating the lacZ reporter genes in yeast. To test this, we inserted the BclI–BglII fragment of pACT-11 containing the 24-kDa polypeptide (Fig. 2), and the BglII–BglII fragment containing the 24-kDa polypeptide plus a small portion of the 25S rRNA sequence (Fig. 2), into the yeast expression vector pDB20.1. Plasmids containing either insert in the same 5′-to-3′ orientation as the ADC1 promoter stimulated synthesis of β-galactosidase when transformed into yeast strains carrying the lacZ reporter gene fused to a wild-type COR15a C-repeat/DRE (Fig. 3). In contrast, the plasmids did not stimulate synthesis of β-galactosidase when transformed into yeast strains carrying the lacZ reporter gene fused to a mutated COR15a C-repeat/DRE (Fig. 3). These data indicated that the 24-kDa polypeptide could bind to the wild-type C-repeat/DRE and activate expression of the lacZ reporter gene in yeast. Additional experiments indicated that the 24-kDa polypeptide could also activate expression of the lacZ reporter gene when it was fused to a wild-type COR78 C-repeat/DRE (dimer of MT66) or a wild-type COR15b C-repeat/DRE (dimer of MT68; data not shown); COR15b is a homolog of COR15a (14). A plasmid containing the BclI–BglII fragment (which encodes only the 24-kDa polypeptide) cloned in opposite orientation to the ADC1 promoter did not stimulate synthesis of β-galactosidase in reporter strains carrying either the wild-type or mutant C-repeat/DRE sequence fused to lacZ (Fig. 3). However, a plasmid carrying the BglII–BglII fragment (containing the 24-kDa polypeptide plus some 25S rRNA sequences) cloned in opposite orientation to the ADC1 promoter did produce significant levels of β-galactosidase in reporter strains carrying the wild-type COR15a C-repeat/DRE (Fig. 3). Thus, sequences located upstream of the 24-kDa polypeptide were able to serve as a cryptic promoter in yeast, a result that offered an explanation for the expression of the 24-kDa polypeptide in the original pACT-11 plasmid.
Figure 3.
Activation of reporter genes in yeast by the 24-kDa polypeptide. “Expression constructs” were transformed into yeast strains carrying the lacZ reporter gene fused to direct repeat dimers of either the wild-type COR15a C-repeat/DRE (oligonucleotide MT50) or the mutant M3COR15a C-repeat/DRE (oligonucleotide MT125). β-Galactosidase activity (nanomoles of o-nitrophenol produced per minute × milligrams of protein) ± SD was determined from cultures grown in triplicate.
Gel Shift Analysis Indicates that the 24-kDa Polypeptide Binds to the C-Repeat/DRE.
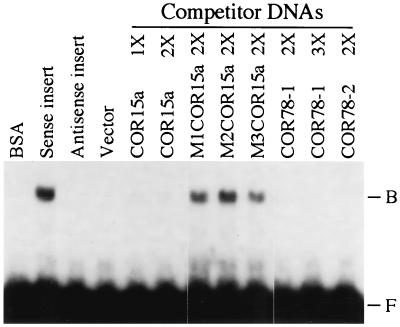
Gel shift experiments were conducted to demonstrate further that the 24-kDa polypeptide bound to the C-repeat/DRE. Specifically, the ORF for the 24-kDa polypeptide was inserted into the pET-28a(+) bacterial expression vector (see Materials and Methods), and the resulting fusion protein was expressed at high levels in E. coli (data not shown). Protein extracts prepared from E. coli expressing the recombinant protein produced a gel shift when a wild-type COR15a C-repeat/DRE was used as probe (Fig. 4). No shift was detected with BSA or E. coli extracts prepared from strains harboring the vector alone or the vector with an antisense insert for the 24-kDa polypeptide. Oligonucleotides encoding wild-type C-repeat/DRE sequences from COR15a or COR78 competed effectively for binding to the COR15a C-repeat/DRE probe, but mutated versions of the COR15a C-repeat/DRE did not (Fig. 4). These in vitro results corroborated the in vivo yeast expression studies, indicating that the 24-kDa polypeptide binds to the C-repeat/DRE sequence. The 24-kDa polypeptide was thus designated CBF1, and the gene encoding it was named CBF1.
Figure 4.
Gel shift assays demonstrating that CBF1 binds to the C-repeat/DRE. The C-repeat/DRE probe (1 ng) used in all reactions was a 32P-labeled dimer of the oligonucleotide MT50, a wild-type C-repeat/DRE from COR15a (Table 1). Shown in the first four lanes are gel shift assays using either BSA or protein extracts prepared from E. coli harboring either the pET-28a(+) expression vector alone (vector) or the vector plus an insert encoding the 24-kDa polypeptide in sense (sense insert) or antisense (antisense insert) orientation. Shown in the next eight lanes are the E. coli protein extract prepared from the “sense insert” strain plus the indicated competitor C-repeat/DRE sequences (100 ng). The 1×, 2×, and 3× refer to whether the oligonucleotides were monomers, dimers, or trimers, respectively, of the indicated C-repeat/DRE sequences.
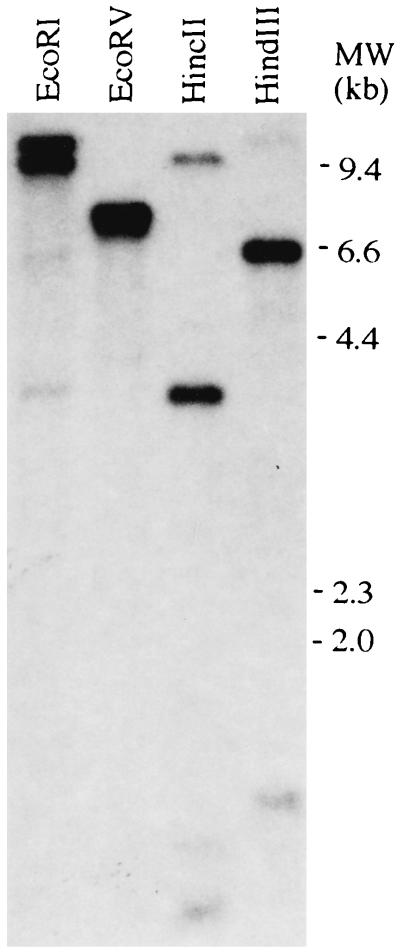
CBF1 Is a Unique or Low Copy Number Gene.
Southern transfers of Arabidopsis DNA that had been digested with various restriction endonucleases were prepared and hybridized at either high (Fig. 5) or low (data not shown) stringency using the entire CBF1 coding sequence as probe. A small number of bands were observed for each DNA digest, indicating that CBF1 is either a unique or low copy number gene (both EcoRV and HincII cut once within the CBF1 cDNA). The same patterns were obtained when the 3′ end of the CBF1 coding sequence (corresponding to the acidic region of CBF1) was used as the hybridization probe (data not shown).
Figure 5.
CBF1 is a unique or low copy number gene. Arabidopsis DNA (≈1 μg) was digested with the indicated restriction endonucleases, and Southern transfers were prepared and hybridized with a 32P-labeled probe encoding the entire CBF1 polypeptide.
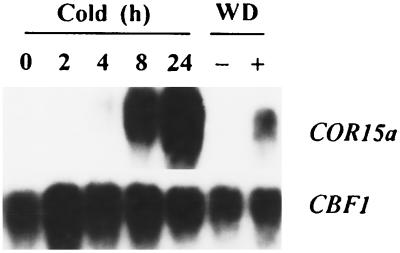
CBF1 Transcript Levels Do Not Change Appreciably in Response to Low Temperature or Water Deficit.
The transcript levels of CBF1 were about the same in control and cold-treated plants and in control and dehydrated leaves (Fig. 6). In contrast, the transcript levels for COR15a increased dramatically in response to both low temperature and water deficit (Fig. 6). Under each environmental condition tested, a single hybridizing band was observed for CBF1 at either high or low stringency with probes for either the entire CBF1 coding sequence or the 3′ acidic region of the protein (data not shown). The size of the CBF1 transcripts was about 1.0 kb.
Figure 6.
Effects of low temperature and water deficit on CBF1 and COR15a transcript levels. Northern transfers were prepared from total RNA (20 μg) isolated from Arabidopsis plants treated at low temperature for the indicated time or detached leaves that either were (+) or were not (−) subjected to a water deficit (WD) treatment. The transfers were hybridized with 32P-labeled probes for either the entire CBF1 coding sequence or the COR15a gene. After autoradiography, the transfers were hybridized a second time with the constitutively expressed eIF-4A (eukaryotic Initiation Factor 4A) gene (32) to confirm that RNA loading and transfer were uniform between the samples (data not shown). The autoradiograms for the COR15a and CBF1 probes were exposed for 1 h and 5 days, respectively. Longer exposures for the COR15a probe indicated that COR15a transcript levels begin to increase by 2 h.
DISCUSSION
The results presented indicate that CBF1 binds to the C-repeat/DRE DNA regulatory element both in vitro (gel shift assays) and in vivo (yeast expression assays) and can function as a transcriptional activator. Thus, it is probable that CBF1 binds to C-repeat/DRE elements in Arabidopsis plants and has a role in regulating transcription in response to low temperature and water deficit. Having available a cDNA clone for CBF1 makes possible a number of strategies to directly test this hypothesis, such as using “antisense” technology to decrease the levels of CBF1 in transgenic plants and determining what effects this has on cold- and dehydration-regulated gene expression. It will also be extremely interesting to determine whether altering the level of CBF1 affects the freezing and drought tolerance of Arabidopsis plants.
CBF1 is a unique or low copy number gene in Arabidopsis. However, the CBF1 protein contains a 60-aa motif, the AP2 domain, that is evolutionarily conserved in plants (30). It is present, for instance, in the APETALA2 (31), AINTEGUMENTA (26, 33), and TINY (27) proteins of Arabidopsis; the EREBPs of tobacco (25); and numerous proteins of unknown function in B. napus, Ricinus communis, and rice (indicated by cDNA sequences deposited in the GenBank expressed sequence tagged cDNA database). The results of Ohme-Takagi and Shinshi (25) indicate that the function of the AP2 domain is DNA binding; this region of the putative tobacco transcription factor EREBP2 is responsible for its binding to the cis-acting ethylene response DNA element referred to as the GCC-repeat. As discussed by Ohme-Takagi and Shinshi (25), the DNA-binding domain of EREBP2 (the AP2 domain) contains no significant amino acid sequence similarities or obvious structural similarities with other known transcription factors or DNA binding motifs. Thus, the domain appears to be a novel DNA-binding motif that, to date, has only been found in plant proteins.
Presumably, the binding of CBF1 to the C-repeat/DRE involves the AP2 domain. In this regard, it is germane to note that the tobacco ethylene response element, AGCCGCC, closely resembles the C-repeat/DRE sequences present in the promoters of the Arabidopsis genes COR15a, GGCCGAC, and COR78/RD29A, TACCGAC. An intriguing possibility thus raised is that CBF1, the EREBPs, and perhaps other AP2 domain proteins are members of a superfamily of DNA binding proteins that recognize a family of cis-acting regulatory elements having, potentially, CCG as a common core sequence. Differences in the sequence surrounding the CCG core element might result in recruitment of distinct AP2 domain proteins that are integrated into signal transduction pathways activated by different environmental, hormonal, and developmental cues.
The yeast transformation experiments indicate that CBF1 has a domain that can serve as a transcriptional activator. The most likely candidate for this domain is the acidic C-terminal half of the polypeptide. Indeed, random acidic amino acid peptides from E. coli have been shown to substitute for the GAL4 acidic activator domain of GAL4 in yeast (34). Moreover, acidic activator domains have been found to function across phylogenic kingdoms (29). The yeast GAL4 acidic activator, for instance, can activate transcription in tobacco (35). It has also been shown that certain plant transcription factors, such as VP1 (36), have acidic domains that function as transcriptional activators in plants. Significantly, the acidic activation domains of the VP16 and GCN4 transcription factors require the “adaptor” proteins ADA2, ADA3, and GCN5 for full activity in yeast (37). Intriguingly, genetic studies indicate that CBF1 also requires ADA2, ADA3, and GCN5 to function optimally in yeast (E.J.S., Steven J. Triezenberg, and M.F.T., unpublished data). The fundamental question thus raised is whether plants have homologs of ADA2, ADA3, and GCN5 and whether these adaptors are required for CBF1 function (and function of other transcription factors with acidic activator regions) in Arabidopsis.
A final point regards the possible regulation of CBF1 activity. In contrast to what occurs in yeast, transcription of CBF1 in Arabidopsis under normal growth conditions does not cause appreciable activation of promoters containing C-repeat/DRE sequences (i.e., COR genes are expressed at very low levels in nonstressed plants). Thus, assuming that CBF1 can bind to C-repeat/DRE sequences in Arabidopsis plants, as it does both in vitro and in yeast, this finding would seem to indicate that CBF1 is “activated” by low temperature and water deficit in Arabidopsis. A number of potential mechanisms could account for CBF1 activation, such as its becoming modified, perhaps by phosphorylation, in response to low temperature or water deficit. Such modification could result in either stabilization of the protein, translocation of the protein from the cytoplasm to the nucleus, or activation of either the DNA binding domain or the activation domain of the protein. Again, having available the CBF1 cDNA makes possible a number of strategies to test these and alternative hypotheses.
Acknowledgments
We thank Joachim Li, Stan Fields, and Steve Triezenberg for providing the yeast strains and yeast cloning vectors used in this study; Pam Green for the eIF-4A gene probe; and Steve Triezenberg, Kevin O’Connell, and an anonymous reviewer for suggestions on how to improve the manuscript. This research was supported by grants to M.F.T. from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (92–37100-7351), the Consortium for Plant Biotechnology Research (U.S. Department of Agriculture Grant 92–34190-6941, Subgrant 5930130–11), and the Michigan Agricultural Experiment Station.
Footnotes
References
- 1.Steponkus P L, Uemura M, Webb M S. In: Advances in Low Temperature Biology. Steponkus P L, editor. Vol. 2. London: JAI; 1993. pp. 211–312. [Google Scholar]
- 2.Guy C, Haskell D, Neven L, Klein P, Smelser C. Planta. 1992;188:265–270. doi: 10.1007/BF00216823. [DOI] [PubMed] [Google Scholar]
- 3.Siminovitch D, Cloutier Y. Cryobiology. 1983;20:487–503. doi: 10.1016/0011-2240(83)90037-8. [DOI] [PubMed] [Google Scholar]
- 4.Thomashow M F. In: Plant Responses to Cellular Dehydration During Environmental Stress. Close T L, Bray E A, editors. Rockville, MD: Am. Soc. Plant Physiol.; 1993. pp. 137–143. [Google Scholar]
- 5.Artus N N, Uemura M, Steponkus P L, Gilmour S J, Lin C, Thomashow M F. Proc Natl Acad Sci USA. 1996;93:13404–13409. doi: 10.1073/pnas.93.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaguchi-Shinozaki K, Shinozaki K. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker S S, Wilhelm K S, Thomashow M F. Plant Mol Biol. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- 8.Jiang C, Iu B, Singh J. Plant Mol Biol. 1996;30:679–684. doi: 10.1007/BF00049344. [DOI] [PubMed] [Google Scholar]
- 9.Welin B V, Olson Å, Nylander M, Palva E T. Plant Mol Biol. 1994;26:131–144. doi: 10.1007/BF00039526. [DOI] [PubMed] [Google Scholar]
- 10.Bray E A. Plant Physiol. 1993;103:1035–1040. doi: 10.1104/pp.103.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordin K, Heino P, Palva E T. Plant Mol Biol. 1991;16:1061–1071. doi: 10.1007/BF00016077. [DOI] [PubMed] [Google Scholar]
- 12.Gilmour S J, Thomashow M F. Plant Mol Biol. 1991;17:1233–1240. doi: 10.1007/BF00028738. [DOI] [PubMed] [Google Scholar]
- 13.Gilmour S J, Hajela R K, Thomashow M F. Plant Physiol. 1988;87:745–750. doi: 10.1104/pp.87.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilhelm K S, Thomashow M F. Plant Mol Biol. 1993;23:1073–1077. doi: 10.1007/BF00021822. [DOI] [PubMed] [Google Scholar]
- 15.Li J J, Herskowitz I. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 16.Strathern J N, Higgens D R. Methods Enzymol. 1991;194:319–329. doi: 10.1016/0076-6879(91)94024-7. [DOI] [PubMed] [Google Scholar]
- 17.Becker D M, Guarente L. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- 18.Schiestl R H, Gietz R D. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 19.Rose M, Botstein D. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- 20.Berger S L, Piña B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Hajela R K, Horvath D P, Gilmour S J, Thomashow M F. Plant Physiol. 1990;93:1246–1252. doi: 10.1104/pp.93.3.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stockinger E J, Mulinix C A, Long C M, Brettin T S, Iezzoni A F. J Hered. 1996;87:214–218. doi: 10.1093/oxfordjournals.jhered.a022987. [DOI] [PubMed] [Google Scholar]
- 24.Walling L L, Chang Y C, Demmin D S, Holzer F M. Nucleic Acids Res. 1988;16:10477–10492. doi: 10.1093/nar/16.22.10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohme-Takagi M, Shinshi H. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klucher K M, Chow H, Reiser L, Fischer R L. Plant Cell. 1996;8:137–153. doi: 10.1105/tpc.8.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson K, Long D, Swinburne J, Coupland G. Plant Cell. 1996;8:659–671. doi: 10.1105/tpc.8.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raikhel N. Plant Physiol. 1992;100:1627–1632. doi: 10.1104/pp.100.4.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahn S. Cell. 1993;72:481–483. doi: 10.1016/0092-8674(93)90064-w. [DOI] [PubMed] [Google Scholar]
- 30.Weigel D. Plant Cell. 1995;7:388–389. doi: 10.1105/tpc.7.4.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jofuku K D, den Boer B G W, Van Montagu M, Okamuro J K. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metz A M, Timmer R T, Browning K S. Gene. 1992;120:313–314. doi: 10.1016/0378-1119(92)90112-3. [DOI] [PubMed] [Google Scholar]
- 33.Elliot R C, Betzner A S, Huttner E, Oakes M P, Tucker W Q J, Gerentes D, Perez P, Smyth D R. Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma J, Ptashne M. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 35.Ma J, Przibilla E, Hu J, Bogorad L, Ptashne M. Nature (London) 1988;334:631–633. doi: 10.1038/334631a0. [DOI] [PubMed] [Google Scholar]
- 36.McCarty D R, Hattori T, Carson C B, Vasil V, Lazar M, Vasil I K. Cell. 1991;66:895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- 37.Guarente L. Trends Biochem Sci. 1995;20:517–521. doi: 10.1016/s0968-0004(00)89120-3. [DOI] [PubMed] [Google Scholar]