Abstract
Juvenile hormone analog (JHA) insecticides are relatively nontoxic to vertebrates and offer effective control of certain insect pests. Recent reports of resistance in whiteflies and mosquitoes demonstrate the need to identify and understand genes for resistance to this class of insect growth regulators. Mutants of the Methoprene-tolerant (Met) gene in Drosophila melanogaster show resistance to both JHAs and JH, and previous biochemical studies have demonstrated a mechanism of resistance involving an intracellular JH binding-protein that has reduced ligand affinity in Met flies. We cloned the Met+ gene by transposable P-element tagging and found reduced transcript level in several mutant alleles, showing that underproduction of the normal gene product can lead to insecticide resistance. Transformation of Met flies with a Met+ cDNA resulted in susceptibility to methoprene, indicating that the cDNA encodes a functional Met+ protein. MET shows homology to the basic helix-loop-helix (bHLH)-PAS family of transcriptional regulators, implicating MET in the action of JH at the gene level in insects. This family also includes the vertebrate dioxin receptor, a transcriptional regulator known to bind a variety of environmental toxicants. Because JHAs include a diverse array of chemicals with JH activity, a mechanism whereby they can exert effects in insects through a common pathway is suggested.
Juvenile hormone (JH) is involved in a variety of critical functions in insects, including development, reproduction, caste determination, and behavior (1, 2). Chemical analogs of JH (JHA) have been developed, and many of them have insecticidal activity against certain insects (3). Because JHAs have effects on insects that are similar to those of exogenously applied JH (4), they act as JH agonists, and several have proven useful as such in physiological studies (5). JHA insecticides have an additional advantage of low vertebrate toxicity (6).
Initially, it was predicted that insects would have difficulty evolving resistance to a compound resembling one of their own hormones (7). However, insect cross-resistance to methoprene shortly thereafter demonstrated that resistance to this class of insecticides is possible (8), and other instances of either cross-resistance or laboratory-selected resistance have been subsequently reported (9). Recently, field resistance to methoprene (isopropyl [2E, 4E]-11-methoxy-3,7,11-trimethyl-2,4-dodecadienoate) in mosquitoes (D. Dame, unpublished data) and to another JHA, pyriproxyfen (2-[1-methyl-2-(4-phenoxyphenoxy)ethoxy]pyridine), in whiteflies (10) after application of these JHAs to susceptible populations has been detected, suggesting that insects can evolve resistance de novo to these compounds.
To understand the genes involved in resistance to JHAs, we mutagenized Drosophila melanogaster and selected methoprene-resistant mutants in the F1 generation (11). Genetic complementation studies showed that these mutants were alleles at a locus that was termed Methoprene-tolerant (Met). Met flies are resistant to the toxic and morphogenetic effects of JH and several JHAs, but not to other classes of insecticides (12). Biochemical studies revealed a target-site resistance mechanism, that of reduced JH binding in cytosolic extracts from either of two JH target tissues in Met flies (13). This property of reduced JH binding was cytogenetically localized to the Met region on the X chromosome and can account for the resistance. Possible identities for this binding protein include either an accessory JH-binding protein in the cytoplasm, similar to the cellular retinoic acid-binding protein in vertebrates (14), or a JH receptor protein involved in the action of JH.
The recovery of two transposable P-element alleles of Met (15) has allowed the Met gene to be cloned. Here we identify the Met gene and its transcripts, and we report homology of Met to the basic helix-loop-helix (bHLH)-PER-AHR/ARNT-SIM (PAS) transcription factor family. Homology to these proteins has implications for MET function in JH action as well as for a mechanism by which chemically diverse JH analog insecticides can function as JH agonists.
EXPERIMENTAL PROCEDURES
D. melanogaster Stocks and Culture.
The alleles of Met used in this study were independently recovered from mutagenesis screens (11) with ethyl methanesulfonate (EMS), cobalt-60 (3,000 R), or P-elements (15) as mutagen. The parent stock was vermilion (v) for all alleles except for the EMS-induced alleles Met1, Met2, and Met3 (11, 12). The v gene is closely linked to Met and is a convenient genetic marker. Identity of newly recovered resistant strains as Met alleles was further substantiated by methoprene-testing flies that were made heterozygous for each allele with a deficiency chromosome Df(1)m259–4, which is deficient for the 10C2–10D4 Met cytogenetic region and uncovers the resistance phenotype (12). Each of the alleles recovered is viable and fertile, and each has been maintained as a homozygous stock in the absence of JHA-selective pressure with no loss in resistance. Both the v strain and the wild-type strain Oregon-RC were obtained from the Mid-America Stock Center, Bowling Green, OH. Flies were raised at 25°C on a standard agar–molasses–yeast extract diet with propionic acid added to retard mold growth.
DNA Cloning and Transformation.
DNA manipulations were done according to standard methods (16). P-element-bearing DNA fragments were isolated from lambda phage from MetA3 and MetK17 genomic libraries as previously described (17). Fragments were subcloned into Bluescript (Stratagene) and used to probe a genomic library constructed from the iso-1 wild-type strain. Several clones were recovered, and one of these included the genomic region shown in Fig. 1 and was used for further characterization. A cDNA library prepared from vitellogenic ovaries from the wild-type strain Canton S was probed with a genomic fragment extending from KpnI to HpaI (Fig. 1), and positive cDNA clones were inserted into Bluescript for further characterization and DNA sequencing.
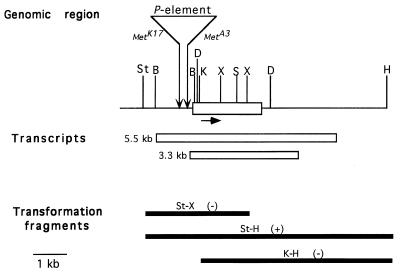
Figure 1.
Met gene region. Genomic organization of an 8-kb region including the Met ORF (boxed). P-element insertional sites in the MetA3 and MetK17 alleles are shown at the downward facing arrows. The solid arrow represents the direction of transcription of Met. The locations of the transcripts as deduced from cDNA sequencing (3.3-kb transcript) and RT-PCR analysis (5.5-kb transcript) are noted below the map. The genomic transformation fragments are indicated below the transcriptional units; those that did not rescue the resistance phenotype are noted (−) and the fragment that produced methoprene susceptibility in transformant flies is noted (+). The fragments are designated by the restriction enzyme sites. D , HindIII; S, SalI; K, KpnI; St, StuI; B, BamHI; X, XhoI; H, HpaI.
For expression of the Met+ ORF from the 3.3-kb cDNA, PCR amplification of a fragment carrying the Met+ ORF plus 580 bp of flanking sequence was carried out by using a 3.3-kb cDNA as template. A forward primer 5′-ACAAGGCAGTAACTC-3′, which began amplification 42 bp upstream of the ATG start codon, and a reverse primer 5′-GTAAAGCCAACTCATTATAC-3′, which ended amplification 538 bp downstream from the TGA stop codon, were synthesized and used for amplification. The amplified fragment was ligated into T-Easy vector (Promega) and subcloned. An EcoRI fragment containing the entire D. melanogaster sequence was excised from the subcloned fragment and ligated into the transformation vector pP{CaSpeR-Hsp70/SV40} [Flybase (l997) ftp.bio.indiana.edu]. Orientation of the fragment in the vector was determined by restriction site analysis.
For germ-line transformation, three genomic fragments shown in Fig. 1 were prepared by excision with the indicated restriction enzymes and were either subcloned into Bluescript, then excised with an EcoRI-NotI double digest and subcloned into the pCaSpeR 4 transformation vector (18), or were subcloned directly into the pCaSpeR 4 vector. The Met+ cDNA ORF fragment described above was used for transformation in the pP{CaSpeR-Hsp70/SV40} vector. Purified plasmids together with pπ25.1wc transformation “helper” DNA in a ratio of 2–3:1 were injected as described (19) into dechorionated w v Met3 embryos. Both vectors carry a functional copy of the white + (w+) gene, capable of restoring eye color to w mutants. Go progeny were individually crossed with w v Met3, and transformants were recognized by restoration of eye color ranging from light orange to red.
To test each DNA fragment for Met+ activity, we utilized a bioassay for a morphogenetic effect of methoprene on D. melanogaster. JH and JHA treatment of Met+/Met+ and Met+/Met larvae results in abnormal adult sternite bristle patterns, consisting of missing and abnormally shaped bristles, particularly on the posterior sternites (12, 20, 21). Flies homozygous for any of the Met alleles recovered to date are completely resistant to this morphogenetic effect of methoprene at even the highest sublethal doses (12, 21). Met/Met+ flies show a level of resistance intermediate between that of Met+/Met+ and Met/Met (12, 21). This morphogenetic effect of methoprene represents the most sensitive method for distinguishing Met/Met, Met+/Met, and Met+/Met+ genotypes.
Light-color-eyed transformant flies having a single ectopic copy of the transforming DNA fragment were crossed with w v Met3 and their F1 progeny were tested on methoprene. Those progeny with colored eyes had a single ectopic copy of the transforming DNA fragment and, thus, were either Met+/Met if carrying a functional Met+ gene or Met/Met if not. White-eyed w v Met3 siblings served as controls. Both transformant and nontransformant progeny that eclosed were examined for sternite bristle defects after methoprene treatment as larvae.
RNA Procedures.
Total RNA was isolated with TriReagent (Molecular Research Center, Cincinnati) from staged animals, subjected to denaturing gel electrophoresis in a formaldehyde-agarose gel, and blotted onto Hybond-N membrane. Following cross-linking, membranes were prehybridized in a solution containing 5× SSPE, 5× Denhardt’s, 0.5% SDS, 50% formamide, and 100 mg/ml yeast tRNA for 5–7 hr at 65°C. Membranes were then hybridized in the same solution at 65°C for 15–17 hr with a [32P]UTP-labeled riboprobe (Promega) synthesized from DNA produced by PCR amplification of a fragment of the Met ORF extending from nucleotide 771 to 1,102. The amplified fragment was subcloned into T-vector (Invitrogen), linearized with SstII, and transcribed from the T7 promoter to produce a 331-bp antisense riboprobe. The membranes were washed with 2× SSC + 0.1% SDS at 22°C for 20 min, followed by two washes with 0.1× SSC + 0.1% SDS at 65°C for 15 min each. Each membrane was placed against x-ray film and subjected to autoradiography at −70°C for 24 hr and developed. Control loading was evaluated by stripping the membrane and reprobing with a [32P]dCTP random-primed cDNA of the ribosomal protein-49 (Rp49) gene (22).
RESULTS
Previous results described the molecular cloning of a 40-kb region including the P-element insertions for both of the P-element alleles, MetA3 and MetK17 (17). Each of these alleles conferred resistance to both the toxic and morphogenetic effects of JH and methoprene, and susceptible revertants could be recovered by standard genetic means (15), demonstrating that the resistance phenotype likely results from the presence of the P-element. Lambda phage clones containing P-elements were recovered from MetA3 and MetK17 genomic libraries. DNA was sequenced bidirectionally from the point of insertion of the P-element in each clone, and the sequence was subjected to computer analysis. An ORF located 273 bp from the P-element insertion site in the MetA3 allele and 424 bp in the MetK17 allele was found (Fig. 1). Because this ORF is transcribed in a direction away from the insertion sites of the P-elements, interruption of transcription by the P-element is possible, as has been found with other P-element mutants in D. melanogaster (23).
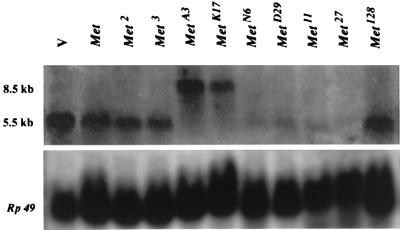
To identify any transcript(s) from this ORF, mRNA was prepared from methoprene-susceptible Oregon-RC late third-instar larvae. This stage in development represents high susceptibility to both the toxic and morphogenetic effects of JH and JHAs (24). A 32P-labeled DNA probe to this ORF failed to detect any transcript(s) on a Northern blot of RNA from these larvae, but an RNA probe (riboprobe) recognized a 5.5-kb transcript (Fig. 2). This transcript was unchanged in size or intensity in three EMS-induced alleles of Met but was reduced in abundance in several x-ray-induced alleles, especially Met27, which appears as a null allele (Fig. 2). MetA3 and MetK17 larvae showed a transcript approximately 3 kb larger than the 5.5-kb transcript, suggesting that transcriptional run-on of the 2.9-kb P-element occurred in these flies, as has been shown with a P-element insertional allele of the yellow mutant (25). These results identify this ORF with at least a portion of the Met gene.
Figure 2.
Northern blot of total RNA isolated from wandering third-instar larvae homozygous for v or any of various Met alleles and probed with a 32P-labeled riboprobe of the Met gene. Each lane was loaded with 40 μg of total RNA, subjected to denaturing gel electrophoresis in a formaldehyde-agarose gel, and blotted onto Hybond-N membrane. Control loading (Lower) was evaluated by stripping the blot and reprobing with a [32P]dCTP random-primed cDNA of Rp49. Met, Met2, and Met3 are ethyl methanesulfonate-induced alleles; MetA3 and MetK17 are P-element alleles, and the remaining alleles were gamma-ray induced from methoprene-susceptible v flies. Met128 has consistently shown overproduction of transcript on Northern blots.
To define the Met gene functionally, P-element-mediated germ-line transformation (19) was carried out with DNA fragments isolated from phage clones from a wild-type genomic library. One of these fragments, designated St-H because of the restriction enzymes used to isolate it, carries a genomic fragment that includes the entire 5.5-kb transcript region. Each of the other two fragments, St-X and K-H, carries genomic DNA that represents an incomplete ORF (Fig. 1). When flies carrying Met3, a strong EMS-induced allele (12), were transformed with fragments St-X and K-H (Fig. 1), the level of methoprene resistance in progeny of the transformants as judged by resistance to sternite bristle disruption was undiminished, indicating no rescue of the mutant phenotype (Table 1). However, when Met3 flies were transformed with fragment St-H, resistance in their progeny to sternite bristle disruption by methoprene was lost (Table 1), indicating that a functional Met+ gene is contained in this sequence. The genomic DNA region contained in the St-H fragment corresponds well with the size and location of the 5.5-kb transcript (Fig. 1).
Table 1.
Drosophila transformant adults having abnormal sternal bristle patterns after methoprene treatment
| Genotype | Methoprene concentration, μl/food vial
|
|||
|---|---|---|---|---|
| 0.01 | 0.005 | 0.0025 | 0.0 | |
| w v Met3 [St-X] | 0 | 0 | 0 | 0 |
| w v Met3 [K-H] | 0 | 0 | 0 | 0 |
| w v Met3 [St-H] | 92* | 78* | 18 | 0 |
| w v Met3 [cMet+] | 70* | 66* | 6 | 0 |
| w v Met3 | 0 | 0 | 0 | 0 |
| v | NS | NS | 54 | 0 |
Values given as percentages of adult females eclosing with at least two missing or defective bristles on the most methoprene-sensitive sternite 7. Nontransformant siblings of each genotype, recognized by white eyes, were completely resistant as expected and are presented as the w v Met3 row for simplicity. Fifty adult females of each genotype were evaluated. Transformant and nontransformant adult responses to each concentration of methoprene were compared (Fisher’s exact test) and were significantly different (P < 0.001, designated by ∗) for the two highest methoprene concentrations. Responses are also shown for v adult females (having two Met+ gene doses) for comparison. NS, nonsurvival to adult stage.
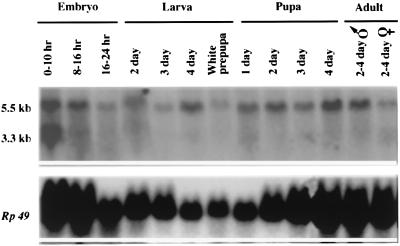
A Northern analysis was carried out to determine the abundance and temporal appearance of the transcript during development (Fig. 3). During the first half of embryonic development, an additional transcript of 3.3 kb is detected. This transcript decays to an undetectable level during the latter half of embryonic development. It is also present in adult female ovaries and is probably contributed to the embryo as a maternal RNA. Neither transcript is abundant during development. Because the methoprene-sensitive period is late third-instar larval through early pupal development (24), then only the larger transcript is developmentally correlated with resistance. Taken together, these data implicate this ORF as the Met gene responsible for resistance.
Figure 3.
Developmental Northern blot analysis of total RNA isolated from the methoprene-susceptible Oregon-RC strain at various times in development and probed with a [32P]UTP-labeled 331-bp riboprobe for Met. Each lane was loaded with 40 μg of total RNA, and the blot was probed with the Met gene riboprobe followed by a DNA probe for the Rp49 gene as described for Fig. 2. The decreased levels of Rp49 in pupae reflect decreased expression during this stage of development (22). Embryos were collected from overnight cultures and either frozen in liquid nitrogen or maintained at 25°C until the desired age. Larvae were staged from timed embryo collections; the indicated times are ±8 hr. Pupae were staged from the white prepupal stage, which lasts for about 1 hr. Adult males consistently show only the 5.5-kb transcript; females show both and, when fully gravid, show more of the 3.3-kb transcript than appears on this blot.
cDNAs corresponding to the 3.3-kb transcript were isolated as apparent full-length cDNAs from a D. melanogaster wild-type Canton-S ovary cDNA library and were sequenced to establish the relationship of this transcript with the genomic sequence. The probable transcriptional start site for this transcript begins 213 bp upstream of the ATG start codon of Met and ends 981 bp downstream from an in-frame TGA termination codon. The DNA sequence (CAAAATG) preceding this ATG corresponds well with a D. melanogaster translation start site consensus sequence (26). Sequence analysis showed the size of the ORF to include 716 aa with a calculated molecular weight of 78,683 (Fig. 4A). The boundaries of the 5.5-kb transcript have not been precisely determined, but they have been inferred by reverse transcription (RT)-PCR to include a transcriptional start site located about 1,100 bp upstream of the ATG site and a termination point located about 2,200 bp downstream from the stop codon of the Met ORF.
Figure 4.
(A) Nucleotide sequence and predicted amino acid sequence of the Met ORF, derived from sequence determination of 3.3-kb cDNA clones. The bHLH domain is underlined, the PAS-A domain is boldly underlined, and the PAS-B domain is double underlined. Single-letter abbreviations for the amino acid residues are: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. (B) Amino acid sequence alignment of the homologous regions for the bHLH, PAS-A, and PAS-B domains of MET with those of human AHR and ARNT. Number in parenthesis indicates the position of the initial amino acid of each domain in the respective protein. Identical sequences are boxed.
To further confirm that this ORF is the Met gene, a DNA fragment consisting of the entire ORF plus flanking sequence was prepared from a subcloned 3.3-kb cDNA molecule. This fragment was prepared by PCR amplification and inserted into the pP{CaSpeR-Hsp70/SV40} transformation vector. This vector carries an hsp70 promoter to drive expression of the inserted DNA, and low-level constitutive expression occurs in transformant flies raised at 25°C [Flybase (l997) ftp.bio.indiana.edu]. When larvae carrying one ectopic copy of this transformation fragment were treated with methoprene and examined after metamorphosis, resistance to methoprene was lost (Table 1), demonstrating that a functional copy of Met+ is contained in the ORF. This result also demonstrates that although the 3.3-kb transcript is not expressed during the late larval–early pupal stage when methoprene sensitivity is found, it nevertheless carries functional Met+ sequence.
Sequence comparison of the Met ORF to sequences deposited in the Genpept and Swiss-Prot databases was carried out by using the fasta program (27). MET shows three regions of homology to members of a family of transcriptional activators known as bHLH-PAS proteins (ref. 28; Fig. 4B). The bHLH-PAS gene family was named for the three founding members: Period (per) gene (29), Aryl hydrocarbon receptor [Ahr (30)], and Single-minded (sim) gene (28). Per is a biological clock gene, and sim is a transcription factor gene, both originally isolated from D. melanogaster. Ahr has been isolated from several vertebrates, and its product functions to bind xenobiotic compounds and a partner protein, the product of the Ah receptor nuclear translocator [arnt (31)] gene. Most of the bHLH-PAS proteins function either as transcription factors or as interactive partners with transcription factor proteins (32). MET generally has higher homology to the vertebrate bHLH-PAS proteins than to those identified in D. melanogaster. A D. melanogaster ARNT-like gene has recently been cloned (33), and DARNT has higher homology to vertebrate ARNT than does MET, suggesting that DARNT, not MET, may function like ARNT in flies. MET homology to these proteins (Fig. 4B) includes the bHLH region that is involved in DNA binding (30–38% identity), the PAS-A region (28–40%), and the PAS-B region (22–35%). The arrangement of these domains in the Met gene is the same as for other bHLH-PAS genes (Fig. 4A).
DISCUSSION
JHAs are important insect growth regulator insecticides that are particularly useful for control of certain insect pests of humans, livestock, companion animals, and stored crops (3). Because of the nontoxicity of JHAs to vertebrates, these compounds have readily found approval for use near humans and domesticated animals.
JHAs have been in use for two decades, but, until recently, resistance to these compounds generally has not been problematic. Mechanisms of resistance that render an insecticide less effective include altered penetration, tissue sequestration, excretion, metabolic detoxification, or target tissue binding of the insecticide (34). Studies that have addressed JHA resistance in pest insects have shown it to be primarily due to enhanced metabolism (35). The resistance mechanism(s) that underlies the control failure recently reported for methoprene and pyriproxyfen (10) is unknown. Our previous study of Met flies experimentally ruled out four mechanisms for Met resistance, including enhanced metabolism, and showed reduced JHIII binding in two target tissues as the probable mechanism of resistance (13). Reduced binding of insecticide by target tissues can result in strong resistance and has been demonstrated in field resistance in insects to other insecticides (36). Although reduced binding has been demonstrated for JHA resistance only in D. melanogaster, there is no reason to rule out participation of a Met homolog in field resistance in pest insects. To this end, the availability of the D. melanogaster Met gene for isolating and characterizing homologous genes in pest insects could lead to monitoring field resistance at the molecular level.
Our finding of MET homology to the bHLH-PAS family of proteins narrows the focus of Met function to transcriptional regulation, with particular attention to the dioxin receptor partners, AHR and ARNT. AHR resides in the cytoplasm of target tissues, where it can bind to a variety of environmental toxicants, including dioxin. After translocation to the nucleus, the AHR-ligand complex binds with ARNT to form a heterodimeric transcription factor capable of binding to a dioxin-responsive element lying 5′ upstream of genes that mediate the biological effects of these compounds (37). Several of these genes have been identified as encoding cytochrome P450 enzymes that carry out detoxification reactions (37). The PAS regions are important in the dimerization of proteins between members of the PAS protein family (38). It is of interest that a dioxin receptor has been previously implicated in insecticide resistance in the moth Helicoverpa zea. In this work (39) dioxin binding in H. zea extract could be competitively inhibited by DDT [1,1,1-trichloro-2,2-bis(4-chlorophenyl)ethane], JHI, or methoprene, leading to the hypothesis that a JH receptor may be involved in resistance to a variety of insecticides. Because the vertebrate dioxin receptor functions as a transcriptional activator of genes involved in the metabolism of xenobiotic compounds (37), it is tempting to invoke a similar action for Met involvement in inducing metabolism of JHAs and other insecticides. However, we have no evidence for this role: the mechanism of Met resistance clearly does not involve increased metabolism (13) as would be predicted from the H. zea results, and Met flies are not resistant to other classes of insecticides (12).
There are several implications of MET as a member of this family. First, Met can now be viewed with greater certainty as a participant in JH action at the gene level instead of controlling a cytoplasmic JH binding protein, an alternate interpretation of the JH binding results (40). Although we have shown homology of MET to bHLH-PAS proteins, we have not demonstrated MET function as a transcriptional regulator. These experiments are complicated by a lack of understanding of the molecular action of JH (41). Transcriptional regulation (42) of certain genes has been correlated with JH appearance during development or after application of exogenous JH, but more direct involvement of this hormone in gene activity has been only occasionally reported (43), and evidence exists for JH acting at the membrane level in adult insects (44). No D. melanogaster genes have been identified that are directly regulated transcriptionally by JH; therefore, designing a reporter gene system to test Met as encoding a functional transcriptional regulator must await future work.
Second, homology to the Ah receptor family suggests that MET may act in a similar manner to bind a variety of ligands having JH activity and subsequently to regulate JH-responsive target genes. Thus, the sundry chemicals having JH activity (3) may owe their activity to a MET protein with promiscuous ligand binding. Interestingly, mice carrying the dioxin receptor Ahrd allele are more resistant to toxic Ah receptor agonists because of decreased binding affinity of the AHR (45), a result that parallels our finding that Met resistance to toxic levels of JH is associated with decreased JH binding affinity.
Third, identity of MET as a PAS protein suggests that a partner is involved in MET function. A partner could be necessary for the presumed function of MET in gene regulation, as is the case in the partner protein composition of the ecdysone receptor complex (46). More significant is the possibility of MET interaction with a partner protein associated with the ecdysone response, similar to the involvement of two other bHLH-PAS proteins, SRC-1 and p/CIP, in nuclear receptor function in vertebrates (47, 48). This is an appealing possibility to explain JH–ecdysone interaction for control of insect metamorphosis (1, 4).
This work isolates a gene involved in resistance to JH and the JH-analog class of insecticides. Further study of Met may help elucidate JHA resistance in pest insects as well as the molecular basis of JH action.
Acknowledgments
We thank J. Herbers and A. Reddy for comments on the manuscript, J. Hooper for teaching us transformation techniques, and J. Tamkun and P. Tolias for the libraries. G. Thomas provided excellent technical support for some of the experiments. This work was supported by grants from the National Science Foundation, the U.S. Department of Agriculture Competitive Grants Program, and American Cyanamid Co. to T.G.W. The sequence of the Met gene has been submitted to GenBank and can be accessed by using the accession number AF034859.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: JH, juvenile hormone; JHA, JH analog.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AF034859 (Methoprene-tolerant gene)].
A commentary on this article begins on page 2725.
References
- 1.Riddiford L M. In: Comprehensive Insect Physiology, Biochemistry, and Pharmacology. Kerkut G, Gilbert L I, editors. Vol. 8. New York: Pergamon; 1985. pp. 37–84. [Google Scholar]
- 2.Nijhout H F, Wheeler D E. Q Rev Biol. 1982;57:109–133. [Google Scholar]
- 3.Staal G B. Annu Rev Entomol. 1975;20:417–460. doi: 10.1146/annurev.en.20.010175.002221. [DOI] [PubMed] [Google Scholar]
- 4.Riddiford L M. Adv Insect Physiol. 1994;24:213–274. [Google Scholar]
- 5.Zhang J, Saleh D S, Wyatt G R. Mol Cell Endocrinol. 1996;122:15–20. doi: 10.1016/0303-7207(96)03884-1. [DOI] [PubMed] [Google Scholar]
- 6.Wright J E. Environ Health Perspect. 1976;14:127–132. doi: 10.1289/ehp.7614127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams C M. Sci Amer. 1967;217:13–17. doi: 10.1038/scientificamerican0767-13. [DOI] [PubMed] [Google Scholar]
- 8.Cerf D C, Georghiou G P. Nature (London) 1972;239:401–402. doi: 10.1038/239401a0. [DOI] [PubMed] [Google Scholar]
- 9.Sparks T C, Hammock B D. In: Pest Resistance to Pesticides. Georghiou G P, Saito T, editors. New York: Plenum; 1983. pp. 615–668. [Google Scholar]
- 10.Ishaaya I, Horowitz A R. Pestic Sci. 1995;43:227–232. [Google Scholar]
- 11.Wilson T G, Fabian J. In: Molecular Entomology. Law J, editor. Vol. 49. New Series: UCLA Symposia on Molecular and Cellular Biology; 1987. pp. 179–188. [Google Scholar]
- 12.Wilson T G, Fabian J. Dev Biol. 1986;118:190–201. doi: 10.1016/0012-1606(86)90087-4. [DOI] [PubMed] [Google Scholar]
- 13.Shemshedini L, Wilson T G. Proc Natl Acad Sci USA. 1990;87:2072–2076. doi: 10.1073/pnas.87.6.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blomhoff R, Green M H, Berg T, Norum K R. Science. 1990;250:399–404. doi: 10.1126/science.2218545. [DOI] [PubMed] [Google Scholar]
- 15.Wilson T G, Turner C. In: Molecular Mechanisms of Insecticide Resistance. Mullin C, Scott J, editors. Vol. 505. Washington, DC: Am. Chem. Soc.; 1992. pp. 99–112. [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Turner C, Wilson T G. Arch Insect Biochem Physiol. 1995;30:133–147. doi: 10.1002/arch.940300205. [DOI] [PubMed] [Google Scholar]
- 18.Thummel C S, Pirrotta V. Drosophila Inf Serv. 1990;71:150. [Google Scholar]
- 19.Spradling A C. In: Drosophila, a Practical Approach. Roberts D B, editor. Oxford: IRL; 1986. pp. 175–197. [Google Scholar]
- 20.Riddiford L M, Ashburner M. Gen Comp Endocrinol. 1991;82:172–183. doi: 10.1016/0016-6480(91)90181-5. [DOI] [PubMed] [Google Scholar]
- 21.Wilson T G. Arch Insect Biochem Physiol. 1996;32:641–649. doi: 10.1002/(SICI)1520-6327(1996)32:3/4<641::AID-ARCH35>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 22.O’Connell P, Rosbash M. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Searles L L, Greenleaf A L, Kemp W E, Voelker R A. Mol Cell Biol. 1986;6:3312–3319. doi: 10.1128/mcb.6.10.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Postlethwait J H. Drosophila Inf Serv. 1973;50:135–138. [Google Scholar]
- 25.Howes G, O’Connor M, Chia W. Nucleic Acids Res. 1988;16:3039–3052. doi: 10.1093/nar/16.7.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavener D R. Nucleic Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Crews S T, Thomas J B, Goodman C S. Cell. 1988;52:143–151. doi: 10.1016/0092-8674(88)90538-7. [DOI] [PubMed] [Google Scholar]
- 29.Reddy P, Zehring W A, Wheeler D A, Pirrotta V, Hadfield C, Hall J C, Rosbash M. Cell. 1984;38:701–710. doi: 10.1016/0092-8674(84)90265-4. [DOI] [PubMed] [Google Scholar]
- 30.Ema M, Sogawa K, Watanabe N, Chujoh Y, Matsushita N, Gotoh O, Funae Y, Fujii-Kuriyama Y. Biochem Biophys Res Commun. 1992;184:246–253. doi: 10.1016/0006-291x(92)91185-s. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman E C, Reyes H, Chu F-F, Sander F, Conley L H, Brooks B A, Hankinson O. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 32.Hogenesch J B, Chan W K, Jackiw V H, Brown R C, Gu Y-Z, Pray-Grant M, Perdew G H, Bradfield C A. J Biol Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 33.Zelzer E, Wappner P, Shilo B-Z. Genes Dev. 1997;11:2079–2089. doi: 10.1101/gad.11.16.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feyereisen R. Toxicol Lett. 1995;82:83–90. doi: 10.1016/0378-4274(95)03470-6. [DOI] [PubMed] [Google Scholar]
- 35.Hammock B D, Mumby S M, Lee P W. Pest Biochem Physiol. 1977;7:261–272. [Google Scholar]
- 36.ffrench-Constant R H, Roush R T, Mortlock D, Dively G P. J Econ Entomol. 1990;83:1733–1737. doi: 10.1093/jee/83.5.1733. [DOI] [PubMed] [Google Scholar]
- 37.Swanson H I, Bradfield C A. Pharmacogenetics. 1993;3:213–230. doi: 10.1097/00008571-199310000-00001. [DOI] [PubMed] [Google Scholar]
- 38.Huang Z J, Edery I, Rosbash M. Nature (London) 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 39.Muehleisen D P, Plapp F W, Jr, Benedict J H, Carino F A. Pest Biochem Physiol. 1989;35:50–57. [Google Scholar]
- 40.Shemshedini L, Lanoue M, Wilson T G. J Biol Chem. 1990;265:1913–1918. [PubMed] [Google Scholar]
- 41.Jones G. Annu Rev Entomol. 1995;40:147–169. doi: 10.1146/annurev.en.40.010195.001051. [DOI] [PubMed] [Google Scholar]
- 42.Venkataraman V, O’Mahony P J, Manczak M, Jones G. Dev Genet. 1994;15:391–400. doi: 10.1002/dvg.1020150502. [DOI] [PubMed] [Google Scholar]
- 43.Berger E M, Goudie K, Klieger L, Berger M, DeCato R. Dev Biol. 1992;151:410–418. doi: 10.1016/0012-1606(92)90181-f. [DOI] [PubMed] [Google Scholar]
- 44.Wyatt G R, Davey K G. Adv Insect Physiol. 1996;26:1–155. [Google Scholar]
- 45.Poland A, Glover E. Mol Pharmacol. 1980;17:86–94. [PubMed] [Google Scholar]
- 46.Yao T-P, Segraves W A, Oro A E, McKeown M, Evans R M. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]
- 47.Yao T-P, Ku G, Zhou N, Scully R, Livingston D M. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]