Abstract
In animal models, STAT3 action in the hypothalamus and liver appears essential for normal body weight and glucose homeostasis in response to insulin. We hypothesised that variation in the STAT3 gene may be associated with body fat and/or insulin resistance in the general population. Five tagging SNPs spanning the STAT3 gene, rs8074524, rs2293152, rs2306580, rs6503695 and rs7211777 were genotyped in 2776 Caucasian female twins (mean age 47.4±12.5 years) from the St Thomas' UK Adult Twin Registry (Twins UK). Minor allele frequencies were as follows: rs8074524 (0.19), rs2293152 (0.37), rs2306580 (0.06), rs6503695 (0.35) and rs7211777 (0.34). The minor allele of rs2293152 was associated with higher HOMA index of insulin resistance (P=0.013) in the full cohort and confirmed in sib-TDT: (P= 0.015; n=60). However, there were no associations with fasting serum insulin or glucose or with obesity variables. Although defective STAT3 action results in obesity and insulin resistance in animal models, we failed to establish any indicative associations with common SNPs in this human study.
Keywords: fat distribution, insulin resistance, genetic susceptibility, signal transduction
Signal transducers and activators of transcription (STAT) are a family of latent cytoplasmic transcription factors that are produced in many cell types. They share a DNA-binding domain, a Src homology 2 (SH2) domain and a putative Src homology 3 (SH3) domain and are activated by tyrosine phosphorylation and dimerization in response to a wide variety of extracellular ligands, such as cytokines and growth factors [1]. It is well known that STAT3 is involved in the regulation of energy homeostasis through the adipose tissue cytokine leptin [2]. Mice with a neural-specific disruption of the Stat3 gene are hyperphagic, obese, diabetic, infertile and hyperleptinemic, suggesting a leptin-resistant condition [3]. Other pathways which converge and function through STAT3 may have similar effects. Mice lacking Stat3 specifically in the liver exhibit insulin resistance and glucose intolerance when fed a high-fat diet and restoration of hepatic STAT3 expression corrects the abnormalities [4]. It has recently been shown that intracerebroventricular insulin infusion stimulates tyrosine phosphorylation of STAT3 in the liver and that inhibition of hepatic glucose production and expression of gluconeogenic genes induced by insulin is impaired in mice with liver-specific STAT3 deficiency [5].
STAT3 action in the mouse hypothalamus and liver therefore appears essential for normal body weight and glucose homeostasis in response to the actions of both leptin and insulin. We hypothesized that variants in the STAT3 gene may influence body fat mass and insulin sensitivity in humans. In this first gene-wide association study of STAT3 variation in relation to body fat and insulin action, we have used tagging SNPs (tSNPs) to test associations with phenotypes in a large sample of Caucasian female twins (n=2771, mean age 47.4±12.5 years). tSNPs effectively capture information of most common variants by taking into account patterns of linkage disequilibrium (LD) across the gene [6].
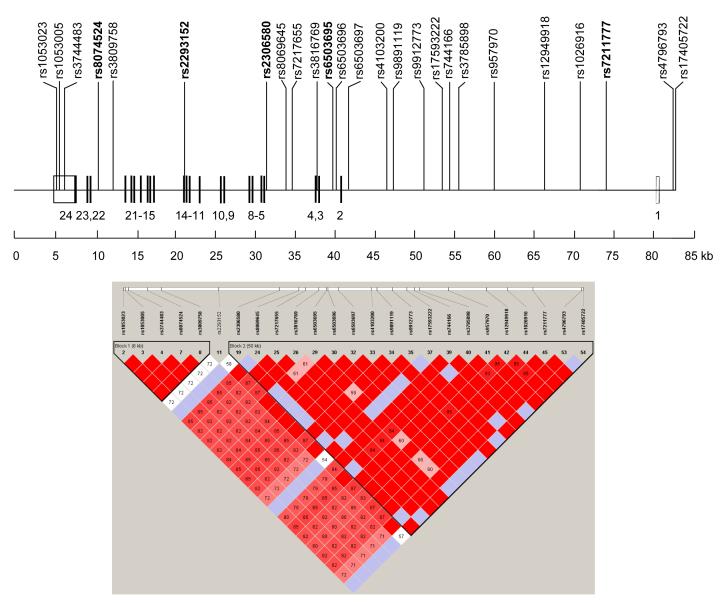
Characteristics of subjects in the Twins UK study sample are shown in Table 1. The 25 common (MAF>0.05) SNPs in the selected 83170 bp region including the 75171 bp STAT3 gene are situated in two blocks of strong LD (r2>0.8). SNP rs2292152 sited between the blocks is not in LD with any other common SNP in the gene (Fig. 1). Five tSNPs were selected: rs8074524, rs2293152, rs2306580, rs6503695 and rs7211777, all of which are in non-coding regions. Table 2 shows the genotype and allele frequencies of the tSNPs in the Twins UK cohort, based on one MZ and both DZ twins genotyped for each pair.
Table 1.
General characteristics of subjects
| Variable | n | Mean ± SD |
|---|---|---|
| Age, years | 2776* | 47.4±12.5 |
| Postmenopausal, % | 2453 | 47.7 |
| Obesity-related variables: | ||
| Leptin, ng/ml | 2776 | 16.5±12.0 |
| BMI, kg/m2 | 2759 | 24.7±4.4 |
| Weight, kg | 2760 | 65.3±11.8 |
| Waist, cm | 2708 | 78.3±10.2 |
| Total fat, kg | 2719 | 23.4±8.8 |
| Total fat, % | 2678 | 35.6±8.0 |
| Central fat, kg | 2696 | 1.33±0.73 |
| Central fat, % | 2696 | 31.2±11.5 |
| Insulin sensitivity:† | ||
| Fasting glucose, mmol/L | 2317 | 4.49±0.55 |
| Fasting insulin, μU/mL | 1984 | 6.96±5.96 |
| HOMA | 1791 | 1.37±1.31 |
| 2h-glucose, mmol/L | 738 | 5.18±1.10 |
| 2h-insulin, μU/mL | 738 | 34.2±25.4 |
| SiM | 738 | 88.5±68.8 |
Number of subjects (846 MZ, 1930 DZ) with leptin data and genotype data on at least 1 SNP
Non-fasting subjects, patients with either type 1 or type 2 diabetes and patients on any anti-diabetic drugs were all excluded.
Type 2 diabetes: Diagnosed patients or subjects with fasting glucose>7.8mmol/L or 2h-glucose>11.1mmol/L.
Fig. 1.
Linkage disequilibrium plot of STAT3 region HapMap SNPs
On gene map, numbers refer to exons, with coding exons as solid boxes and untranslated exons as open boxes. tSNPs are shown in bold type. Note reverse strand orientation. Genotype data for SNPs of MAF>0.05 in the region Chr17:40835507 – 40918677 in 90 CEU subjects were downloaded from http://www.hapmap.org (Phase II HapMap Release January 2006). The pairwise LD plot (r2) was obtained using the integral Tagger program. SNP rs2293152 lies between two blocks and does not tag any other common SNPs in this gene.
Table 2.
STAT3 SNP genotype and allele distributions
| HapMap | rs# | Genotype | Total* | Minor allele freq. and 95% CI | H-W test† | MAF in | |||
|---|---|---|---|---|---|---|---|---|---|
| # | 11 | 12 | 22 | χ 2 | P | HapMap | |||
| 7 | rs8074524 | 1377 | 650 | 81 | 2108 | 19.3 (18.1-20.5) | 0.15 | NS | 15.8 |
| 11 | rs2293152 | 797 | 995 | 267 | 2059 | 37.1 (35.7-38.6) | 1.21 | NS | 40.4 |
| 19 | rs2306580 | 1939 | 260 | 9 | 2208 | 6.3 (5.6-7.1) | 0.21 | NS | 11.2 |
| 29 | rs6503695 | 915 | 1050 | 240 | 2205 | 34.7 (33.3-36.1) | 0.78 | NS | 34.8 |
| 45 | rs7211777 | 979 | 1010 | 249 | 2238 | 33.7 (32.3-35.1) | 1.82 | NS | 35.8 |
Includes one of each MZ pair and both DZ pairs
Tested in one of each MZ pair, one of each DZ pair and all singleton DZ twins.
We found no associations between any of the five tSNPs and general or central obesity scores, so individual variables were not tested further. The only association for a 2 df overall genotypic test (i.e. a codominant model) showing borderline nominal significance (P=0.052) was between the intronic SNP rs2293152 and the HOMA index of insulin resistance. We performed follow-up analyses to determine the best model for all insulin resistance related phenotypes and found a recessive association between rs2293152 and HOMA in subjects with available data (n=1791) (Table 3). Subjects homozygous for the minor allele had significantly higher HOMA (P=0.013), in comparison to subjects homozygous or heterozygous for the major allele, explaining 0.34% of variance. Association with HOMA was confirmed in sib-TDT (P= 0.015; n=60), which is based on dizygous twins drawn from the full cohort who are discordant for rs2293152 genotype, suggesting that population stratification did not account for the association in the full cohort. However, we found no significant association between rs2293152 and fasting insulin and glucose levels used in calculation of the HOMA score. No tSNPs were associated with insulin sensitivity measure SiM based on 738 subjects. Haplotype analyses based on rs7211777 and rs6503695 were also tested, however, no significant effects were observed for any phenotype.
Table 3.
Nominal significant associations of STAT3 rs2293152 with insulin sensitivity phenotypes
| GEE |
sTDT |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Number (11+12/22) | Mean (SD) |
P | Explained variance | No. of pairs | Mean (SD) |
P | |||
| 11+12 | 22 | 11+12 | 22 | ||||||
| HOMA | 1361/193 | 1.35 (1.29) | 1.57 (1.63) | 0.013 | 0.34% | 60 | 1.13 (0.67) | 1.36 (0.96) | 0.015 |
| Fasting glucose, mmol/L | 1762/254 | 4.48 (0.55) | 4.55 (0.59) | NS | --- | 79 | 4.43 (0.42) | 4.52 (0.54) | NS |
| Fasting insulin, μU/mL | 1508/210 | 6.91 (5.89) | 7.57 (7.42) | NS | --- | 69 | 6.26 (4.30) | 7.13 (5.25) | 0.048 |
| SiM | 554/76 | 88.2 (69.0) | 92.0 (71.9) | NS | --- | 22 | 100.9 (46.7) | 116.9 (96.0) | NS |
The findings are generalisable, as few differences were found between Twins UK subjects and singletons in the UK female population, other than that MZ twins had a slightly lower weight and a smaller variance for weight than DZ twins and singletons [7]. The main strengths of our study lie in the large number of subjects with measures of regional fat distribution and insulin-resistance and comprehensive coverage of variation in this 75 kb gene using tagging SNPs. The test of association with obesity variables involving 1930 DZ (865 pairs) and 846 MZ (413 pairs) provided power in excess of 80% (and α = 0.05) to detect a locus effect of 0.5%. Although there were fewer subjects than this available with insulin resistance measures, the current study had 80% (α = 0.05) power to detect a locus effect explaining 0.65% of the variance in HOMA index (n=1791) and 1.5% of the variance in SiM (n=738). The apparent association of rs2293152 with HOMA in the full cohort accounted for 0.34% of variance, however, the sample of 738 was underpowered to detect an association of comparable strength with SiM. The rs2293152 genotyping success rate was the lowest at 87%, but the association with HOMA cannot be dismissed as spurious on the grounds of sample size, as 2059 genotypes still represents a powerful study. Also, the effect of reduced sample size on power was in some way offset by the relatively high minor allele frequency (0.37).
Inoue et al. originally showed that Stat3 −/− mice develop insulin resistance associated with increased glucose production [4] and more recently that insulin acting in the hypothalamus down-regulates gluconeogenic gene expression in the liver by stimulating IL-6 signalling via STAT3 [5]. Gogorawa et al. have found insulin secretion to be impaired in pancreatic beta cell-specific Stat3 −/− mice [8]. We found a significant increase in HOMA in STAT3 rs2293152 rare homozygotes compared to the rest. However, fasting glucose concentration, which reflects basal hepatic glucose production and fasting insulin, which reflects beta cell competence, showed no significant changes. It therefore seems that the STAT3 variant has minimal effect on gluconeogenesis or insulin secretion and the modest association with HOMA may not be genuine. We would not expect profound effects on phenotype matching those of animal knockout models as a result of SNP variation potentially resulting in small changes in the level of STAT3 gene expression or minor changes in protein structure associated with carriage of a particular allele. However, the expectation would be for a trend toward a comparable effect, which might only become significant in a much larger study sample.
The absence of any association with obesity variables is interesting in view of a recent discovery that mice deficient in interleukin 18, a cytokine that signals via STAT3, are hyperphagic, obese and insulin resistant [9]. Hepatic insulin resistance in the Il18−/− mice involved an enhanced expression of genes associated with gluconeogenesis in the liver, which resulted from defective phosphorylation of STAT3. Resistance seemed to be secondary to increased food intake and obesity (specifically accumulation of body fat), because the effect only emerged after 6 months.
In tagging only common SNPs, we excluded the possibility of discovering any substantial effect associated with low frequency SNPs (MAFs<0.05), which could be functional. However, evidence from animal models suggests that defective STAT3 activity remains a strong candidate in the development of obesity, with insulin resistance emerging as a secondary effect.
Methods
Study design
The Twins UK Registry comprises unselected, Caucasian mostly female volunteers ascertained from the general population through national media campaigns in the UK [10]. The study sample comprised 2776 subjects (423 MZ pairs, 940 DZ pairs, and 50 singletons) with available leptin data. No difference was observed in the distributions of age and menopausal status between subjects with and without leptin data (n=362). The number of individuals in the study cohort with data on other phenotypic variables is shown in Table 1. Means and ranges of quantitative phenotypes in Twins UK are similar to an age-matched sample of the UK female population [7]. Informed consent was obtained from all participants before they entered the studies, which were approved by the local research ethics committee.
Zygosity, body composition and biochemical analyses
Zygosity was determined by standardised questionnaire and confirmed by DNA fingerprinting. Height was measured to the nearest 0.5 cm using a wall-mounted stadiometer. Weight (light clothing only) was measured to the nearest 0.1 kg using digital scales. BMI was used as a measure of general adiposity and calculated as weight divided by height squared (kg/m2). Waist circumference (cm) was measured at the level midway between the lower rib margin and the iliac crest. Body composition was measured by dual emission X-ray absorptiometry (Hologic QDR-2000, Vertec, Waltham, MA, USA). Serum leptin concentration was determined after an overnight fast using a radioimmunoassay (Linco Research, St Louis, MO, USA). Fasting insulin was measured by immunoassay (Abbott Laboratories Ltd., Maidenhead, UK) and glucose was measured on an Ektachem 700 multichannel analyser using an enzymatic colorimetric slide assay (Johnson and Johnson Clinical Diagnostic Systems, Amersham, UK). A random sub-sample of 738 subjects underwent an oral glucose tolerance test (OGTT) for which glucose and insulin levels were measured before and 2 h after a 75-g oral glucose load.
Selection of tSNPs
The STAT3 gene spans over 75 kb and contains 24 exons. 25 polymorphic SNPs with minor allele frequency (MAF) >0.05 in an 83170 bp region including the STAT3 gene are listed on the HapMap database (HapMap Data Release #20/Phase II Jan 2006; http://www.hapmap.org) (ESM Table I). Genotypes of 90 CEU parent-offspring trio subjects were downloaded from HapMap into Haploview (http://www.broad.mit.edu/mpg/haploview/). The integral Tagger program (http://www.broad.mit.edu/tagger) was then used to select five tSNPs which capture common variants (MAF>5%) at a minimum r2 of 0.8, using the aggressive multi-marker tagging mode. This provides not only individual tSNPs, but also multi-marker combinations (haplotypes composed of 2 or 3 markers) which capture additional variants. As shown in ESM Table II, 9 of the 25 common variants are tagged by the haplotypes constructed by rs7211777 and rs6503695.
Genotyping in cohorts
The tSNPs were genotyped by Pyrosequencing, (Biotage, Uppsala, Sweden). Genotyping accuracy as assessed by inclusion of duplicates (50 pairs of monozygous (MZ) twins) in the arrays was approx. 98% and negative controls (water blanks) were included on each plate. Genotyping success rate for each tSNP varied between 87.0% and 94.8%. Primers and PCR conditions for tSNP genotyping in the full cohort are given in ESM Table II.
Statistical analyses
Hardy-Weinberg equilibrium for genotype was tested by a χ2 test with 1 df in one twin of each pair chosen at random to prevent inflated significance. Phenotypes significantly (P<0.05) deviating from normal were log transformed to obtain normal distributions prior to analysis. Factor analysis was used to combine strongly correlated indices of obesity into two measures: one for general obesity (serum leptin, BMI, weight, total fat mass and % total fat) and one for central obesity (waist circumference, central fat mass and % central fat). Insulin-resistance measures HOMA and SiM described previously [11] were available for subsets of subjects with leptin data. Preliminary association analyses were performed using STATA 8 (StataCorp, College Station, Texas). To reduce the likelihood of generating false positive associations through multiple testing, single variables characterizing obesity were analysed only if initial tests with the general and central obesity scores yielded a positive association for at least one of these combined variables. This strategy was also used for the two indices of insulin sensitivity. Association analyses were performed using Generalized Estimating Equations (GEE) [12], which allows for the relatedness between twins and yields unbiased standard errors and P-values. To limit the number of models tested for individual SNP association analyses, we first performed a 2-df overall test of genotypic association. Only in the presence of a significant association were additive, dominant and recessive models (all 1-df) further tested to find the best mode of inheritance. To control for population stratification, dizygous (DZ) twin pairs discordant for genotype were also used in sib-TDT association analysis to confirm the results of regular association tests, as described elsewhere [11]. Age and menopausal status were included as covariates in the models. The program developed by Purcell et al. [13] was used to calculate the genetic power of the study. Assuming a sibling correlation of 0.3, a sample of 840 DZ pairs is adequate to detect a locus effect of 0.5% with 80% power (and α = 0.05). The current study of 940 DZ pairs with additionally 423 MZ pairs and 50 singletons provided even greater power.
Supplementary Material
Acknowledgements
This study was funded by the Wellcome Trust, Project grant No. 073142. The Twin Research and Genetic Epidemiology Unit received support from the Wellcome Trust, Arthritis Research Campaign, the Chronic Disease Research Foundation and the European Union 5th Framework Programme Genom EU twin no. QLG2-CT-2002-01254 and EuroClot project LSHM-CT-2004-005268.
References
- 1.Darnell JE. STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Vaisse C, Halaas JL, Horvath CM, Darnell JE, Jr, Stoffel M, Friedman JM. Leptin activation of Stat3 in the hypothalamus of wild-type and ob/ob mice but not db/db mice. Nat Genet. 1996;14:95–97. doi: 10.1038/ng0996-95. [DOI] [PubMed] [Google Scholar]
- 3.Gao Q, Wolfgang MJ, Neschen S, et al. Disruption of neural signal transducer and activator of transcription 3 causes obesity, diabetes, infertility, and thermal dysregulation. Proc Natl Acad Sci USA. 2004;101:4661–4666. doi: 10.1073/pnas.0303992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inoue H, Ogawa W, Ozaki M, et al. Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nat Med. 2004;10:168–174. doi: 10.1038/nm980. [DOI] [PubMed] [Google Scholar]
- 5.Inoue H, Ogawa W, Asakawa A, et al. Role of hepatic STAT3 in brain-insulin action on hepatic glucose production. Cell Metab. 2006;3:267–275. doi: 10.1016/j.cmet.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Chapman JM, Cooper JD, Todd JA, Clayton DG. Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum Hered. 2003;56:18–31. doi: 10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- 7.Andrew T, Hart DJ, Snieder H, de Lange M, Spector TD, MacGregor AJ. Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res. 2001;4:464–477. doi: 10.1375/1369052012803. [DOI] [PubMed] [Google Scholar]
- 8.Gorogawa S, Fujitani Y, Kaneto H, et al. Insulin secretory defects and impaired islet architecture in pancreatic beta-cell-specific STAT3 knockout mice. Biochem Biophys Res Commun. 2004;319:1159–1170. doi: 10.1016/j.bbrc.2004.05.095. [DOI] [PubMed] [Google Scholar]
- 9.Netea MG, Joosten LA, Lewis E, et al. Deficiency of interleukin-18 in mice leads to hyperphagia, obesity and insulin resistance. Nat Med. 2006;12:650–656. doi: 10.1038/nm1415. [DOI] [PubMed] [Google Scholar]
- 10.Spector TD, MacGregor AJ. The St. Thomas' UK Adult Twin Registry. Twin Res. 2002;5:440–443. doi: 10.1375/136905202320906246. [DOI] [PubMed] [Google Scholar]
- 11.Jamshidi Y, Snieder H, Wang X, Spector TD, Carter ND, O'Dell SD. Common polymorphisms in SOCS3 are not associated with body weight, insulin sensitivity or lipid profile in normal female twins. Diabetologia. 2006;49:306–310. doi: 10.1007/s00125-005-0093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trégouët D-A, Ducimetère P, Tiret L. Testing association between candidate-gene markers and phenotype in related individuals, by use of estimating equations. Am J Hum Genet. 1997;61:189–199. doi: 10.1086/513895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.