Abstract
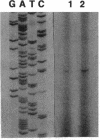
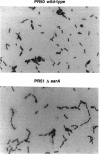
We have utilized transposon mutagenesis to obtain insertional mutations in Providencia stuartii that activate the chromosomal aac(2')-la gene. Two closely linked mini-Tn5Cm insertions were obtained in a locus designated aarA, and a single insertion was obtained in a separate locus, aarC. Nucleotide sequence analysis, complementation studies, and localization of the sites of mini-Tn5Cm insertion have allowed the identification of the aarA coding region. The deduced AarA protein had a molecular mass of 31,086 kDa and displayed characteristics of an integral membrane protein. A strain deleted for the aarA gene by allelic exchange showed at least a fourfold increase in the accumulation of aac(2')-la mRNA and an eightfold increase in aminoglycoside resistance. Mutations in aarA were pleiotrophic and also resulted in loss of pigmentation and a deficiency in cell separation during division.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartowsky E., Normark S. Purification and mutant analysis of Citrobacter freundii AmpR, the regulator for chromosomal AmpC beta-lactamase. Mol Microbiol. 1991 Jul;5(7):1715–1725. doi: 10.1111/j.1365-2958.1991.tb01920.x. [DOI] [PubMed] [Google Scholar]
- Chou J. H., Greenberg J. T., Demple B. Posttranscriptional repression of Escherichia coli OmpF protein in response to redox stress: positive control of the micF antisense RNA by the soxRS locus. J Bacteriol. 1993 Feb;175(4):1026–1031. doi: 10.1128/jb.175.4.1026-1031.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. P., Hächler H., Levy S. B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993 Mar;175(5):1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. P., McMurry L. M., Hooper D. C., Wolfson J. S., Levy S. B. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989 Aug;33(8):1318–1325. doi: 10.1128/aac.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. P., McMurry L. M., Levy S. B. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol. 1988 Dec;170(12):5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha M. A., Kropinski A. M. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol. 1990 Jun;172(6):3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambino L., Gracheck S. J., Miller P. F. Overexpression of the MarA positive regulator is sufficient to confer multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1993 May;175(10):2888–2894. doi: 10.1128/jb.175.10.2888-2894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay P., Le Coq D., Steinmetz M., Berkelman T., Kado C. I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985 Nov;164(2):918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George A. M., Levy S. B. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983 Aug;155(2):531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Honoré N., Nicolas M. H., Cole S. T. Regulation of enterobacterial cephalosporinase production: the role of a membrane-bound sensory transducer. Mol Microbiol. 1989 Aug;3(8):1121–1130. doi: 10.1111/j.1365-2958.1989.tb00262.x. [DOI] [PubMed] [Google Scholar]
- Kaniga K., Delor I., Cornelis G. R. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991 Dec 20;109(1):137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- Kolibachuk D., Greenberg E. P. The Vibrio fischeri luminescence gene activator LuxR is a membrane-associated protein. J Bacteriol. 1993 Nov;175(22):7307–7312. doi: 10.1128/jb.175.22.7307-7312.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R., Inuzuka M., Helinski D. R. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978 Dec;15(4):1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- Korfmann G., Sanders C. C. ampG is essential for high-level expression of AmpC beta-lactamase in Enterobacter cloacae. Antimicrob Agents Chemother. 1989 Nov;33(11):1946–1951. doi: 10.1128/aac.33.11.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S., Galleni M., Lindberg F., Normark S. Signalling proteins in enterobacterial AmpC beta-lactamase regulation. Mol Microbiol. 1989 Aug;3(8):1091–1102. doi: 10.1111/j.1365-2958.1989.tb00259.x. [DOI] [PubMed] [Google Scholar]
- Lindquist S., Lindberg F., Normark S. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC beta-lactamase gene. J Bacteriol. 1989 Jul;171(7):3746–3753. doi: 10.1128/jb.171.7.3746-3753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S., Weston-Hafer K., Schmidt H., Pul C., Korfmann G., Erickson J., Sanders C., Martin H. H., Normark S. AmpG, a signal transducer in chromosomal beta-lactamase induction. Mol Microbiol. 1993 Aug;9(4):703–715. doi: 10.1111/j.1365-2958.1993.tb01731.x. [DOI] [PubMed] [Google Scholar]
- Miller V. L., Mekalanos J. J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988 Jun;170(6):2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller V. L., Taylor R. K., Mekalanos J. J. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell. 1987 Jan 30;48(2):271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- Normark S., Boman H. G., Matsson E. Mutant of Escherichia coli with anomalous cell division and ability to decrease episomally and chromosomally mediated resistance to ampicillin and several other antibiotics. J Bacteriol. 1969 Mar;97(3):1334–1342. doi: 10.1128/jb.97.3.1334-1342.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrovsky de Spicer P., O'Brien K., Maloy S. Regulation of proline utilization in Salmonella typhimurium: a membrane-associated dehydrogenase binds DNA in vitro. J Bacteriol. 1991 Jan;173(1):211–219. doi: 10.1128/jb.173.1.211-219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather P. N., Orosz E., Shaw K. J., Hare R., Miller G. Characterization and transcriptional regulation of the 2'-N-acetyltransferase gene from Providencia stuartii. J Bacteriol. 1993 Oct;175(20):6492–6498. doi: 10.1128/jb.175.20.6492-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw K. J., Rather P. N., Sabatelli F. J., Mann P., Munayyer H., Mierzwa R., Petrikkos G. L., Hare R. S., Miller G. H., Bennett P. Characterization of the chromosomal aac(6')-Ic gene from Serratia marcescens. Antimicrob Agents Chemother. 1992 Jul;36(7):1447–1455. doi: 10.1128/aac.36.7.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Hötte B., Klauke B., Kosier B. Isolation and characterization of insertion sequence elements from gram-negative bacteria by using new broad-host-range, positive selection vectors. J Bacteriol. 1991 Feb;173(4):1502–1508. doi: 10.1128/jb.173.4.1502-1508.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomanen E., Lindquist S., Sande S., Galleni M., Light K., Gage D., Normark S. Coordinate regulation of beta-lactamase induction and peptidoglycan composition by the amp operon. Science. 1991 Jan 11;251(4990):201–204. doi: 10.1126/science.1987637. [DOI] [PubMed] [Google Scholar]
- Watson N., Dunyak D. S., Rosey E. L., Slonczewski J. L., Olson E. R. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol. 1992 Jan;174(2):530–540. doi: 10.1128/jb.174.2.530-540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S. Z., Larson T. J. Structures of the promoter and operator of the glpD gene encoding aerobic sn-glycerol-3-phosphate dehydrogenase of Escherichia coli K-12. J Bacteriol. 1988 Sep;170(9):4209–4215. doi: 10.1128/jb.170.9.4209-4215.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Herrero M., Jakubzik U., Timmis K. N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990 Nov;172(11):6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]