Abstract
In humans, loss-of-function mutations in the gene encoding Wnt1 inducible signaling pathway protein 3 (WISP3) cause the autosomal-recessive skeletal disorder progressive pseudorheumatoid dysplasia (PPD). However, in mice there is no apparent phenotype caused by Wisp3 deficiency or overexpression. Consequently, the in vivo activities of Wisp3 have remained elusive. We cloned the zebrafish ortholog of Wisp3 and investigated its biologic activity in vivo using gain-of-function and loss-of-function approaches. Overexpression of zebrafish Wisp3 protein inhibited bone morphogenetic protein (BMP) and Wnt signaling in developing zebrafish. Conditioned medium–containing zebrafish and human Wisp3 also inhibited BMP and Wnt signaling in mammalian cells by binding to BMP ligand and to the Wnt coreceptors low-density lipoprotein receptor–related protein 6 (LRP6) and Frizzled, respectively. Wisp3 proteins containing disease-causing amino acid substitutions found in patients with PPD had reduced activity in these assays. Morpholino-mediated inhibition of zebrafish Wisp3 protein expression in developing zebrafish affected pharyngeal cartilage size and shape. These data provide a biologic assay for Wisp3, reveal a role for Wisp3 during zebrafish cartilage development, and suggest that dysregulation of BMP and/or Wnt signaling contributes to cartilage failure in humans with PPD.
Introduction
Mutations in the secreted protein Wnt1 inducible signaling pathway protein 3 (Wisp3) cause the autosomal-recessive human skeletal disease progressive pseudorheumatoid dysplasia (PPD) (1). PPD is a spondylo-epiphyseal dysplasia that becomes clinically apparent between 3 and 8 years of age and leads to joint failure by young adulthood (2–5). Histologic examination of cartilage at the time of joint replacement surgery reveals changes that resemble common, end-stage osteoarthritis (2). Over 20 different mutations in WISP3 have been identified in patients with PPD, including exon-deletion, frameshift, nonsense, and missense mutations (1, 6, 7), which suggests that PPD results from a loss of WISP3 protein function.
Wisp3 is a member of the CCN family of secreted growth factors. This family is composed of 6 proteins in mammals: connective tissue growth factor (Ctgf), cysteine-rich protein 61 (Cyr61), nephroblastoma overexpressed (Nov), Wisp1, Wisp2, and Wisp3 (8, 9). The Wisp proteins derive their names from the initial identification of WISP1 as a gene whose expression increased in a Wnt1-transfected colon cancer cell line (10); WISP2 and WISP3 were identified by DNA sequence homology searching (10). All CCN family members have several conserved structural domains (Figure 1A). These domains share homology with other proteins including IGF-binding proteins (IGFBPs), von Willebrand factor (VWC; also known as a cysteine-rich domain), thrombospondin 1, and proteins that contain carboxyterminal cystine knots (CTs), such as Noggin, SOST, WISE, Cerberus, and members of the TGF-β superfamily (11–16). To date, only WISP3 among CCN family members has been linked to a Mendelian genetic human disease, although the phenotypes in mice with targeted disruption of Ctgf and Cyr61 have been reported previously (17, 18).
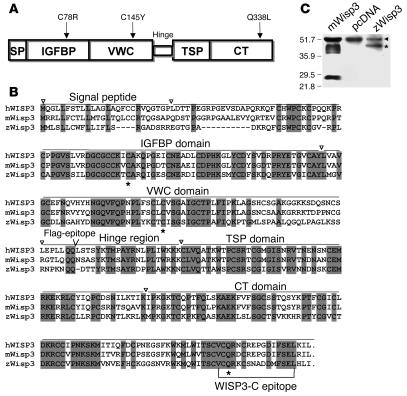
Figure 1. Structure and sequence of Wisp3.
(A) Domain structure of Wisp3, with locations of the 3 tested disease-causing PPD missense mutations noted. Signal peptide (SP), IGFBP, VWC, thrombospondin 1 (TSP), and CT domains are shown. (B) Alignment of Wisp3 amino acid sequence for hWISP3, mWisp3, and zWisp3. Fully conserved residues across these species are shaded. Asterisks denote the residues affected by PPD-causing missense mutations. Inverted triangles denote approximate domain boundaries. Bracketed region indicates the mouse polypeptide epitope against which the WISP3-C antibody was generated. (C) Western blot of HEK293T cell conditioned media containing recombinant mWisp3, control (pcDNA), or zWisp3, separated by 12% SDS-PAGE under reducing conditions and probed with the WISP3-C antibody. WISP3-C antibody detected mWisp3 and zWisp3 (asterisk) as well as a background band (arrowhead) unique to the HEK293T culture media. A single zWisp3 band (~45 kDa) was detected, whereas 3 mWisp3 isoforms (due to N-linked glycosylation) were detected along with a carboxyterminal cleavage fragment (~28 kDa).
Ex vivo studies have been performed with human Wisp3 (hWISP3) to try to identify a biologic activity (19–26). Human chondrocytic cell lines that transiently expressed wild-type hWISP3 had increased expression of type II collagen, aggrecan, and Sox9 mRNAs, as determined by semiquantitative RT-PCR, and increased production of type II collagen, as determined by Western blot (19). These same investigators used transient transfection and/or addition of purified recombinant protein to demonstrate an ability of WISP3 to regulate superoxide dismutase activity and the accumulation of reactive oxygen species (23). Another group has studied the role of WISP3 in cancer cells. Stably transfected mammary epithelial cancer cell lines expressing hWISP3 exhibited reduced proliferation, reduced anchorage-independent growth, and less activity in a cell invasion assay (22) and had altered responses to IGF and RhoC signaling (20, 21). When hWISP3-expressing tumor cell lines were implanted into nude mice, the tumors that formed were less aggressive (22). Stable inhibition of hWISP3 expression in human mammary epithelial cells using RNA inhibition caused the opposite effects (24). Most recently, human primary chondrocytes recovered from a single patient with PPD at the time of joint replacement surgery were noted to have increased cell proliferation and abnormal matrix metalloproteinase processing compared with wild-type chondrocytes from a single control patient (25). Whether any of these previously reported ex vivo biologic activities reflect endogenous in vivo functions of hWISP3 has not been tested.
To identify in vivo biologic activities for Wisp3, we cloned the ortholog in zebrafish (zwisp3). Herein we report that zwisp3 mRNA and its corresponding protein, zWisp3, were expressed in zebrafish embryos and larvae. We describe the phenotypes that resulted from RNA-mediated zWisp3 overexpression and morpholino-mediated zWisp3 depletion. We show that when overexpressed, wild-type zWisp3 antagonized BMP and canonical Wnt signaling, whereas zWisp3 mutants that cause PPD had either no or reduced inhibitory activity. We found that zWisp3 inhibited BMP signaling by binding to the ligand and modulated Wnt signaling by binding to the coreceptor complex. Finally, we report that zWisp3 morphant larvae had altered integrity of their pharyngeal cartilage, suggesting an in vivo link between zWisp3 function in zebrafish and humans.
Results
Cloning of zwisp3.
Using the hWISP3 protein sequence to query the zebrafish genome led to the identification of zWisp3 (Figure 1B). zwisp3 cDNA was cloned as an RT-PCR product from embryos at 24 hours post-fertilization (hpf). The predicted protein contained a signal peptide and other domains found in mammalian Wisp3 (Figure 1, A and B). zWisp3 and hWISP3 have 49% identity and 54% similarity at the amino acid level. zWisp3 has the greatest similarity to mammalian Wisp3 and much lower similarity to other CCN family members. Only a single zwisp3 locus was observed among the zebrafish genomic DNA and expressed sequence tag (EST) sequences available at GenBank. All PPD-causing missense mutations that have been identified to date (C78R in the IGFBP domain, C145Y in the VWC domain, and Q338L in the CT domain) alter amino acid residues that are highly evolutionarily conserved (Figure 1, A and B). A polyclonal antibody, WISP3–C, that recognizes a highly conserved epitope in hWISP3 and mouse Wisp3 (mWisp3) (Figure 1B), was also able to immunodetect zWisp3 (Figure 1C and Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI32001DS1).
zWisp3 is expressed during zebrafish development.
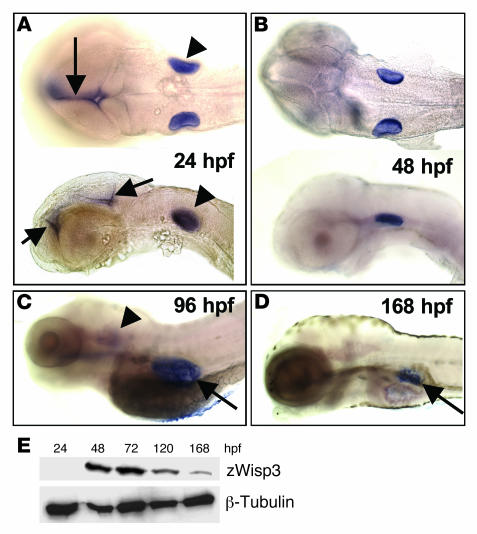
The zwisp3 mRNA was first detected by RT–PCR at 6 hpf (data not shown) and by whole-mount in situ hybridization at 24 hpf in the otic vesicles and the brain (Figure 2A). Expression of zwisp3 persisted in the otic vesicles until 96 hpf (Figure 2C); however, expression in the brain disappeared after 48 hpf (Figure 2B). Expression of zwisp3 was also observed in the swim bladder at 96 hpf and persisted in this structure until at least 168 hpf (Figure 2, C and D). These results contrast with previous studies in humans and mice in which mRNA expression was only detected by RT-PCR (1, 10, 27). While zwisp3 mRNA was detected by in situ hybridization at 24 hpf, zWisp3 protein expression was detected by Western blot at 48 hpf (Figure 2E). These results also contrast with previous studies in mice in which endogenous mWisp3 protein was undetectable prenatally and postnatally (27).
Figure 2. Expression of endogenous zWisp3.
(A–D) Whole-mount in situ hybridization with an antisense zwisp3 RNA probe during early embryonic development. (A) Dorsal (top) and lateral (bottom) views of 24-hpf embryos demonstrated zwisp3 mRNA expression in the otic vesicle (arrowheads) and midline brain (arrows). (B) Ventral and lateral views of 48-hpf embryos demonstrated persistent zwisp3 expression in the otic vesicles. (C) Lateral view of 96-hpf larva showed diminished expression in the otic vesicle (arrowhead) and new expression in the developing swim bladder (arrow). (D) Lateral view of 168-hpf larva demonstrated persistent expression in the swim bladder (arrow). No zwisp3 expression was observed by in situ hybridization in embryos prior to 24 hpf (not shown). (E) Western blot of zWisp3 protein extracted from deyolked embryos and larvae. Protein (60 μg) was separated by 10% SDS-PAGE under reducing conditions and immunodetected using WISP3-C antibody or anti–β-tubulin antibody as a control. zwisp3 can be detected by RT-PCR at 6 hpf (not shown), by in situ hybridization at 24 hpf, and zWisp3 by Western blot at 48 hpf.
Morpholino-induced depletion of zWisp3 affects mandibular and pharyngeal cartilage development.
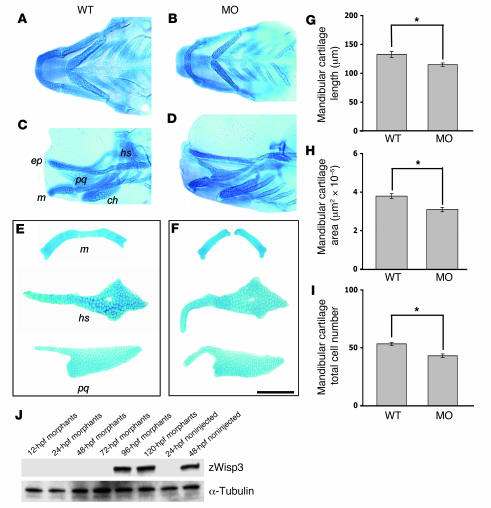
A zwisp3 antisense morpholino that targets the translation start site and a zwisp3 splice-site morpholino that targets zwisp3 mRNA splicing each interfered with the normal production of zWisp3. Each morpholino caused zWisp3 to be undetectable by Western blot until at least 72 hpf (Figure 3J), whereas a mismatched control morpholino had no effect (data not shown). Knockdown of zWisp3 reduced the size of the mandibular cartilage and altered other pharyngeal cartilages (Figure 3, A–I). Of note, the pharyngeal cartilages appeared normal in situ in the morphants (Figure 3, B and D) but assumed abnormal shapes and were noticeably more fragile when recovered during microdissection (Figure 3F).
Figure 3. Pharyngeal cartilage changes in 108-hpf morphants and quantitative morphometric analysis of mandibular cartilage from wild-type and morphant larvae.
(A–D) Ventral (A and B) and lateral (C and D) images of Alcian blue–stained pharyngeal cartilages in intact wild-type and morphant larva (24 ng zwisp3 translation blocking morpholino; MO). The ethmoid plate (ep), mandibular (m), palatoquadrate (pq), hyosympletic (hs), and ceratohyal (ch) cartilages are indicated. (E and F) Representative images of dissected Alcian blue–stained mandibular, palatoquadrate, and hyosympletic cartilages from wild-type and morphant larvae. Note zWisp3 morphant larvae had a smaller mandibular cartilage and abnormally shaped palatoquadrate and hyosympletic cartilages. During microdissection, these cartilages were noticeably more fragile in morphant compared with wild-type larvae. Similar findings were observed for larvae injected with the zwisp3 splice-site morpholino. Scale bar: 100 μm. (G) Mandibular length in morphants significantly decreased compared with wild-type fish. (H) Mandibular surface area in morphants significantly decreased compared with wild-type fish. (I) The number of chondrocytes per hemimandible was significantly fewer in morphants compared with wild-type fish. (J) Western blot of zWisp3 protein extracted from deyolked embryos and larvae. Protein (60 μg) was separated by 10% SDS-PAGE under reducing conditions and immunodetected using WISP3-C antibody or anti–α-tubulin antibody as a control. Note the morphants had no immunodetectable zWisp3 until 96 hpf, while zWisp3 was detected by 48 hpf in noninjected controls. *P < 0.01.
zWisp3 overexpression disrupts dorsal-ventral patterning in developing zebrafish.
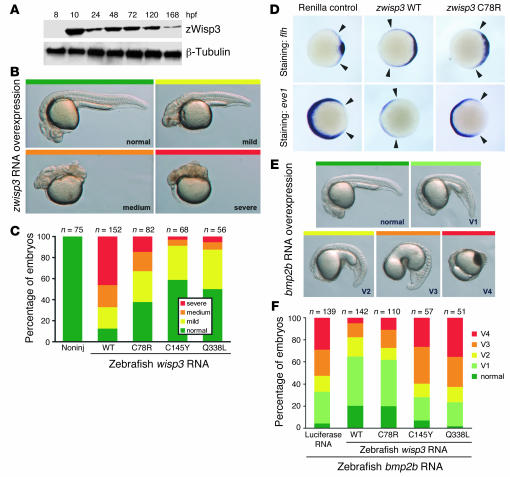
The results of the morpholino experiments indicated a function for zWisp3 in pharyngeal cartilage development, but did not suggest a specific biologic pathway. To identify pathways in which zWisp3 might function, we overexpressed zWisp3. Injection of 150 pg zwisp3 RNA into fertilized eggs produced zWisp3 expression before endogenous protein normally appears (Figure 4A and Supplemental Figure 1A). Intriguingly, zWisp3 overexpression caused a dramatic decrease of ventral, and a concomitant expansion of dorsal, cell fates (Figure 4, B and C).
Figure 4. Overexpression of zWisp3 in zebrafish inhibits BMP signaling.
(A) Western blot of zWisp3 protein extracted from deyolked embryos and larvae injected with 150 pg zwisp3 RNA. Protein (40 μg) was separated by 10% SDS-PAGE under reducing conditions and detected using WISP3-C or anti–β-tubulin antibody. zWisp3 protein was detectable by 10 hpf when overexpressed. (B) Photographs of live 24-hpf embryos depicting the phenotypic classes of dorsalization. Embryos classified as mild correspond to convergence-extension classes C1 and C2, medium to C3 and C4, and severe to C5, as previously described (30). (C) Phenotype frequencies (shown in B) caused by injection of wild-type, C78R, C145Y, and Q338L zwisp3 RNA. Mutants had reduced biologic activity compared with wild-type zWisp3. (D) Photographs of 60% epiboly embryos oriented with their dorsal domains to the right. Arrowheads indicate extent of strong expression. Expression of flh was strongly expanded in wild-type zwisp3–injected embryos and less dramatically so in zwisp3 C78R–injected embryos. Expression of eve1 was strongly reduced in wild-type zwisp3–injected embryos and less dramatically reduced in C78R-injected embryos. (E) Photographs of live 24-hpf embryos depicting phenotypic classes of ventralization caused by injection of zbmp2b RNA as previously reported (30). (F) Phenotype frequencies (shown in E) caused by injection of zbmp2b RNA plus equimolar amounts of control (luciferase), zwisp3 wild-type, or zwisp3 missense mutant RNAs. C145Y and Q338L did not inhibit BMP signaling, while wild-type and C78R RNAs substantially rescued the defects caused by zbmp2b overexpression.
This dorsalizing effect was less severe when zwisp3 RNAs containing PPD-associated missense mutations (C78R, C145Y, and Q338L) were injected (Figure 4C), even though the expression of the mutant proteins was comparable to that of wild-type zWisp3 (Supplemental Figure 1B); this indicates that the biologic activity of zWisp3 was mediated at the protein level and not at the mRNA level. To confirm that zWisp3 overexpression changed patterning of the developing embryo along the dorsal-ventral axis, we assayed expression of genes specific for dorsal and ventral domains at early gastrulation, when the dorsal-ventral axis is established. zWisp3 overexpression caused a severe expansion of dorsal cell fates, as seen by expanded expression of floating head (flh), and a dramatic reduction of ventral cell fates, as indicated by reduced expression of even-skipped-like1 (eve1; Figure 4D).
zWisp3 inhibits BMP signaling.
Patterning of the embryo along the dorsal-ventral axis during gastrulation is controlled by BMP and Wnt signaling pathways (28). In particular, loss of BMP signaling causes very similar phenotypes (29, 30) to those of zWisp3 overexpression, suggesting that zWisp3 may regulate BMP signaling. We tested this by coexpressing zWisp3 and zbmp2b in developing zebrafish. Injection of zbmp2b RNA caused expansion of ventral and loss of dorsal cell fates (Figure 4E) (30). Coinjection of zwisp3 with zbmp2b markedly reduced the severity of the phenotype induced by zbmp2b alone (Figure 4F). The missense mutations in the VWC and CT domains (C145Y and Q338L, respectively) had little BMP inhibitory activity, whereas the IGFBP domain missense mutant (C78R) retained some ability to dorsalize embryos (Figure 4C) and was better able to prevent the ventralizing effect of zbmp2b overexpression (Figure 4F). We concluded that Wisp3 can inhibit BMP signaling.
zWisp3 inhibits canonical Wnt signaling.
Wisp3 contains a CT domain, as do Cerberus, WISE, and SOST, which have previously been shown to regulate canonical Wnt signaling (14, 16, 31), as have 2 CCN family members, Cyr61 and Ctgf (32, 33). Therefore, we tested whether zWisp3 inhibits Wnt signaling in addition to BMP signaling. Others have previously reported that overexpression of zWnt8 in developing zebrafish antagonizes the formation of anterior head structures (34–36). We confirmed this effect (Figure 5, C and D) and also found that overexpression of wild-type human low-density lipoprotein receptor–related protein 6 (hLRP6) or a constitutively active form of this receptor (hLRP6ΔN) caused a similar phenotype (Figure 5, A and B).
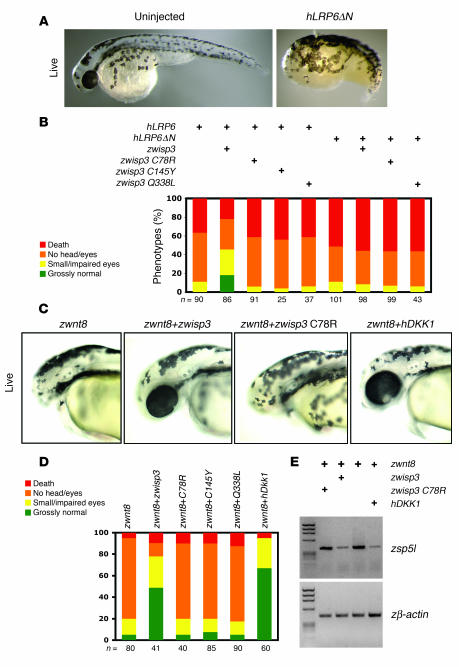
Figure 5. Overexpression of zWisp3 in zebrafish modulates canonical Wnt signaling.
(A) Photograph of a 30-hpf live embryo injected with an RNA encoding a constitutively active form of hLRP6 (hLRP6ΔN). Note the deficient formation of anterior structures compared with the uninjected control. (B) Quantification of the phenotypic classes that result from injecting wild-type hLRP6 or hLRP6ΔN with wild-type, C78R, C145Y, or Q338L forms of zwisp3. Note the severity of the hLRP6-induced phenotype was reduced when wild-type zWisp3, but not missense mutant zWisp3, was coexpressed. Also note that wild-type zWisp3 did not reduce phenotypic severity caused by expression of hLRP6ΔN. (C) Photographs of live 30-hpf embryos that had been co-injected with zwnt8 RNA and either wild-type or C78R mutant zwisp3 RNA, or hDKK1 RNA. Note wild-type zwisp3 and hDKK1 rescued the zWnt8-induced loss-of-anterior-structure phenotype. (D) Quantification of the phenotypic effects of overexpressing zWnt8 with or without wild-type or missense mutant zWisp3 or hDKK1. Note the severity of the zWnt8-induced phenotype was reduced by coexpressing wild-type zWisp3 or hDKK1, but not missense mutant zWisp3 (C78R, C145Y, and Q338L). (E) Agarose gels containing RT-PCR amplimers of zebrafish sp5l (zsp5l) and zebrafish β-actin (zβ-actin) using template RNA extracted from 80% epiboly-stage embryos injected with zWnt8 and either wild-type or C78R zwisp3 or hDKK1. Note that zWnt8-induced expression of zsp5l was reduced only when wild-type zwisp3 or hDKK1 were co-injected.
Coinjection of wild-type zwisp3 with hLRP6 or with zwnt8 rescued the phenotype, whereas coinjection of the 3 PPD-associated missense mutants did not (Figure 5, B and D). To determine the site of zWisp3 inhibition, we coinjected wild-type zwisp3 with a constitutively active form of hLRP6 that lacks the extracellular domain (hLRP6ΔN). In this experiment the head formation defect was not rescued (Figure 5B), indicating that zWisp3 likely affects extracellular rather than intracellular canonical Wnt signaling components. We also tested whether zWisp3 interfered with the ability of Wnt8 to induce expression of zsp5l, which is directly regulated by canonical Wnt signaling (37). Indeed, coinjection of wild-type zwisp3 and zwnt8 decreased the level of zsp5l expression compared with that in embryos that were injected only with zwnt8 (Figure 5E). Taken together, the results of these experiments show that zWisp3 was able to inhibit BMP and Wnt signaling in zebrafish.
Wisp3 inhibits BMP and Wnt signaling in mammalian cells.
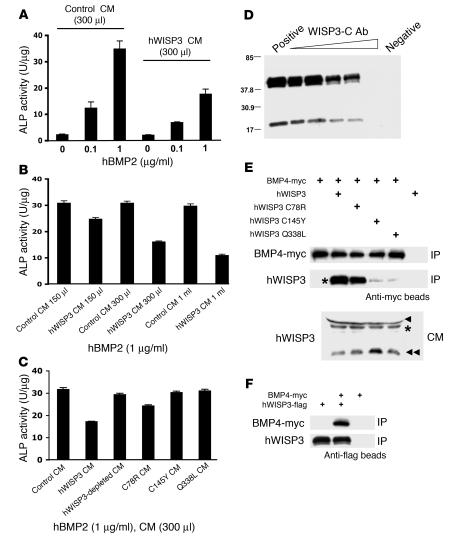
To determine whether the BMP inhibitory activity of Wisp3 observed in zebrafish occurs in mammalian cells, we added recombinant hBMP2 to mouse 10T1/2 cells grown in control conditioned medium (CM) or in CM containing hWISP3. It was previously shown that 10T1/2 cells have increased alkaline phosphatase (ALP) activity following hBMP2 stimulation (38). Consistent with Wisp3 being a BMP inhibitor, hWISP3-containing CM, but not control CM, inhibited the hBMP2-induced increase in ALP activity (Figure 6, A–C). The ability of hWISP3-containing CM to inhibit BMP signaling was reduced when hWISP3 was immunodepleted from the medium (Figure 6, C and D), indicating that hWISP3 is a direct inhibitor of BMP signaling. We performed co-IP experiments to determine whether hWISP3 inhibited signaling by physically interacting with BMP, similar to the mechanism by which Noggin acts as a BMP inhibitor (39). Wild-type hWISP3 efficiently coprecipitated with mBMP4, whereas the hWISP3 missense mutants C145Y and Q338L, which had the least dorsalizing activity in the zebrafish BMP assays, did not (Figure 6, E and F). Interestingly, the C78R missense mutant retained some dorsalizing activity in the zebrafish assay and also retained the ability to coprecipitate mBMP4 (Figure 6E).
Figure 6. hWISP3 inhibits BMP2 signaling in mammalian cells and physically interacts with mBMP4.
(A) ALP activity in 10T1/2 cells cultured with recombinant hBMP2. hBMP2 induced ALP activity in a dose-dependent manner, and this induction was reduced when hWISP3-containing CM versus control CM was used. (B) hBMP2-mediated ALP induction in the presence of increasing amounts of hWISP3-containing CM. hWISP3 inhibited ALP induction dose dependently. (C) ALP activity in the presence of hBMP2 and 300 μl control CM, hWISP3 CM, immunodepleted hWISP3 CM, or PPD-associated mutant CM. The largest reduction in ALP activity occurred with hWISP3 CM. (D) Western blot of hWISP3 supernatants from CM mixed with protein G beads coated with increasing amounts of WISP3-C antibody. WISP3-C antibody dose-dependently immunodepleted hWISP3 protein. Positive control is supernatant after beads uncoated with antibody were used; negative control is eluate from these uncoated beads. (E) Western blots of protein precipitated with anti-myc beads probed with either anti-myc or WISP3-C antibody. Wild-type hWISP3 and the C78R missense mutant efficiently coprecipitated with mBMP4, whereas the C145Y and Q338L mutants did not. Input hWISP3 containing CM used in the co-IP experiments is shown at bottom. Asterisks indicate full-length hWISP3; arrowhead indicates a cross-reacting band observed only in CM from HEK293T cells; and double arrowhead indicates a hWISP3 cleavage product found in CM from HEK293T cells. The cleavage product did not coprecipitate with mBMP4 (not shown). (F) Western blots of protein precipitated with anti-flag beads probed with either anti-myc or WISP3-C antibody. mBMP4 coprecipitated with wild-type hWISP3.
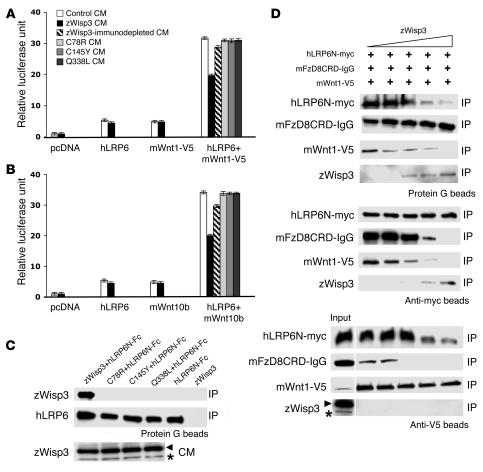
To determine whether Wisp3 also has Wnt-inhibitory activity in mammalian cells, we transiently transfected human HEK293T cells with expression plasmids TopFlash (firefly luciferase under the control of a canonical Wnt signaling–responsive promoter), hLRP6, and either mWnt1 tagged with V5 (mWnt1-V5) or mWnt10b and then tested whether the Wnt-induced increase in luciferase differed among cells cultured in control CM, zWisp3-containing CM, or CM containing either of 3 PPD-associated missense mutants. CM containing wild-type zWisp3, but not CM containing the 3 PPD-associated missense mutants or control CM, reduced luciferase activity (Figure 7, A and B). Because zWisp3 was unable to inhibit a constitutively active form of hLRP6 in zebrafish, we hypothesized that Wisp3 would inhibit Wnt signaling by binding either to Wnt ligand or to the Wnt coreceptors. To further characterize how Wisp3 inhibits Wnt signaling, we performed co-IP experiments using zWisp3 and components of the Wnt co-receptor complex: Wnt1-v5, mouse frizzled 8 (mFzD8CRD-IgG), and hLRP6N-myc, similar to what has previously been reported for the inhibitors DKK1 and WISE (14, 40). zWisp3 did not interact directly with mWnt1-V5 (Figure 7D); however, it interacted under low-stringency conditions (washing in TBST) with mFzD8CRD-IgG (Figure 7D) and interacted under high-stringency conditions (washing in RIPA buffer) with hLRP6N-Fc and hLRP6N-myc (Figure 7, C and D). None of the 3 PPD-associated missense mutants interacted with the extracellular domain of hLRP6 (Figure 7C). We also mixed CM containing hLRP6N-myc and mFzD8CRD-IgG with mWnt1-V5 in the presence of increasing amounts of zWisp3 (Figure 7D). Each component was then immunoprecipitated and coprecipitation of other components was assessed. zWisp3 interfered with the ability of mWnt1-V5 to form a trimeric complex with hLRP6N-myc and mFzD8CRD-IgG (Figure 7D). These data suggest that zWisp3 inhibits canonical Wnt signaling by binding to hLRP6 and mFzD8, preventing them from serving as Wnt receptors. We also tested the possibility that Wisp3 could interfere with Wnt signaling by binding to and promoting the internalization of LRP6 from the cell surface with another transmembrane protein, Kremen2, similar to the mechanism of action of the protein DKK1 (41). However, in surface biotinylation experiments we found that Wisp3 did not promote LRP6 internalization (Supplemental Figure 2).
Figure 7. zWisp3 physically and biologically interacts with the Wnt coreceptors LRP6 and FzD8.
(A) Ratio of firefly luciferase activity to Renilla luciferase activity in HEK293T cells transfected with TopFlash, pRL-TK, and either empty vector (pcDNA), hLRP6, mWnt1-V5, or hLRP6 plus mWnt1-V5, and then cultured in either control CM, wild-type zWisp3, zWisp3-immunodepleted CM, or PPD-associated missense mutant CM. Wild-type zWisp3 CM, but not the immunodepleted or the 3 zWisp3 mutant CMs, reduced luciferase activity. (B) Same experimental design as in A, except mWnt10b was used instead of mWnt1-V5. (C) Western blots of protein precipitated with protein G beads from mixtures of hLRP6N-Fc and zWisp3 containing CM, and probed with either WISP3-C or anti-mouse IgG antibody. Western blot of CM immunodetected with WISP3-C antibody (bottom) demonstrated comparable expression of wild-type and missense mutant zWisp3. Only wild-type zWisp3 coprecipitated with hLRP6. WISP3-C antibody detected zWisp3 (asterisk) as well as a background band (arrowhead) unique to HEK293T culture medium. (D) Western blots of individual components of the mWnt1-V5/hLRP6N-myc/mFzD8CRD-IgG complex following their IP in the presence of increasing amounts of zWisp3. hLRP6N-myc was immunoprecipitated and immunodetected using an anti-myc antibody. FzD8CRD-IgG was precipated with protein G and immunodetected using anti-mouse IgG antibody. mWnt1-V5 was immunoprecipitated and immunodetected using anti-V5 antibody, and zWisp3 was immunodetected using WISP3-C antibody. Increased zWisp3 interfered with the ability of the mWnt1-V5/hLRP6N-myc/mFzD8CRD-IgG trimeric complex to form. Asterisk in the input column of the lower panel indicates zWisp3, with the upper band representing a background band (arrowhead) unique to HEK293T culture medium.
Discussion
We have identified in vivo biologic activities for Wisp3 using zebrafish. These insights are important because the role of Wisp3 in mammals has been difficult to determine. Although mutations in WISP3 have been found in patients with PPD, a skeletal disease associated with precocious cartilage failure, affected tissue has only become available after it is histologically indistinguishable from common, end-stage osteoarthritis (2). Importantly, we showed that Wisp3 missense mutants that are disease-causing in humans had lost or reduced biologic activities in zebrafish. In mice, neither Wisp3 deficiency nor Wisp3 overexpression caused a discernable phenotype (ref. 27 and Y. Nakamura, unpublished observations). The lack of a mouse model of PPD and our inability to detect endogenous Wisp3 mRNA or Wisp3 protein expression in mammalian tissues has limited our ability to interrogate its biologic function.
zwisp3 mRNA was detectable by whole-mount in situ hybridization and zWisp3 protein was detectable by Western blot during embryonic development (Figure 2). Overexpression of zWisp3 during early zebrafish development revealed an ability to inhibit BMP and Wnt signaling (Figures 4 and 5). We did not observe the opposite phenotype when zWisp3 was depleted using morpholinos, which is not surprising because there was no detectable endogenous zWisp3 protein expression during gastrulation.
The ability of hWISP3 and zWisp3 to inhibit BMP and Wnt signaling was also observed in mammalian cells (Figures 6 and 7). We found that hWISP3 and mBMP4 physically interacted, suggesting an inhibitory mechanism involving direct ligand binding. Consistent with this hypothesis is the finding that the C78R missense mutant retained its ability to bind mBMP4 and its ability to inhibit BMP signaling, whereas the C145Y and Q338L mutants lost both abilities. Another CCN family member, Ctgf, has also previously been shown to physically interact with BMP and to inhibit BMP signaling (42), indicating that this biologic activity may be conserved within the CCN family. Rather than binding Wnt ligand to inhibit signaling, zWisp3 instead bound the coreceptors LRP6 and FzD8, binding most strongly to LRP6 (Figure 7). Two other CCN family members, Cyr61 and Ctgf, have also been shown to modulate canonical Wnt signaling in Xenopus laevis (32, 33), again suggesting conserved biologic activity within the CCN family.
Importantly, we demonstrated that missense mutations in zWisp3, which are associated with the human disease PPD, affected the BMP and Wnt inhibitory functions in zebrafish. Missense mutations in the IGFBP, VWC, and CT domains each had comparable effects on Wnt signaling, implying that proper folding of multiple domains is necessary for the protein’s Wnt-inhibitory function. In contrast, the C78R mutation had a milder effect on BMP signaling, indicating that the IGFBP domain may be dispensable for this biologic activity. However, it is premature to speculate whether this difference points to Wisp3 having a more important role in regulating the Wnt pathway than the BMP pathway in humans.
Depletion of zWisp3 using either translation blocking or a splice-site morpholino caused mandibular cartilage undergrowth and affected the shape and strength of other pharyngeal cartilages (Figure 3). These effects were unexpected because whole-mount in situ hybridization did not identify zwisp3 expression in chondrocytes. Chondrocytes could be expressing zwisp3 at levels detectable only by RT-PCR, similar to what has been observed in humans and mice, or the in situ hybridization protocol may not have had sufficient sensitivity to detect expression in cartilage. BMP signaling is important during zebrafish pharyngeal cartilage morphogenesis, as demonstrated by the malformations observed in zebrafish with mutations in smad5, a cytoplasmic effector of the BMP pathway (43). In mammals, BMP signaling regulates cartilage development (44) and homeostasis (45), the latter being demonstrated by the occurrence of precocious osteoarthritis in mice with postnatal disruption of a BMP receptor (45). In humans, genetic evidence implicates subtle perturbations in the regulation of BMP and related signaling pathways as risk factors for common osteoarthritis (46, 47). Wnt signaling has also been implicated in cartilage differentiation (48, 49) and skeletal homeostasis (50). Mouse models of precocious osteoarthritis in the context of altered Wnt signaling have not yet been described, although disturbed skeletal homeostasis has been reported in mice lacking the Wnt coreceptor Lrp5 (51, 52). In humans, coding variants in secreted Wnt inhibitors have been associated with common osteoarthritis (53, 54), and studies of human articular cartilage explants detect Wnt and BMP signaling responses to mechanical stress (55). Future research is needed to determine whether the pharyngeal cartilage fragility in the zWisp3 morphants relates to the precocious osteoarthritis observed in humans with PPD and whether both are consequences of perturbed BMP and/or Wnt signaling.
Biologic activities for Wisp3 have previously been sought ex vivo using mammalian systems (19–26). Some studies suggest that Wisp3 deficiency might cause PPD by reducing the normal expression of several cartilage matrix proteins or the ability of chondrocytes to handle reactive oxygen species (19, 23). The fragility of the pharyngeal cartilage elements in the morphants could reflect altered abundance of structural matrix components. Formal testing of this hypothesis in zebrafish morphants must await the development of proteomic and/or immunohistochemical methods that can detect differences in tissue structure and composition. Surprisingly, no apparent defects in skeletal growth or matrix production were observed in the Wisp3 knockout mice (27), although there is precedent for mutations that cause recognizable skeletal phenotypes in humans being nonpenetrant or exceedingly subtle in mice (56). Other studies suggested that Wisp3 controls tumor cell growth and invasiveness (20–22, 24). The specificity of these effects is difficult to determine given the absence of increased rates of cancer in humans with PPD and in mice with targeted disruption of mWisp3, although studies using mice with increased predisposition to cancer may be illuminating.
Multiple signaling pathways are involved in complex developmental processes. For example, during gastrulation, Wnt and BMP signaling are required for specification of ventral fates (28, 57). Specification of dorsal cell fates depends on the action of secreted inhibitors of these pathways. In addition, some inhibitors affect multiple pathways, such as Cerberus, which inhibits BMP and Wnt signaling during the formation of the vertebrate head (16). BMP and Wnt signaling are both required during skeletal differentiation, growth, and homeostasis, and we have shown that zWisp3 has the ability to modulate both pathways. Given the ability of zWisp3 to inhibit BMP and Wnt signaling, when overexpressed in zebrafish or when added to mammalian cell cultures, it is also surprising that Wisp3 overexpression in mice causes no phenotype. There are several potential explanations for the interspecies difference in the biologic activity of overexpressed Wisp3 in vivo. These include differences in the protein’s posttranslational modification, abundance, stability, or interacting partners. One inter-species difference we observed in this study is that zWisp3 existed as a full-length protein when expressed in HEK293T cells, whereas hWISP3 or mWisp3 protein was often cleaved, leading to the production of smaller carboxyterminal fragments (Figure 1C and Figure 6, D and E). These smaller fragments, while abundant in CM, did not coprecipitate with either mBMP4 or hLRP6, which suggests that they are not biologically active inhibitors. Similar cleavage of overexpressed protein in transgenic mice may render the protein nonfunctional. Whereas studies in mice have not been revealing, the zebrafish assay will be useful in determining whether other missense mutations in patients suspected of having PPD are disease-causing or rare, benign polymorphisms.
Methods
Zebrafish maintenance.
Developing and adult zebrafish were maintained using standard methods (58). Embryos from natural matings were staged as described previously (59). Wild-type matings used the Oregon AB line. To suppress pigmentation, embryos were raised in zebrafish aquatic system water containing 1-phenyl-2-thiourea (0.003%; Sigma-Aldrich). Zebrafish care and experiments were approved by the Institutional Animal Care and Use Committee of Case Western Reserve University.
Cloning of zWisp3.
The hWISP3 protein sequence (GenBank accession no. AAC96323.1) was used to query zebrafish genomic DNA sequences in GenBank using the program tblastx (www.ncbi.nlm.nih.gov/BLAST/). A zebrafish bacterial artificial chromosome (BAC; GenBank accession no. AL954744) that contains exons encoding a protein with high amino acid similarity to hWISP3 was identified. Of note, the genes tube1 and lama5, which are syntenic with WISP3 in humans and mice, were also present on this zebrafish BAC. The sequence from the BAC was used to design PCR primers to amplify zwisp3 cDNA using mRNA from 24-hpf wild-type embryos as template for RT-PCR. Total RNA was extracted using Isogen (Nippon Gene Co.) and converted to cDNA using the TaKaRa RNA Kit version 3.0 (Takara Bio Inc.) according to the manufacturer’s protocol. Primers used to amplify the full-length zwisp3 cDNA coding sequence were 5′-CTCCTTTGCAAGCCGTGT-3′ (forward) and 5′-GCATTGTGTTCTTCCCGTTT-3′ (reverse), the 3′ ends of which correspond to immediately upstream of the putative translation start site and 5 bases downstream of the putative translation stop site. The PCR product of the full-length zwisp3 cDNA was cloned into the TOPO cloning vector (Invitrogen). Individual clones were sequenced and compared with each other and with zebrafish EST and BAC sequences in GenBank. This enabled us to identify PCR-induced mutations and high-throughput sequencing errors and to determine the correct zwisp3 sequence.
Monitoring mRNA expression by RT-PCR and whole-mount in situ hybridization.
Total RNA from 0-, 5-, 6-, 8-, 11-, 19-, and 24-hpf embryos was extracted as described above. Forward and reverse primers to amplify between exons of zwisp3 were used to detect spliced transcripts. Primers used to amplify zebrafish sp5l were 5′-CCCAGCTCATCTCCAGAAAG-3′ (forward) and 5′-GAGCTCTTCCTGAGGGGTTT-3′ (reverse), and those for zebrafish β-actin were 5′-GTTTTCCCCTCCATTGTTGGAC-3′ (forward) and 5′-CAGGATCTTCATCAGGTAGTCTGTCA-3′ (reverse).
Whole-mount in situ hybridization was carried out with 4-, 9-, 14-, 24-, 48-, 72-, 96-, 120-, and 168-hpf embryos and larvae as previously described (60). Primers used to synthesize sense and antisense zwisp3 probes were 5′-GCTATGAGGTCGGCATCTGT-3′ (forward) and 5′-GGACCAAACAGTTCTTCTTCCA-3′ (reverse). The origin of eve1 and flh cDNA probes are described by the Zebrafish Model Organism Database (http://zfin.org). Digoxigenin-labeled RNA probes were generated using a T7 and T3 labeling kit (Roche) according to the manufacturer’s instructions. The signals were detected with an ALP-conjugated anti-digoxigenin antibody (Roche) and visualized using 4-nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate. The reaction was stopped by replacing substrate with several rinses of PBT (PBS plus 0.1% Tween 20); embryos were stored in PBS containing 0.02% sodium azide in the dark at 4°C until they were photographed.
Generation of expression constructs and in vitro transcription of RNAs.
Full-length zwisp3 was cloned into the pCS2+ expression vector. Three different amino acid substitutions, corresponding to human mutations found in PPD (C78R, C145Y, and Q338L), were incorporated into the expression vector by site-directed mutagenesis (Figure 1, A and B, and Supplemental Figure 1). These same mutations were also incorporated into a hWISP3 expression vector by site-directed mutagenesis. All clones were sequenced to confirm that only the intended mutations were incorporated into the constructs.
Other cDNA constructs used in this study included full-length hLRP6 (61), a chimeric protein containing the extracellular domain of hLRP6 linked to the Fc fragment of the IgG heavy chain (hLRP6N-Fc) (62), the extracellular domain of hLRP6 linked to a myc epitope tag (61), a constitutively active form of hLRP6 lacking the extracellular domain (hLRP6ΔN) (61, 63), MESD-C2 (64), zwnt8 (37), hDKK1 (40), TopFlash, pRL-TK, mWnt1-V5 (64), mWnt10b (64), mKremen2 (from B. Williams, Van Andel Research Institute, Grand Rapids, Michigan, USA), mouse IgG-tagged Frizzled 8 (mFzD8CRD-IgG) (40), mouse Frizzled 4 (mFzD4) (65), myc-tagged mBMP4 (mBMP4-myc) (66), and mSPC4 (66). Constructs were cloned into pcDNA3.1 and/or pCS2+ for mammalian cell and zebrafish assays, respectively.
RNAs were synthesized from NotI-digested pCS2+ plasmids using the sp6 mMessage mMachine kit (Ambion). Phenol red (0.1%) was added to the RNA solution as a tracer, and RNAs were injected into 1-cell-stage embryos. Following injection, embryos were cultured in aquatic system water and analyzed until 168 hpf.
Immunodetection of zWisp3.
A rabbit polyclonal antibody, WISP3-C (27), that had been generated against a highly conserved polypeptide epitope within the CT domains of hWISP3 and mWisp3 was tested for cross-reactivity to zWisp3. mWisp3 and zWisp3 cDNAs were cloned into the pcDNA3.1 expression vector (pcDNA; Invitrogen) and transfected into 80% confluent HEK293T cells (ATCC) using LipofectAmine Plus Reagent (Invitrogen) following the manufacturer’s protocol. At 4 hours after transfection, 6.5 ml DMEM (Cellgro) supplemented with 20% FBS was added to the cultures. The following day, 10 ml DMEM containing 10% FBS was added for 24 hours, followed by 24 hours incubation in 5 ml serum free-DMEM. Conditioned serum-free medium was spun at 16,000 g for 5 minutes. Supernatant (125 μl) was acetone precipitated (5:1 acetone/medium ratio) overnight at –20°C. Following centrifugation at 16,000 g for 5 minutes, the acetone was decanted and the pellet permitted to dry prior to being resuspended in 50 μl of 1× SDS-PAGE loading buffer at 55°C for 10 minutes. β-Mercaptoethanol (BME) was added to the samples (5% v/v) prior to boiling for 5 minutes. Each sample (25 μl) was separated by SDS-PAGE, transferred to Immobilon P membrane (Millipore Corp.), and blocked with 4% nonfat dry milk in Tris-buffered saline (TBS) at 4°C overnight. The blot was then incubated in 4% nonfat dry milk in TBS, 0.05% Tween 20 (TBST) containing WISP3-C antibody. Goat anti-rabbit HRP-conjugated antibody was used as a secondary antibody (Pierce). ECL plus Western blotting Detection System (Amersham Biosciences) and X-OMAT AR film (Eastman Kodak Company) were used to detect signal.
Immunodetection of zWisp3 in zebrafish embryo extracts.
Embryos were dechorionated and deyolked and then flash frozen in a methanol/dry ice bath for 5 minutes. Ten embryos were pooled together and then lysed on ice for 20 minutes in 200 μl Triton X-100 lysis buffer (20 mM Tris-HCl, pH 8.0, 137 mM NaCl, 1% Triton X-100, 1 mM EGTA, 10% glycerol, 1.5 mM MgCl2, 1 mM dithiothreitol, 1 mM sodium vanadate, 50 mM sodium fluoride, and proteinase inhibitor mixture; Roche). The protein concentration of the lysates was determined by Bradford assay (67) (Bio-Rad). Protein (40 or 60 μg) was then separated by SDS-PAGE and immunodetected using the WISP3-C antibody, as described above.
Morpholinos to knock down endogenous zWisp3 expression.
An antisense morpholino oligonucleotide (5′-CAGCACAGTAGTGATAGCATCATAC-3′) against the translation initiation codon of zwisp3 and a splice-site blocking morpholino (5′-GTAAGTAGATAGGCTCACTCACAAG-3′) were designed and produced by Gene Tools LLC. As a negative control, a 5-base-mispaired morpholino (5′-CACCACACTACTGATACCATGATAC-3′) was used. Morpholinos were dissolved in 1× Danieau’s buffer (68) to concentrations of 1–12 mg/ml. Then, 1 or 2 nl of morpholino was injected into 1-cell-stage embryos.
Visualization of zebrafish phenotypes.
Following injection of morpholino or RNA, the embryos were maintained at 28.5°C in aquatic system water and then removed at different time points for visual inspection and photography. Photographs were obtained by anesthetizing embryos and larvae with 0.02% tricaine (MS222; Sigma-Aldrich) and then transferring them onto glass slides containing 3% methylcellulose. A Zeiss Axioplan 2 imaging microscope with an Axiocam camera and Open Lab software were used, and embryos and larvae were returned to the incubator in aquatic system water and allowed to develop further until they were again removed for visualization and photography.
Cartilage staining and analysis.
Anesthetized 108-hpf larvae were fixed in 4% paraformaldehyde at 4°C overnight. Larvae were rinsed in several washes of PBS and transferred into a 0.1% Alcian blue solution dissolved in 80% ethanol and 20% glacial acetic acid at room temperature overnight. After staining, the larvae were rinsed in ethanol and rehydrated gradually into PBS. They were then transferred into a solution of 1% KOH and 3% H2O2 for 1 hour to bleach all pigment. After another wash with PBS, tissues were cleared in fresh 0.05% trypsin dissolved in saturated sodium borate for 1 hour. The specimens were stored in 50% glycerol, 50% PBS, and 0.25% KOH at room temperature until they became well cleared. After photography of the entire larvae, the mandibular arch cartilages were dissected free from surrounding tissue, mounted in 75% glycerol, and sealed under coverslips. The images of the stained, flat-mounted cartilages were captured using a Leica DMLB upright microscope and Spot camera (Diagnostic Instruments Inc.). The Spot image analysis program was used to measure the length and surface area of the individual cartilages. The total number of chondrocytes per mandibular cartilage element was counted by visual inspection. Changes in the thickness of the cartilage plate were not quantified.
Generation of Wisp3-containing CM.
Wild-type hWISP3- and zWisp3-containing CM were generated by transiently transfecting HEK293T cells as previously described. CM was stored at –20°C until use. CM was similarly generated for the missense mutants (C78R, C145Y, and Q338L) in the human and zebrafish proteins and using an empty vector alone (control CM).
Bioassay for BMP signaling in mammalian cells.
BMP signaling bioassays were performed using 10T1/2 cells as previously described (38). Briefly, cells were plated in 6-well plates (5 × 104 cells/well), and recombinant hBMP2 (R&D Systems) was added to the culture medium. Cells were then cultured in 1.7 ml DMEM (10% FBS) and 300 μl CM obtained from empty vector–transfected or hWISP3-transfected HEK293T cells. This culture medium was changed every other day until the cells were harvested on day 6. The 10T1/2 cells were washed twice with ice-cold PBS, lysed in ALP assay buffer (10 mmol/l Tris-HCl, pH 7.5, 1 mmol/l MgCl2, and 0.5% NP-40), and centrifuged at 1,000 g for 10 minutes; the supernatants were then used for ALP and protein assays. ALP activity was measured with the Lab Assay ALP kit (Wako Chemicals USA) per the manufacturer’s protocol. Total protein content was determined with the BCA Protein Assay kit (Pierce) using bovine albumin as a standard. Results are expressed as units ALP activity per μg total cell protein. Experiments were performed in triplicate on 2 separate occasions.
A second BMP inhibition experiment was performed by culturing 10T1/2 cells in the presence of a single concentration of hBMP2 (1 μg/ml) and increasing amounts of control CM (150 μl, 300 μl, or 1 ml per 2 ml total culture medium) or hWISP3-containing CM (150 μl, 300 μl, or 1 ml per 2 ml total culture medium). Medium was again changed every other day, and cells were harvested and ALP activity was determined as described above.
Reporter assay for canonical Wnt signaling in mammalian cells.
HEK293T cells were plated at 2 × 105 cells/well in 24-well plates 24 hours prior to transfection. Cells were transfected by LipofectAmine Plus (Invitrogen) with the expression vectors TopFlash (100 ng), pRL-TK (5 ng), MESD-C2 (20 ng), hLRP6 (20 ng), and either mWnt1-V5 (100 ng) or mWnt10b (100 ng) in 250 μl serum-free medium according to the manufacturer’s protocol. As needed, empty vector was added to make a total transfected DNA amount of 295 ng. At 3 hours following transfection, 150 μl of control CM or CM containing wild-type, C78R, C145Y, or Q338L zWisp3 was added to the these cells along with 50 μl serum-free DMEM and 50 μl 100% FCS. Cells were harvested 21 hours later for determination of protein expression and luciferase activity. Cells were lysed in 100 μl passive lysis buffer (Promega), and firefly (TopFlash) luciferase activity and Renilla (pRL-TK) luciferase activity were measured using the dual luciferase assay kit (Promega) in a luminometer (Molecular Devices) following the manufacturers’ protocols. Renilla luciferase activity served as an internal control for transfection efficiency. Wnt-induced signaling via the canonical pathway is depicted as the ratio between firefly and Renilla luciferase activity. All experiments were performed in triplicate and repeated on 3 separate occasions.
Immunodepletion of hWISP3 from CM.
Protein G beads (50 μl) were incubated and rotated with 0 μl, 20 μl, 100 μl, 200 μl, 500 μl, or 1 ml of affinity purified WISP3-C polyclonal antibody at 4°C overnight. The next day, after washing the beads twice with PBS, 500 μl of hWISP3 CM was added to each sample, followed by rotation at 4°C overnight. After bead removal by centrifugation at 10,000 g for 5 minutes, the remaining supernatants were used for the assays. Immunodepletion was confirmed by testing aliquots of the supernatants for hWISP3 by Western blot.
Co-IP.
For BMP co-IP, COS7 cells were cultured in DMEM supplemented with 10% FBS. Cells were plated at 2 × 105 cells/well in 6-well culture plates 24 hours prior to transfection. Cells were cotransfected with 2 μg mBMP4-myc plasmid and 0.5 μg mSPC4 plasmid as described previously (66). At 24 hours after transfection, the culture medium was changed to 1 ml OPTIMEM (Invitrogen) containing 1% Knockout SR (Invitrogen). CM was collected 48 hours later and stored frozen at –20°C until use. mBMP4-myc–containing CM (500 μl) was mixed with 1 ml wild-type, C78R, C145Y, or Q338L hWISP3-containing CM and 65 μl anti-myc beads (Santa Cruz Biotechnology Inc.). The samples were rocked for more than 16 hours at 4°C. Beads were pelleted, washed 4 times at 4°C for 5 minutes each in RIPA buffer (50 mM Tris, pH 8, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, and 0.1% SDS), and then resuspended in 65 μl 2× SDS-PAGE loading buffer with 5% BME. After boiling for 5 minutes, 20 μl were separated by SDS-PAGE, transferred to Immobilon P, and immunodetected using anti-myc and WISP3-C antibodies. In addition, an hWISP3 construct containing a Flag epitope in the hinge region was used to generate hWISP3-Flag–containing CM in HEK293T cells. hWISP3-Flag-containing CM (1 ml) was mixed with 500 μl mBMP4-myc CM and 100 μl beads coated with anti-Flag M2 monoclonal antibody (Sigma-Aldrich). The samples were rocked for 4 hours at 4°C, and the beads were processed as described above.
For hLRP6N-Fc/zWisp3 and mWnt1-V5/hLRP6N-myc/mFzD8CRD-IgG/zWisp3 co-IP, HEK293T cells were cultured in DMEM supplemented with 10% FCS. Cells were plated at 2 × 105 cells/well in 6-well culture plates 24 hours prior to transfection. Cells were transfected with either 1 μg each of expression plasmids for hLRP6N-Fc and MESD-C2; hLRP6N-myc and MESD-C2; wild-type or C78R, C145Y, or Q338L missense mutant zWisp3; or mFzD8CRD-IgG using LipofectAmine Plus per the manufacturer’s protocol (Invitrogen). At 24 hours after transfection, the culture medium was changed to 1 ml serum-free DMEM. CM was collected 24 hours later and stored frozen at –20°C until use. hLRP6N-Fc-containing CM (100 μl) was mixed with 100 μl of CM containing either wild-type zWisp3 or the missense mutant forms of zWisp3, and 50 μl of protein G Sepharose beads (Amersham Pharmacia Biotech). mWnt1-V5–containing CM was generated similarly, except that FuGENE6 (Roche) was used and L cells were transfected (64). hLRP6N-myc-containing CM (1 ml), 1.6 ml mWnt1-V5–containing CM, and 600 μl mFzD8CRD-IgG–containing CM were mixed and then 0 μl, 100 μl, 500 μl, 1 ml, or 5 ml of CM containing wild-type zWisp3 was added. To equalize the total volume in the IP assay, control CM was added. After the conditioned media were mixed, components were immunoprecipitated by adding either 50 μl protein G Sepharose beads (Amersham Pharmacia Biotech), 65 μl anti-myc antibody–coated beads (Santa Cruz Biotechnology Inc.), or 25 μl anti-V5 antibody–coated beads (Bethyl Laboratory Inc.). Samples were rocked at 4°C for 3 hours. The antibody-coated beads were then pelleted and washed 3 times at 4°C for 5 minutes in either RIPA buffer (high stringency) or TBST (low stringency).
Beads were pelleted and resuspended in 60 μl 2× SDS-PAGE loading buffer containing 5% BME. After boiling for 5 minutes, 20 μl were then resolved by SDS-PAGE. Following transfer to Immobilon P, wild-type zWisp3 protein was probed by WISP3-C antibody, hLRP6N-Fc by HRP-conjugated goat anti-mouse IgG antibody (Pierce), hLRP6-myc protein by HRP-conjugated anti-myc antibody (Santa Cruz Biotechnology), mFzD8CRD-IgG protein by goat anti-mouse IgG antibody (Pierce), and mWnt1-V5 protein by anti-V5-HRP antibody (Invitrogen). Immunoreactive bands were detected using the ECL detection kit as described above.
Internalization assay for hLRP6.
A human LRP6-EGFP–expressing HEK293T stable cell line (kindly provided by T. Biechele, University of Washington School of Medicine, Seattle, Washington, USA) was seeded onto 6-well plates coated with poly-L-lysine (BD) at ~90% confluence. The following day the cells were transfected with plasmids encoding mKremen2 (0.5 μg), full-length mFzD4 (0.5 μg), and mWnt1-V5 (0.5 μg). The total DNA amount for all transfections was brought to 1.5 μg with empty pcDNA3.1, and transfections were performed using LipofectAmine following the manufacturer’s recommendation (Invitrogen). At 24 hours later, the culture medium was switched to medium containing 500 μl hDDK1-flag CM, wild-type zWisp3 or C145Y zWisp3 CM, 100 μl FCS, and 400 μl serum-free DMEM. Cells were harvested from individual wells after 0, 10, and 40 minutes. Cells were washed 3 times in ice-cold PBS, and cell surface proteins were biotinylated with 0.5 mg/ml noncleavable sulfo-NHS-biotin in PBS (Pierce) for 30 minutes at room temperature. Unreacted biotin was removed by briefly washing the cells 3 times with TBS containing 10% w/v glycine. Membrane proteins were extracted in 250 μl cold RIPA buffer, and protein concentration was determined by Bradford assay. Total cellular hLRP6-EGFP in each sample was determined by SDS-PAGE separation of 10 μg of protein from the whole cell lysate followed by immunodetection using an anti-GFP antibody (Sigma-Aldrich). As a loading control, blots were reprobed with an anti–β-tubulin antibody (Sigma-Aldrich). From the remaining lysates, 250 μg total protein from each sample was immunoprecipitated using 5 μg/ml anti-GFP antibody and 50 μl protein G–sepharose beads (Amersham Pharmacia Biotech). Immunoprecipitates were washed 5 times with RIPA buffer and eluted with 40 μl SDS-PAGE sample buffer. Eluates (20 μl) were separated by SDS-PAGE and immunodetected with an anti-biotin monoclonal antibody (Sigma-Aldrich). Blots were reprobed with the anti-GFP antibody.
Statistics.
A 1-tailed Student’s t test, with a P value of less than 0.01, was used when considering differences in mandibular cartilage length, surface area, and chondrocyte number between wild-type and morphant larva.
Supplementary Material
Acknowledgments
We thank Daniel B. Constam for providing BMP4-myc and SPC4 plasmids; Xi He for providing hLRP6 and hLRP6ΔN plasmids; Bart Williams for providing mouse Kremen 2 plasmid; Travis Biechele for sharing the LRP6-EGFP stable cell line; members of the Warman, Liang, and Moon laboratories for sharing their scientific expertise; and Bruce B. Riley and Richard Harland for their thoughtful suggestions. K. Tamai was a postdoctoral fellow in Xi He’s laboratory and supported by a NIH grant to Xi He. M.L. Warman is a recipient of a Clinician Scientist Award in Translational Research from the Burroughs Wellcome Fund. Y. Nakamura and G. Weidinger are associates and R.T. Moon and M.L. Warman are investigators of the Howard Hughes Medical Institute.
Footnotes
Nonstandard abbreviations used: ALP, alkaline phosphatase; BMP, bone morphogenetic protein; CM, conditioned medium; CT, carboxyterminal cystine knot; Ctgf, connective tissue growth factor; Cyr61, cysteine-rich protein 61; h-, human; hpf, hour post-fertilization; IGFBP, IGF-binding protein; LRP6, low-density lipoprotein receptor–related protein 6; m-, mouse; PPD, progressive pseudorheumatoid dysplasia; VWC, von Willebrand factor; Wisp, Wnt inducible signaling pathway protein; z-, zebrafish.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:3075–3086 (2007). doi:10.1172/JCI32001
Yukio Nakamura and Matthew L. Warman’s present address is: Howard Hughes Medical Institute, Orthopaedic Research Laboratories, and Department of Orthopaedic Surgery, Children’s Hospital Boston, Boston, Massachusetts, USA.
References
- 1.Hurvitz J.R., et al. Mutations in the CCN gene family member WISP3 cause progressive pseudorheumatoid dysplasia. Nat. Genet. 1999;23:94–98. doi: 10.1038/12699. [DOI] [PubMed] [Google Scholar]
- 2.
- 3.Spranger J., Albert C., Schilling F., Bartsocas C. Progressive pseudorheumatoid arthropathy of childhood (PPAC): a hereditary disorder simulating juvenile rheumatoid arthritis. Am. J. Med. Genet. 1983;14:399–401. doi: 10.1002/ajmg.1320140224. [DOI] [PubMed] [Google Scholar]
- 4.Wynne-Davies R., Hall C., Ansell B.M. Spondylo-epiphysial dysplasia tarda with progressive arthropathy. A “new” disorder of autosomal recessive inheritance. J. Bone Joint Surg. Br. 1982;64:442–445. doi: 10.1302/0301-620X.64B4.6807993. [DOI] [PubMed] [Google Scholar]
- 5.el-Shanti H.E., Omari H.Z., Qubain H.I. Progressive pseudorheumatoid dysplasia: report of a family and review. J. Med. Genet. 1997;34:559–563. doi: 10.1136/jmg.34.7.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehl S., et al. Clinical, radiographic, and genetic diagnosis of progressive pseudorheumatoid dysplasia in a patient with severe polyarthropathy. Rheumatol. Int. 2004;24:53–56. doi: 10.1007/s00296-003-0341-0. [DOI] [PubMed] [Google Scholar]
- 7.Delague V., et al. Molecular study of WISP3 in nine families originating from the Middle-East and presenting with progressive pseudorheumatoid dysplasia: identification of two novel mutations, and description of a founder effect. Am. J. Med. Genet. A. 2005;138:118–126. doi: 10.1002/ajmg.a.30906. [DOI] [PubMed] [Google Scholar]
- 8.Rachfal A.W., Brigstock D.R. Structural and functional properties of CCN proteins. Vitam. Horm. 2005;70:69–103. doi: 10.1016/S0083-6729(05)70003-0. [DOI] [PubMed] [Google Scholar]
- 9.Leask A., Abraham D.J. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J. Cell Sci. 2006;119:4803–4810. doi: 10.1242/jcs.03270. [DOI] [PubMed] [Google Scholar]
- 10.Pennica D., et al. WISP genes are members of the connective tissue growth factor family that are up-regulated in wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14717–14722. doi: 10.1073/pnas.95.25.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brigstock D.R. The CCN family: a new stimulus package. J. Endocrinol. 2003;178:169–175. doi: 10.1677/joe.0.1780169. [DOI] [PubMed] [Google Scholar]
- 12.Perbal B. CCN proteins: multifunctional signalling regulators. Lancet. 2004;363:62–64. doi: 10.1016/S0140-6736(03)15172-0. [DOI] [PubMed] [Google Scholar]
- 13.Brunkow M.E., et al. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am. J. Hum. Genet. 2001;68:577–589. doi: 10.1086/318811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itasaki N., et al. Wise, a context-dependent activator and inhibitor of Wnt signalling. Development. 2003;130:4295–4305. doi: 10.1242/dev.00674. [DOI] [PubMed] [Google Scholar]
- 15.Avsian-Kretchmer O., Hsueh A.J. Comparative genomic analysis of the eight-membered ring cystine knot-containing bone morphogenetic protein antagonists. Mol. Endocrinol. 2004;18:1–12. doi: 10.1210/me.2003-0227. [DOI] [PubMed] [Google Scholar]
- 16.Piccolo S., et al. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature. 1999;397:707–710. doi: 10.1038/17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivkovic S., et al. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development. 2003;130:2779–2791. doi: 10.1242/dev.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mo F.E., et al. CYR61 (CCN1) is essential for placental development and vascular integrity. Mol. Cell. Biol. 2002;22:8709–8720. doi: 10.1128/MCB.22.24.8709-8720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sen M., Cheng Y.H., Goldring M.B., Lotz M.K., Carson D.A. WISP3-dependent regulation of type II collagen and aggrecan production in chondrocytes. Arthritis Rheum. 2004;50:488–497. doi: 10.1002/art.20005. [DOI] [PubMed] [Google Scholar]
- 20.Kleer C.G., Zhang Y., Pan Q., Merajver S.D. WISP3 (CCN6) is a secreted tumor-suppressor protein that modulates IGF signaling in inflammatory breast cancer. Neoplasia. 2004;6:179–185. doi: 10.1593/neo.03316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleer C.G., et al. WISP3 and RhoC guanosine triphosphatase cooperate in the development of inflammatory breast cancer. Breast Cancer Res. 2004;6:R110–R115. doi: 10.1186/bcr755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleer C.G., et al. WISP3 is a novel tumor suppressor gene of inflammatory breast cancer. Oncogene. 2002;21:3172–3180. doi: 10.1038/sj.onc.1205462. [DOI] [PubMed] [Google Scholar]
- 23.Miller D.S., Sen M. Potential role of WISP3 (CCN6) in regulating the accumulation of reactive oxygen species. Biochem. Biophys. Res. Commun. 2007;355:156–161. doi: 10.1016/j.bbrc.2007.01.114. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y., Pan Q., Zhong H., Merajver S.D., Kleer C.G. Inhibition of CCN6 (WISP3) expression promotes neoplastic progression and enhances the effects of insulin-like growth factor-1 on breast epithelial cells. Breast Cancer Res. 2005;7:R1080–R1089. doi: 10.1186/bcr1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou H.D., et al. Cellular and molecular responses in progressive pseudorheumatoid dysplasia articular cartilage associated with compound heterozygous WISP3 gene mutation. J. Mol. Med. 2007 doi: 10.1007/s00109-007-0193-2. In press. [DOI] [PubMed] [Google Scholar]
- 26.Davis L., Chen Y., Sen M. WISP-3 functions as a ligand and promotes superoxide dismutase activity. Biochem. Biophys. Res. Commun. 2006;342:259–265. doi: 10.1016/j.bbrc.2006.01.132. [DOI] [PubMed] [Google Scholar]
- 27.Kutz W.E., Gong Y., Warman M.L. WISP3, the gene responsible for the human skeletal disease progressive pseudorheumatoid dysplasia, is not essential for skeletal function in mice. Mol. Cell. Biol. 2005;25:414–421. doi: 10.1128/MCB.25.1.414-421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schier A.F., Talbot W.S. Molecular genetics of axis formation in zebrafish. Annu. Rev. Genet. 2005;39:561–613. doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- 29.Hild M., et al. The smad5 mutation somitabun blocks Bmp2b signaling during early dorsoventral patterning of the zebrafish embryo. Development. 1999;126:2149–2159. doi: 10.1242/dev.126.10.2149. [DOI] [PubMed] [Google Scholar]
- 30.Kishimoto Y., Lee K.H., Zon L., Hammerschmidt M., Schulte-Merker S. The molecular nature of zebrafish swirl: BMP2 function is essential during early dorsoventral patterning. Development. 1997;124:4457–4466. doi: 10.1242/dev.124.22.4457. [DOI] [PubMed] [Google Scholar]
- 31.Semenov M.V., He X. LRP5 mutations linked to high bone mass diseases cause reduced LRP5 binding and inhibition by SOST. J. Biol. Chem. 2006;281:38276–38284. doi: 10.1074/jbc.M609509200. [DOI] [PubMed] [Google Scholar]
- 32.Mercurio S., Latinkic B., Itasaki N., Krumlauf R., Smith J.C. Connective-tissue growth factor modulates WNT signalling and interacts with the WNT receptor complex. Development. 2004;131:2137–2147. doi: 10.1242/dev.01045. [DOI] [PubMed] [Google Scholar]
- 33.Latinkic B.V., et al. Xenopus Cyr61 regulates gastrulation movements and modulates Wnt signalling. Development. 2003;130:2429–2441. doi: 10.1242/dev.00449. [DOI] [PubMed] [Google Scholar]
- 34.Lekven A.C., Thorpe C.J., Waxman J.S., Moon R.T. Zebrafish wnt8 encodes two wnt8 proteins on a bicistronic transcript and is required for mesoderm and neurectoderm patterning. Dev. Cell. 2001;1:103–114. doi: 10.1016/s1534-5807(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 35.Hashimoto H., et al. Zebrafish Dkk1 functions in forebrain specification and axial mesendoderm formation. Dev. Biol. 2000;217:138–152. doi: 10.1006/dbio.1999.9537. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu T., Bae Y.K., Muraoka O., Hibi M. Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev. Biol. 2005;279:125–141. doi: 10.1016/j.ydbio.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Thorpe C.J., Weidinger G., Moon R.T. Wnt/beta-catenin regulation of the Sp1-related transcription factor sp5l promotes tail development in zebrafish. Development. 2005;132:1763–1772. doi: 10.1242/dev.01733. [DOI] [PubMed] [Google Scholar]
- 38.Katagiri T., et al. The non-osteogenic mouse pluripotent cell line, C3H10T1/2, is induced to differentiate into osteoblastic cells by recombinant human bone morphogenetic protein-2. Biochem. Biophys. Res. Commun. 1990;172:295–299. doi: 10.1016/s0006-291x(05)80208-6. [DOI] [PubMed] [Google Scholar]
- 39.Zimmerman L.B., De Jesus-Escobar J.M., Harland R.M. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 40.Semenov M.V., et al. Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr. Biol. 2001;11:951–961. doi: 10.1016/s0960-9822(01)00290-1. [DOI] [PubMed] [Google Scholar]
- 41.Mao B., Niehrs C. Kremen2 modulates Dickkopf2 activity during Wnt/LRP6 signaling. Gene. 2003;302:179–183. doi: 10.1016/s0378-1119(02)01106-x. [DOI] [PubMed] [Google Scholar]
- 42.Abreu J.G., Ketpura N.I., Reversade B., De Robertis E.M. Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat. Cell Biol. 2002;4:599–604. doi: 10.1038/ncb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimmel C.B., Miller C.T., Moens C.B. Specification and morphogenesis of the zebrafish larval head skeleton. Dev. Biol. 2001;233:239–257. doi: 10.1006/dbio.2001.0201. [DOI] [PubMed] [Google Scholar]
- 44.Yoon B.S., et al. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5062–5067. doi: 10.1073/pnas.0500031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rountree R.B., et al. BMP receptor signaling is required for postnatal maintenance of articular cartilage. PLoS Biol. 2004;2:e355. doi: 10.1371/journal.pbio.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kizawa H., et al. An aspartic acid repeat polymorphism in asporin inhibits chondrogenesis and increases susceptibility to osteoarthritis. Nat. Genet. 2005;37:138–144. doi: 10.1038/ng1496. [DOI] [PubMed] [Google Scholar]
- 47.Miyamoto Y., et al. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat. Genet. 2007;39:529–533. doi: 10.1038/2005. [DOI] [PubMed] [Google Scholar]
- 48.Spater D., et al. Wnt9a signaling is required for joint integrity and regulation of Ihh during chondrogenesis. Development. 2006;133:3039–3049. doi: 10.1242/dev.02471. [DOI] [PubMed] [Google Scholar]
- 49.Hill T.P., Spater D., Taketo M.M., Birchmeier W., Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev. Cell. 2005;8:727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 50.Gong Y., et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 51.Clement-Lacroix P., et al. Lrp5-independent activation of Wnt signaling by lithium chloride increases bone formation and bone mass in mice. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17406–17411. doi: 10.1073/pnas.0505259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kato M., et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J. Cell Biol. 2002;157:303–314. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loughlin J., et al. Functional variants within the secreted frizzled-related protein 3 gene are associated with hip osteoarthritis in females. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9757–9762. doi: 10.1073/pnas.0403456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lane N.E., et al. Frizzled-related protein variants are risk factors for hip osteoarthritis. Arthritis Rheum. 2006;54:1246–1254. doi: 10.1002/art.21673. [DOI] [PubMed] [Google Scholar]
- 55.Dell’Accio F., et al. Activation of WNT and BMP signaling in adult human articular cartilage following mechanical injury. Arthritis Res. Ther. 2006;8:R139. doi: 10.1186/ar2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aszodi A., Bateman J.F., Gustafsson E., Boot-Handford R., Fassler R. Mammalian skeletogenesis and extracellular matrix: what can we learn from knockout mice? Cell Struct. Funct. 2000;25:73–84. doi: 10.1247/csf.25.73. [DOI] [PubMed] [Google Scholar]
- 57.Ramel M.C., Buckles G.R., Baker K.D., Lekven A.C. WNT8 and BMP2B co-regulate non-axial mesoderm patterning during zebrafish gastrulation. Dev. Biol. 2005;287:237–248. doi: 10.1016/j.ydbio.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 58. Westerfield, M. 1994.The zebrafish book: a guide for the laboratory use of zebrafish. University of Oregon Press. Eugene, Oregon, USA. 385 pp. [Google Scholar]
- 59.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 60.Thisse C., Thisse B., Schilling T.F., Postlethwait J.H. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 61.Tamai K., et al. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 62.Holmen S.L., Salic A., Zylstra C.R., Kirschner M.W., Williams B.O. A novel set of Wnt-Frizzled fusion proteins identifies receptor components that activate beta -catenin-dependent signaling. J. Biol. Chem. 2002;277:34727–34735. doi: 10.1074/jbc.M204989200. [DOI] [PubMed] [Google Scholar]
- 63.Tamai K., et al. A mechanism for Wnt coreceptor activation. Mol. Cell. 2004;13:149–156. doi: 10.1016/s1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- 64.Ai M., et al. Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass-associated missense mutations in LRP5 affect canonical Wnt signaling. Mol. Cell. Biol. 2005;25:4946–4955. doi: 10.1128/MCB.25.12.4946-4955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blitzer J.T., Nusse R. A critical role for endocytosis in Wnt signaling. BMC Cell Biol. 2006;7:28. doi: 10.1186/1471-2121-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Constam D.B., Robertson E.J. Regulation of bone morphogenetic protein activity by pro domains and proprotein convertases. J. Cell Biol. 1999;144:139–149. doi: 10.1083/jcb.144.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 68.Nasevicius A., Ekker S.C. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 2000;26:216–220.. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.