Abstract
Over 2,600 transgenic rice plants in nine strains were regenerated from >500 independently selected hygromycin-resistant calli after Agrobacterium-mediated transformation. The plants were transformed with fully modified (plant codon optimized) versions of two synthetic cryIA(b) and cryIA(c) coding sequences from Bacillus thuringiensis as well as the hph and gus genes, coding for hygromycin phosphotransferase and β-glucuronidase, respectively. These sequences were placed under control of the maize ubiquitin promoter, the CaMV35S promoter, and the Brassica Bp10 gene promoter to achieve high and tissue-specific expression of the lepidopteran-specific δ-endotoxins. The integration, expression, and inheritance of these genes were demonstrated in R0 and R1 generations by Southern, Northern, and Western analyses and by other techniques. Accumulation of high levels (up to 3% of soluble proteins) of CryIA(b) and CryIA(c) proteins was detected in R0 plants. Bioassays with R1 transgenic plants indicated that the transgenic plants were highly toxic to two major rice insect pests, striped stem borer (Chilo suppressalis) and yellow stem borer (Scirpophaga incertulas), with mortalities of 97–100% within 5 days after infestation, thus offering a potential for effective insect resistance in transgenic rice plants.
Keywords: rice transformation, insecticidal proteins, insect resistance, transgenic plants, Bacillus thuringiensis toxin
Rice is one of the world’s most important food crops, and intense efforts, including use of genetic engineering technologies, must be engaged to increase its yield if the impending global rice shortage is to be avoided (1). Engineering rice for pest resistance is a major challenge, one strategy being the introduction of Bacillus thuringiensis (Bt) crystal insecticidal protein (δ-endotoxin) genes (cry genes). These proteins (Bt toxins) are highly toxic to lepidopteran, dipteran, and coleopteran insects (2), among which are important pests of rice such as striped stem borer (SSB), yellow stem borer (YSB), and leaffolder (Cnaphalocrocis medinalis and Marasmia patnalis) that cause annual losses of an estimated 10 million tonnes (3).
Rice plants containing cryIA(b) or cryIA(c) have been obtained by using protoplast (4) or particle bombardment methods (5–7). However, the numbers of plants obtained and levels of the toxin proteins in these studies were unfortunately still very low from a breeder’s point of view. In contrast, >2,600 transgenic plants were produced with the modified cry genes in nine rice strains by using a modified Agrobacterium-based rice transformation procedure (8). Herein we report that high levels of CryIA(b) and CryIA(c) were detected among these transgenic plants, indicating that many candidate transgenics in this large screen may be the result of optimal position effects. Insect feeding assays with R1 plant tissues indicated that the transgenic plants were highly toxic to two major rice insects, SSB and YSB, with near 100% mortality within 5 days. This indicates that Agrobacterium transformation technology can indeed prove to be very effective in improving rice with important agronomic traits.
MATERIALS AND METHODS
Transformation Vectors.
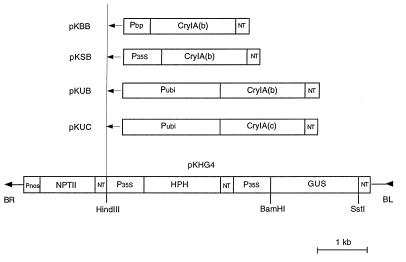
Four vectors used to transform rice were all based on a pBin19-derived binary vector pKHG4 (9). The sequences encoding the insecticidal proteins CryIA(b) and CryIA(c) from B. thuringiensis were resynthesized chemically with optimized codon usage for plants (10). The sequences were placed under the control of the maize ubiquitin promoter (11), CaMV35S promoter (12), a pollen-specific promoter (Bp10 gene promoter) (13), and the nos terminator. These chimeric genes [Ubi-cryIA(b), 4.1 kb; Ubi-cryIA(c), 4.1 kb; 35S-cryIA(b), 2.9 kb; and Bp-cryIA(b), 2.6 kb] were inserted into the HindIII site in pKHG4 as HindIII fragments, resulting in four Bt gene vectors, pKUB, pKUC, pKSB, and pKBB respectively (Fig. 1).
Figure 1.
T-DNA of pKUB, pKUC, pKSB, and pKBB. These vectors were constructed by inserting different Bt genes into the unique HindIII site in binary vector pKHG4. Sequence outside of the border is identical to pBin19. BR, right border; BL, left border; HPH, hygromycin phosphotransferase; NPTII, neomycin phosphotransferase; CryIA(b) and CryIA(c), synthetic insecticidal protein genes from B. thuringiensis; P35S, CaMV 35S promoter; Pubi, maize ubiquitin promoter; Pnos, nopaline synthase promoter; Pbp, Bp10 pollen gene promoter; NT, 3′ termination signal of nopaline synthase.
Rice Transformation.
A transformation method (8) modified from Hiei and coworkers (14) was used to transform callus from mature or immature (Nipponbare) embryos of nine rice strains (Table 1). Agrobacterium LBA4404 (15) and EHA105 (16) were used to transform vigorously growing rice calli (1- to 4-months-old).
Table 1.
Production of hygromycin-resistant calli and plants from A. tumefaciens-inoculated rice callus
| Rice strain | A.t. strain | Amount of callus cocultured, mg | HygR callus isolated, n | HygR callus/1,000 mg cocultured callus |
n
|
Regeneration rate, % B/A | ||
|---|---|---|---|---|---|---|---|---|
| HygR callus for regeneration A | Regenerating callus B | Plants regenerated C | ||||||
| Nipponbare | LBA4404(pKUB) | 2,670 | 131 | 49.1 | 92 | 89 | 444 | 97 |
| Zhong8215 | LBA4404(pKUB) | 3,240 | 105 | 32.4 | 77 | 77 | 733 | 100 |
| 93VA | EHA105(pKUB) | 1,740 | 129 | 74.1 | 106 | 66 | 334 | 62 |
| EHA105(pKUC) | 2,910 | 81 | 27.8 | 45 | 42 | 85 | 93 | |
| ZAU16 | EHA105(pKUC) | 2,210 | 66 | 29.9 | 45 | 31 | 303 | 69 |
| EHA105(pKBB) | 1,430 | 22 | 15.4 | 14 | 13 | 93 | 93 | |
| 91RM | EHA105(pKUB) | 2,430 | 78 | 32.1 | 56 | 29 | 82 | 52 |
| EHA105(pKUC) | 2,520 | 27 | 10.7 | 25 | 25 | 131 | 100 | |
| T8340 | EHA105(pKUB) | 1,180 | 66 | 55.9 | 44 | 41 | 62 | 93 |
| EHA105(pKUC) | 2,280 | 39 | 17.1 | 34 | 12 | 21 | 35 | |
| EHA105(pKSB) | 2,580 | 99 | 38.4 | 26 | 19 | 35 | 53 | |
| EHA105(pKBB) | 1,680 | 86 | 51.2 | 54 | 44 | 162 | 81 | |
| Pin92-528 | EHA105(pKUC) | 670 | 11 | 16.4 | 10 | 9 | 14 | 90 |
| EHA105(pKBB) | 2,350 | 12 | 5.1 | 7 | 7 | 9 | 100 | |
| T90502 | EHA105(pKUC) | 1,320 | 28 | 21.1 | 14 | 12 | 23 | 86 |
| Kaybonnet | EHA105(pKUB) | 790 | 5 | 6.3 | 5 | 4 | 6 | 76 |
| Total or mean | 34,270 | 1,008 | 34.3 | 671 | 533 | 2,603 | 79 | |
Assay for β-Glucuronidase (GUS) Activity.
Expression of the gus gene was assayed following an improved histochemical staining procedure (17).
Assay for CryIA(b) and CryIA(c) Proteins.
Ground samples were extracted with buffer. After vortexing and spinning for 5 min in a microfuge, 2 μl of crude extract was applied to nitrocellulose membrane and subjected to dot blot ELISA by using a polyclonal goat antibody specific for CryIA(b) essentially as described (10). This antibody was found equally reactive to CryIA(b) and CryIA(c). The Bt protein levels were measured from digitized images of the blots by using a scanner interfaced to a desktop computer with aid of sigmagel analysis software (Jandel, San Rafael, CA). Different dilutions of fast protein liquid chromatography purified, trypsin-digested CryIA(c) protein from B. thuringiensis subsp. kurstaki HD-73 were applied to each blot as an internal standard with extract from nontransgenic plants as negative control. Protein determination was performed by using the Bio-Rad protein assay reagents.
DNA and RNA Blot Analysis.
Genomic DNA (1–5 μg) was isolated from leaf tissues by using an Easy-DNA kit (Invitrogen). Digested DNA was fractionated on 0.7% agarose gel, transferred onto a nylon membrane, and hybridized to digoxigenin (DIG)-labeled probes according to manufacturer’s instructions (Boehringer Mannheim). Total RNA (10 μg) was extracted from leaf tissues by using a hot phenol method (18). Transcripts for cryIA(b) and cryIA(c) were analyzed with the standard Northern blotting method (19) by using the DIG-labeled cryIA(b) and cryIA(c) coding sequences as probes.
Progeny Test.
Selfed seeds (R1 generation) of the transformants were sown in solidified half-strength Murashige and Skoog medium with 50 mg/L hygromycin. Hygromycin resistance was scored 10 days after sowing. For GUS and Bt protein assays, leaf tissues taken from the seedlings grown on the same medium with or without hygromycin were used. Progeny that had at least one plant showing GUS or Bt protein expression were recorded as positive.
Insect Bioassays.
Insecticidal activity of the transgenic plants toward two major rice insects SSB and YSB was assayed by using laboratory culture dishes, similar to described methods (6, 7). At the flowering stage, stem cuttings with sheath tissue were taken from R1 plants of three primary transformants. The plants were either homozygous or heterozygous for Ubi-cryIA(b) or Ubi-cryIA(c) genes and were positive for Bt toxin as determined by the dot blot ELISA. Nontransgenic plants, which had no detectable Bt toxins, were selected from segregating populations and were used as negative controls. Insect egg masses were collected from the rice fields at the International Rice Research Institute in The Philippines. One to five days after infestation, the stem segments were dissected and examined for the number of live and dead insects as well as for tissue damage. Only larvae found inside the stems were recorded.
RESULTS
Transformation of Rice.
A summary of the transformation experiments is presented in Table 1. These data indicate that, from 1 g of the inoculated rice callus, 5–74 hygromycin-resistant calli were obtained. Although a remarkable difference in the yield of the hygromycin-resistant callus was seen among different rice and Agrobacterium tumefaciens strains, it could not be attributed simply to a single factor such as genotype or A. tumefaciens strain/plasmid combination. On the other hand, it is clear from these experiments that even the common A. tumefaciens strain LBA4404 could produce hygromycin-resistant callus at a yield comparable to the so-called “super-virulence” strain EHA105.
A total of 2,603 plants was regenerated from 533 callus lines in the nine rice strains. These callus lines had been selected from different callus pieces or well separated regions of the same calli and were therefore considered to be results of independent transformation events. Among them, 2,026 plants were from the calli transformed with vectors containing the cryIA(b) sequence, and the remaining 577 plants were transformed with the cryIA(c) sequence. In the cryIA(b) plants, 1,661 plants were transformed with pKUB; 330 were transformed with pKBB, and the remaining 35 plants were transformed with pKSB. On average, 3.9 plants were regenerated per callus line transferred into regeneration medium. However, notable differences in the regenerating ability were observed not only among the different rice strains but also among different callus lines in the same strains. For example, 0–65 plants were regenerated from various callus lines in rice strain 93VA transformed with EHA105 (pKUB). On the basis of callus lines, overall, 79% of them could regenerate into plants, whereas the rates varied from 35 to 100% in different treatments (Table 1). For further characterization, a total of 892 hygromycin-resistant plants representing all rice and A. tumefaciens strain combinations was grown in a greenhouse. These plants consisted of 405 independent transformants from 212 calli engineered with Ubi-cryIA(b), 93 with Ubi-cryIA(c), 12 with 35S-cryIA(b), and 88 with Bp-cryIA(b) constructs.
Integration of T-DNA [portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells] in the Rice Genome.
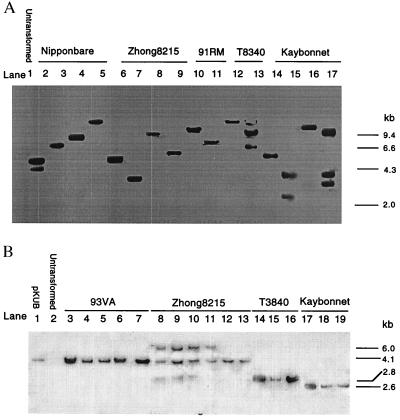
To ascertain the transgenic nature of the regenerated plants, DNA extracted from GUS-positive R0 plants of five rice strains was digested with BamHI and probed with DIG-labeled gus. BamHI cuts only once within the probed transgene (Fig. 1), just upstream of the coding sequence and so provided critical information regarding the insertion position and number of the transgene. Of the 16 plants analyzed, 12 had only one hybridization band ranging in size from 2.3 to >10 kb, whereas the remaining four plants had two to three bands (Fig. 2A), indicating that the transgenic plants analyzed had one to three insertions of the transgene at different locations in the rice genome.
Figure 2.
Southern blot analysis of plants regenerated from hygromycin-resistant calli. DNA (1–5 μg) was digested with appropriate restriction enzymes, separated on 0.7% agarose gel and hybridized to a DIG-labeled gus (A) or cryIA(b)(B) probe. (A) DNA digested with BamHI. Lanes: 2–9, plants transformed with pKUB; 10–11, plants transformed with pKUC; 12–13, plants transformed with pKSB; and 14–17, plants transformed with pKBB. (B) DNA digested with HindIII. Lanes: 1, pKUB digested with HindIII; 2, untransformed rice plant; 3–7, plants (cv. 93VA) transformed with pKUC; 8–13, plants (cv. Zhong8215) transformed with pKUB; 14–16, plants (cv. T8340) transformed with pKSB; and 17–19, plants (cv. Kaybonnet) transformed with pKBB.
To analyze the integrity of the introduced genes, DNA from 107 R0 plants representing all rice strains except Pin92–528 and T90502 was digested with HindIII, which released the Ubi-cryIA(b/c) genes. The analysis was performed on 81 plants for cryIA(b) and 26 plants for cryIA(c). A representative blot of the cry genes is shown in Fig. 2B. After HindIII digestion, DNA bands corresponding to intact chimeric genes Ubi-cryIA(b) (4.1 kb), Ubi-cryIA(c) (4.1 kb), 35S-cryIA(b) (2.8 kb), and Bp10-cryIA(b) (2.6 kb) were detected in 93% (75/81) of the cryIA(b) plants and in 58% (15/26) of the cryIA(c) plants. Moreover, the gus band at the expected 1.8-kb position was observed in all of the analyzed plants. In most plants, only one band of the expected size was seen (data not shown). This accounted for 68% of the cryIA(b)-positive plants, 80% of the cryIA(c)-positive plants, and 92% of the gus-positive plants. In addition to these expected bands, a small proportion of plants also gave rise to hybridization signals of unexpected sizes, which were mostly larger than the expected size.
Expression of cryIA(b) and cryIA(c).
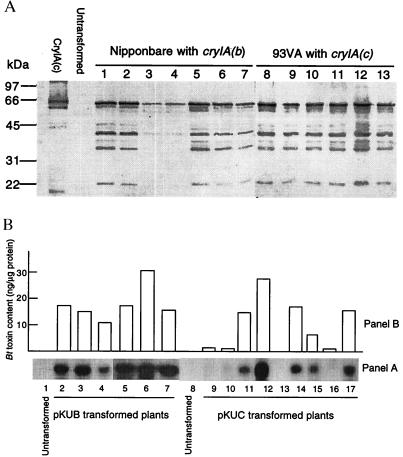
Production of CryIA(b) and CryIA(c) proteins in the regenerated plants was examined immunologically by using a polyclonal antibody against CryIA(b), which also was found to be cross-reactive with CryIA(c) (10). When the proteins were separated electrophoretically on SDS gel, a major band closely corresponding to the purified Bt toxin was detected in plants transformed with the chimeric cryIA(b)&(c) genes (Fig. 3A). Two to three fast-migrating bands also were seen. These additional bands were found to have resulted from protein degradation during the boiling process in the sample preparation (data not shown). No antibody reactive protein was found in untransformed plants.
Figure 3.
Expression of cryIA(b) and cryIA(c) in Agrobacterium-transformed rice plants. (A) Western analysis of Bt toxins in transformed rice plants; 2–4 μg of proteins extracted from untransformed and transformed plants were subjected to 10% SDS/PAGE, transferred to nitrocellulose membrane, and reacted with a polyclonal antibody specific to CryIA(b). Samples from two cultivars Nipponbare (transformed with pKUB, lanes 1–7) and 93VA (transformed with pKUC, lanes 8–13) are shown in the blot, together with the CryIA(c) from B. thuringiensis as standard. (B) Comparison of the levels of the cryIA(b) and cryIA(c) transcripts and Bt toxins. (Panel A) Northern blot analysis of cryIA(b) and cryIA(c) transcripts in plants transformed with pKUB (lanes 2–7) and pKUC (lanes 9–17). Total RNA (10 μg/lane) was extracted from leaf tissues, separated electrophoretically on 1.2% agarose-formaldehyde gel, blotted to Hybond-N nylon membrane, and hybridized to a DIG-labeled cryIA(b) fragment. (Panel B) Corresponding Bt toxin levels in the plants used for Northern analysis. The Bt toxin levels were determined by comparison of intensities of the immunologically developed color from the plant samples with that from the purified CryIA(c).
In the R0 generation, 166 independent transformed plants containing Ubi-cryIA(b), Ubi-cryIA(c), or 35S-cryIA(b) were assayed for Bt protein, and 60% of these were found to be positive. The levels varied greatly from a mere detection limit (0.1 ng/μg protein) to >30 ng/μg (3%) of the soluble protein in different plants. Most of the plants had toxin levels between 0.2 and 2% of the soluble proteins. In 10 35S-cryIA(b) plants determined for their Bt protein levels, they ranged from 0 to 0.15% of soluble protein (five plants = 0, one plant <0.01, two plants = 0.04–0.05, two plants = 0.9–0.15% of soluble protein), which is at least 10 times lower than the levels observed in most of the Ubi-cryIA(b) and Ubi-cryIA(c) plants. As expected, the pollen-specific Bp10 gene promoter (13) did not direct detectable expression of CryIA(b) in leaf tissue.
Northern analysis of 14 selected plants with different Bt toxin levels indicated that high levels of cryIA(b) and cryIA(c) transcripts were present in the leaf tissues, and a positive correlation between the levels of the transcripts and the toxin proteins was apparent (Fig. 3B).
Transmission of Transgenes Through Sexual Generation.
Inheritance of the transgenes (hph, cryIA(b), cryIA(c), and gus) was investigated in the R1 generation from the selfed seeds. Segregations for hygromycin resistance were observed in 18 progenies of 19 primary transformants tested when the seedlings were grown in hygromycin-containing medium (Table 2). The segregating ratios in 68% (13/19) of the lines tested fit the 3:1 model for single dominant gene inheritance. In the other six lines (32%), there were more sensitive plants than expected from the Mendelian model. To investigate the expression of the linked gus and Bt genes in R1 plants, seedlings from 78 primary transformants were grown in hygromycin-free medium and examined for the two transgenic traits. The results summarized in Table 3 indicated that 94% (73/78) of the tested lines expressed at least one of the two transgenic traits in the R0 generation. Of them, 97% (71/73) were able to transmit the transgenic traits to R1 generation, and the remaining 3% (2 lines) could not transmit all of the transgenic traits expressed in R0 generation to R1 generation. In most GUS+/Bt+ progenies, cosegregation of the gus and Bt genes was seen in R1 seedlings, that is, the plants were positive or negative for both traits. However, unlinked gus and Bt expression also was seen in a few progenies.
Table 2.
Segregation of hygromycin resistance in R1 generation
| Transformant | Plants responded to hygromycin, n
|
Ratio | χ2 | |
|---|---|---|---|---|
| Resistant | Sensitive | |||
| Zhong8215 LBA4404(pKUB) | ||||
| 2 | 49 | 15 | 3:1 | 0.08 |
| 4 | 46 | 21 | 3:1 | 1.47 |
| 23 | 22 | 8 | 3:1 | 0.04 |
| Kaybonnet EHA105(pKBB) | ||||
| 3 | 0 | 34 | ||
| 3 | 159 | 45 | 3:1 | 0.94 |
| 6 | 28 | 13 | 3:1 | 0.99 |
| 7 | 44 | 10 | 3:1 | 1.22 |
| 8 | 61 | 28 | 3:1 | 1.95 |
| 11 | 43 | 18 | 3:1 | 0.65 |
| Kaybonnet EHA105(pKUB) | ||||
| 13 | 69 | 24 | 3:1 | 0.33 |
| 91RM EHA105(pKUB) | ||||
| 13 | 2 | 13 | ||
| 30 | 40 | 14 | 3:1 | 0.04 |
| 91RM EHA105(pKUC) | ||||
| 4 | 8 | 16 | ||
| 8 | 7 | 9 | ||
| 22 | 12 | 10 | ||
| 29 | 29 | 13 | 3:1 | 0.79 |
| 32 | 8 | 9 | ||
| 34 | 92 | 26 | 3:1 | 0.55 |
| 44 | 22 | 6 | 3:1 | 0.19 |
Table 3.
Transmission of the transgenic traits to R1 generation
| Vector | R0 plants tested, n | Phenotype in:
|
R1 progenies, n | |||
|---|---|---|---|---|---|---|
| R0
|
R1
|
|||||
| GUS | Bt | GUS | Bt | |||
| pKUB | 24 | + | + | + | + | 24 |
| 8 | + | − | + | − | 8 | |
| 3 | − | − | − | − | 3 | |
| pKUC | 23 | + | + | + | + | 22 |
| + | − | 1 | ||||
| 13 | + | − | + | − | 12 | |
| − | − | 1 | ||||
| 2 | − | − | − | − | 2 | |
| pKSB | 5 | + | + | + | + | 5 |
Insecticidal Activity.
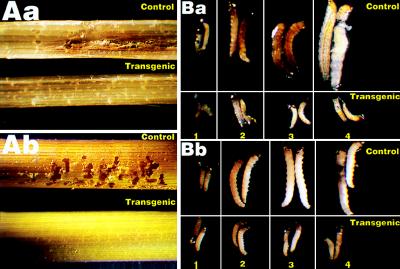
Stem cuttings from three transgenic lines carrying Ubi-cryIA(b) (Zhong8215–4 and Kaybonnet-13) and Ubi-cryIA(c) (91RM-44) were infested with the neonate larvae of SSB and YSB. Four and five days after infestation, the stem segments were dissected and examined. Mortality of 97–100% was observed in the three transgenic lines for both insects (Table 4), whereas in the control tissues, mortality was <5% in most cases. The very few surviving larvae on transgenic stem segments were affected adversely and died shortly. Significant tissue damages were seen in control plants as a result of SSB and YSB feeding, whereas in transgenic tissues, there was little detectable damage (Fig. 4 Aa and Ab).
Table 4.
Insecticidal activity of transgenic rice plants to SSB and YSB
| Insect | Plant | Bt gene | Toxin in % of the total soluble protein | Insect, n
|
Mortality, % | |
|---|---|---|---|---|---|---|
| Found in stem tissue | Surviving | |||||
| SSB | Zhong8215 | Control | 0 | 58 | 55 | 5.1 |
| Zhong8215-4 | Ubi-cryIA(b) | 0.28 | 52 | 0 | 100.0 | |
| Kaybonnet | Control | 0 | 53 | 52 | 1.9 | |
| Kaybonnet-13 | Ubi-cryIA(b) | 0.23 | 57 | 0 | 100.0 | |
| 91RM | Control | 0 | 58 | 57 | 1.7 | |
| 91RM-44 | Ubi-cryIA(c) | 0.31 | 68 | 0 | 100.0 | |
| YSB | Zhong8215 | Control | 0 | 62 | 61 | 1.6 |
| Zhong8215-4 | Ubi-cryIA(b) | 0.28 | 39 | 1 | 97.4 | |
| Kaybonnet | Control | 0 | 36 | 36 | 0.0 | |
| Kaybonnet-13 | Ubi-cryIA(b) | 0.23 | 27 | 0 | 100.0 | |
| 91RM | Control | 0 | 28 | 28 | 0.0 | |
| 91RM-44 | Ubi-cryIA(c) | 0.31 | 19 | 0 | 100.0 | |
Figure 4.
Insecticidal activity of transgenic rice. (A) Tissue damage caused by SSB (Aa) and YSB (Ab) feeding. The stem cuttings of control (upper) and transgenic (K13) (lower) plants were infested with neonate SSB or YSB larvae and pictured 5 days after infestation. (×5.) (B) SSB (Ba) and YSB (Bb) larval development in nontransgenic (upper) and transgenic (K13) (lower) plants. SSB and YSB larvae were allowed to feed on stem cuttings of nontransgenic and transgenic plants and were photographed 1–4 days after infestation. (×10.)
Close examination showed that SSB and YSB larvae reared on transgenic plants began to die 1 and 2 days after infestation, respectively (Fig. 5), and did not grow any more thereafter (Fig. 4 Ba and Bb). Four and five days after infestation, their mortality had reached 100% (Fig. 5; Table 4). In comparison, SSB and YSB larvae fed on control plants had very low mortality (≈5% on average; Fig. 4; Table 4) and developed normally, causing massive tissue damage (Fig. 4), during the bioassay periods.
Figure 5.
Larvicidal activity of transgenic plants. The stem cuttings of control and transgenic (K13) plants were infested with neonate YSB (A) or SSB (B) larvae. Mortality was calculated 1–5 days after infestation.
DISCUSSION
To date, Agrobacterium has not been used to produce transgenic rice plants with agronomically important genes. A. tumefaciens has been used to produce transgenic rice plants, particularly with the aid of a “super-binary” vector, but these plants carried only marker genes, not useful agronomic traits (20–25).
In the present study, a large number of rice plants carrying the modified Bt insecticidal protein genes cryIA(b) and cryIA(c) have been produced in nine rice strains by using a modified Agrobacterium transformation procedure. This result further demonstrates the feasibility and effectiveness of Agrobacterium-mediated transformation in rice. Moreover, it is also clear that the commonly used Agrobacterium strain (LBA4404) and binary vector (Bin19) derivatives are able to transform rice at yields comparable to that of supervirulent strain EHA105 (Table 1) that carried a plasmid derived from super-virulent Ti plasmid pTiBo542 (26), thus broadening the choice of Agrobacterium for monocot transformation.
Southern blot analysis with BamHI-digested DNA suggested that, in most of the analyzed plants, the transgene was inserted only once into the rice genome, although in a few cases, two to three insertions may have taken place (Fig. 2A). The different sizes of hybridization signals also indicated that they resulted from the stable T-DNA integration into the rice genome and not from endophytic Agrobacterium contamination. In the 107 R0 plants analyzed, all contained at least one of the three genes probed [cryIA(b), cryIA(c), and gus]. This result indicates that virtually no escapes occurred in the selection procedure used (8). Lack of Southern hybridization signals for cryIA(b) and particularly cryIA(c) in a small proportion of gus hybridization-positive plants indicated that not all of the transferred genes were inserted into the rice genome as intact T-DNA fragments. However, presence of the expected hybridization signals in the majority of the transformed plants showed that the probed genes [Ubi-cryIA(b) and Ubi-cryIA(c)] and coding sequence of gus remained intact when integrated into the rice genome (Fig. 2B).
Genetic analysis of the R1 generation for the expression of the transgenes further demonstrated the stable incorporation of T-DNA into the rice nuclear DNA. The Bt toxin production was transmitted through the sexual generation to R1 progeny in most of the lines tested, along with hygromycin resistance and GUS activity. Most of the segregation patterns of hygromycin resistance inheritance were formed in a Mendelian fashion (Table 2). In a small proportion of the tested lines, deviation from the expected segregation ratios may have resulted from the chimeric structure of the transgenic plants. The high cosegregation rate of gus and Bt toxin gene expression further confirmed the low degree of DNA rearrangement indicated by Southern analysis.
A number of strategies have been devised to increase the expression of Bt genes. These include the use of Arabidopsis thaliana small subunit leader and transit peptide to increase transcription and translation efficiency (27), the combination of the 35S promoter and the castor bean intron (4), and amplification of the toxin gene in chloroplasts (28) as well as modification of codon usage to match codon preference in plants (4–6, 10, 29, 30). In the present study, we attempted to achieve a high Bt toxin level in rice by using chimeric genes consisting of synthetic and modified Bt coding sequences and the strong maize ubiquitin promoter, which has been shown to direct a high level of reporter gene expression in monocot plants (31, 32). Although varying greatly among individual R0 plants, the toxin levels in 60% of the R0 transgenic plants were immunologically positive for CryIA(b) and CryIA(c). In ≈10% of these plants, the toxin levels were as high as 3% of the total soluble proteins. This is 10 to >100 times higher than the CryIA(b) and CryIA(c) contents in the previously reported transgenic rice plants (4–7). This is a significant advance because such high levels have been proposed as a necessary component of an effective integrated pest management program limiting build-up of insect resistance in transgenic crops (33). It is interesting to note that, in a previous study (6) using the maize ubiquitin promoter, the maximal level of expression achieved was only 0.024% total soluble protein. Some of the reasons for the differential performance of this promoter may include gene transfer methods, gene copy number, host genotype, numbers of transformants screened, and plant growth conditions, which certainly merit further investigation.
Feeding assays with R1 plants from three independent transformants confirmed that CryIA(b) and CryIA(c) proteins produced in the transgenic plants were highly toxic to SSB and YSB larvae. The larvae began to die 1 or 2 days after feeding on the transgenic stem tissues. A mortality of ≈100% was reached 4–5 days after infestation. The toxin levels in these transgenic plants were estimated to be 0.23–0.31% of the total soluble proteins at the time of feeding. Although a considerably lower CryIA(b) content (0.009% of soluble proteins) in transgenic rice was reported to confer 100% YSB and SSB mortality in one study (5), much higher levels of CryIA(b) and CryIA(c) (up to 0.05% and 0.024% of soluble proteins) have only resulted in 10–50% and 76–92% mortality for SSB (4) and YSB (6).
In our bioassays, it was observed that SSB and YSB feeding in the first 1–2 days on the transgenic stem tissues was very limited and did not cause significant damage to the plants. In fact, most of the larvae, particularly those of YSB, were found dead in the sheath tissue before they were able to penetrate into the stem. Taken together, these observations suggest that the toxin levels in these transgenics are sufficient to confer a high degree of SSB and YSB resistance in rice. Further investigation into the relationship between the toxin levels and insect mortality, and consequently insect resistance, is of great importance in establishing a suitable insect management strategy for this primal global food crop.
Acknowledgments
We dedicate this paper to Professor Brian S. Hartley, F.R.S., for his inspiration and guidance. Dr. T. Candresse, Institut National de la Recherche Agronomique Centre de Bordeaux, 33883 Villenave d’Ornon, France, supplied the plasmid pKHG4; Dr. P. H. Quail, Plant Gene Expression Center, U.S. Department of Agriculture, Berkeley, CA, supplied the ubiquitin promoter; Dr. E. E. Hood, Pioneer Hi-Bred International, Des Moines, Iowa, supplied A. tumefaciens strain EHA105; SmithKline Beecham Pharmaceuticals, Worthing, U.K., donated ticarcillin disodium; Rice Breeding Programs, Zhejiang Agricultural University, Hangzhou, China, and Dr. K. A. K. Moldenhauer, University of Arkansas Rice Research and Extension Center, Stuttgart, AR, kindly supplied rice seeds. We are grateful to: Dr. N. B. Carozzi, Ciba-Geigy, Research Triangle Park, NC, for the antibody; Drs. K. Nakamura, Nagoya University, Nagoya, Japan, and T. Komari, Japan Tobacco, Toyoda, Japan, for advice; Dr. M. Cohen, International Rice Research Institute, Los Banos, The Philippines, for SSB and YSB eggs and advice on insect bioassay. Expert technical assistance came from C. Sauder, D. Bowness, L. Vaters, and K. Dang. This work was supported by The Rockefeller Foundation and Natural Sciences and Engineering Research Council. An International Foundation for Science grant (C/1345–2), a Rockefeller Foundation postdoctoral fellowship, and a United Nations Educational, Scientific, and Cultural Organization fellowship to X.C. are greatly appreciated.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: SSB, striped stem borer; YSB, yellow stem borer; GUS, β-glucuronidase; DIG, digoxigenin; T-DNA, portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells.
References
- 1.Xiao J, Grandillo S, Ahn S N, McCouch S R, Tanksley S D. Nature (London) 1996;384:223–224. [Google Scholar]
- 2.Hofte H, Whiteley H R. Microbiol Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herdt R W. In: Rice Biotechnology. Khush G S, Toenniessen G H, editors. Wallingford, U.K.: CAB International; 1991. pp. 19–54. [Google Scholar]
- 4.Fujimoto H, Itoh K, Yamamoto M, Kyozuka J, Shimamoto K. Bio/Technology. 1993;11:1151–1155. doi: 10.1038/nbt1093-1151. [DOI] [PubMed] [Google Scholar]
- 5.Wünn J, Klöti A, Burkhardt P K, Biswas G C G, Launis K, Iglesias V A, Potrykus I. Bio/Technology. 1996;14:171–176. doi: 10.1038/nbt0296-171. [DOI] [PubMed] [Google Scholar]
- 6.Nayak P, Basu D, Das S, Basu A, Ghosh D, Ramakrishnan N A, Ghosh M, Sen S K. Proc Natl Acad Sci USA. 1997;94:2111–2116. doi: 10.1073/pnas.94.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghareyazie B, Alinia F, Menguito C A, Rubia L G, de Palma J M, Liwanag E A, Cohen M B, Khush G S, Bennett J. Mol Breed. 1997;3:401–404. [Google Scholar]
- 8.Cheng X, Sardana R, Altosaar I. In: Methods in Biotechnology: Recombinant Proteins from Plants—Production and Isolation of Clinically Useful Compounds. Cunningham C, Porter A J R, editors. Totowa, NJ: Humana; 1997. , Chapter 1, Vol. 3, pp. 1–9. [Google Scholar]
- 9.Le Gall O, Torregrosa L, Danglot Y, Candresse T, Bouquet A. Plant Sci. 1994;102:161–170. [Google Scholar]
- 10.Sardana R, Dukiandjiev S, Giband M, Cheng X, Cowan K, Sauder C, Altosaar I. Plant Cell Rep. 1996;15:677–681. doi: 10.1007/BF00231923. [DOI] [PubMed] [Google Scholar]
- 11.Christensen A H, Quail P H. Transgenic Res. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- 12.Rothstein S J, Lahners K N, Lotstein R J, Carozzi N B, Jayne S M, Rice D A. Gene. 1987;53:153–161. doi: 10.1016/0378-1119(87)90003-5. [DOI] [PubMed] [Google Scholar]
- 13.Albani D, Sardana R, Robert L S, Altosaar I, Arnison P G, Fabijanski S F. Plant J. 1992;2:331–342. [PubMed] [Google Scholar]
- 14.Hiei Y, Ohta S, Komari T, Kumashiro T. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 15.Hoekema A, Hirsch P R, Hooykaas P J J, Schilperoort R A. Nature (London) 1983;303:179–180. [Google Scholar]
- 16.Hood E E, Helmer G L, Fraley R T, Chilton M-D. J Bacteriol. 1986;168:1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rueb S, Hensgens L A M. Rice Genet Newsl. 1989;6:168–169. [Google Scholar]
- 18.Verwoerd T C, Dekker B M M, Hoekema A. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Chen M-T, Lee T-M, Chang H-H. Plant Cell Physiol. 1992;33:577–583. [Google Scholar]
- 21.Aldemita R R, Hodges T K. Planta. 1996;199:612–617. [Google Scholar]
- 22.Dong J J, Teng W M, Buchholz W G, Hall T C. Mol Breed. 1996;2:267–276. [Google Scholar]
- 23.Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro K. Nat Biotechnol. 1996;14:745–750. doi: 10.1038/nbt0696-745. [DOI] [PubMed] [Google Scholar]
- 24.Park S H, Pinson S R M, Smith R H. Plant Mol Biol. 1996;32:1135–1148. doi: 10.1007/BF00041397. [DOI] [PubMed] [Google Scholar]
- 25.Rashid H, Yokoi S, Toriyama K, Hinata K. Plant Cell Rep. 1996;15:727–730. doi: 10.1007/BF00232216. [DOI] [PubMed] [Google Scholar]
- 26.Jin S, Komari T, Gordon M P, Nester E W. J Bacteriol. 1989;169:4417–4425. doi: 10.1128/jb.169.10.4417-4425.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong E Y, Hironaka C M, Fischhoff D A. Plant Mol Biol. 1992;20:81–93. doi: 10.1007/BF00029151. [DOI] [PubMed] [Google Scholar]
- 28.McBride K E, Svab Z, Schaaf D J, Hongan P S, Stalker D M, Maliga P. Bio/Technology. 1995;13:362–365. doi: 10.1038/nbt0495-362. [DOI] [PubMed] [Google Scholar]
- 29.Perlak F J, Deaton R W, Armstrong T A, Fuchs R L, Sims S R, Greenplate J T, Fischhoff D A. Bio/Technology. 1990;8:939–943. doi: 10.1038/nbt1090-939. [DOI] [PubMed] [Google Scholar]
- 30.Perlak F J, Fuchs R L, Dean D A, McPherson S L, Fischhoff D A. Proc Natl Acad Sci USA. 1991;88:3324–3328. doi: 10.1073/pnas.88.8.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toki S, Takamatsu S, Nojiri C, Ooba S, Anazi H, Iwata W, Christensen A H, Quail P H, Uchimiya H. Plant Physiol. 1992;100:1503–1507. doi: 10.1104/pp.100.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cornejo M J, Luth D, Blankenship K M, Anderson O D, Blechl A E. Plant Mol Biol. 1993;23:567–581. doi: 10.1007/BF00019304. [DOI] [PubMed] [Google Scholar]
- 33.Gould F. Biocontrol Sci Technol. 1994;4:451–461. [Google Scholar]