Abstract
The molecular mechanisms of differential pattern formation along the left/right (L/R) axis in the nervous system are poorly understood. The nervous system of the nematode Caenorhabditis elegans displays several examples of L/R asymmetry, including the directional asymmetry displayed by the two ASE taste receptor neurons, ASE left (ASEL) and ASE right (ASER). Although bilaterally symmetric in regard to all known morphological criteria, these two neurons display distinct chemosensory capacities that correlate with the L/R asymmetric expression of three putative sensory receptor genes, gcy-5, expressed only in ASER, and gcy-6 and gcy-7, expressed only in ASEL. In order to understand the genetic basis of L/R asymmetry establishment, we screened for mutants in which patterns of asymmetric gcy gene expression are disrupted, and we identified a cascade of several symmetrically and asymmetrically expressed transcription factors that are sequentially required to restrict gcy gene expression to either the left or right ASE cell. These factors include the zinc finger transcription factor che-1; the homeobox genes cog-1, ceh-36, and lim-6; and the transcriptional cofactors unc-37/Groucho and lin-49. Specific features of this regulatory hierarchy are sequentially acting repressive interactions and the finely balanced activity of antagonizing positive and negative regulatory factors. A key trigger for asymmetry is the L/R differential expression of the Nkx6-type COG-1 homeodomain protein. Our studies have thus identified transcriptional mediators of a putative L/R-asymmetric signaling event and suggest that vertebrate homologs of these proteins may have similar functions in regulating vertebrate brain asymmetries.
Keywords: C. elegans, left/right asymmetry, transcription factor, homeobox
The overall body plan of most adult animals is largely bilaterally symmetric (Ludwig 1932). However, specific deviations from this bilateral symmetry are apparent at two different levels. First, individual organs such as the heart, stomach, and spleen are placed in a left/right (L/R) asymmetric manner in many organisms (Ludwig 1932). Second, organs or morphological features that display an inherent pattern of bilateral symmetry can show defined deviations from symmetry. Such symmetry breakages are evident in the structure of seemingly bilaterally symmetric nervous systems of several species, from invertebrates to humans. In vertebrates, neuronal L/R asymmetry is evident in the different sizes of bilaterally positioned structures, such as the left and right temporal lobes of the human brain (Galaburda 1991) or the diencephalic habenular nuclei in several amphibian brains (Ludwig 1932). More recently, L/R-specific gene expression patterns have been observed in the brains of several vertebrate species (Ramsdell and Yost 1998; Mercola and Levin 2001) and, in zebrafish, have been shown to correlate with the determination of L/R-specific features of diencephalic structures (Concha et al. 2000; Essner et al. 2000; Liang et al. 2000). Smaller sized invertebrate brains, in which individual cell types can be more easily identified and characterized, also reveal L/R asymmetries in otherwise largely bilaterally symmetric ganglia. These asymmetries include L/R-specific positioning of individual cells, L/R-specific cell death, and L/R-specific gene expression profiles (for review, see Hobert et al. 2002). Of the two most widely used invertebrate model systems that are amenable to genetic studies, Drosophila melanogaster and Caenorhabditis elegans, neuronal L/R asymmetries have only been observed in C. elegans, thus making it the prime choice to genetically dissect the development of neuronal L/R asymmetry.
Evidence in vertebrates and the nematode C. elegans suggests that bilateral neuronal structures may diversify with a L/R-specific bias as a means to increase the functional capacities of the nervous system. In humans, certain brain functions, such as language and attention, are highly lateralized (Davidson and Hugdahl 1994). Nervous system functions in other vertebrate species, including rodents (Glick and Ross 1981) and fish (Miklosi et al. 1997), also display a lateral bias. In C. elegans, laterality has been observed in chemosensory capacities, specifically in the AWC and ASE neuron classes (Pierce-Shimomura et al. 2001; Wes and Bargmann 2001). The AWC odorsensory neuron class consists of two neurons, AWC left and AWC right (AWCL and AWCR, respectively; Fig. 1), whose symmetries extend to many differentiated features of the neurons including cell position, axonal and dendritic morphology, outgrowth and placement, and synaptic connectivity (Fig. 1; White et al. 1986). In addition, both cells express similar sets of genes and are both required for chemotaxis to specific odorants (Bargmann et al. 1993). However, a putative G-protein coupled olfactory receptor, encoded by the str-2 gene, was found to be expressed asymmetrically in these neurons (Fig. 1; Troemel et al. 1999). Induction of str-2 occurs stochastically through a calcium-signaling pathway in either the left or the right cell, but never in both (Troemel et al. 1999; Sagasti et al. 2001). The functional significance of this stochastic distribution is demonstrated by the fact that mutants defective for str-2 asymmetry show odor-discrimination defects (Wes and Bargmann 2001).
Figure 1.
Neural asymmetry in the AWC(L/R) and ASE(L/R) sensory neurons is genetically separable. The top panel is a schematic depiction of the anterior third of a worm, showing the anatomy and gene expression profiles of the AWC(L/R) odorsensory neurons (top left panels) and the ASE(L/R) gustatory neurons (top right panel). The bottom panel shows a list of mutants that were tested for asymmetry defects in each neuron class. Results with mutants that affect asymmetric str-2 expression in AWC(L/R), monitored using the kyIs140 transgene, are taken from Troemel et al. (1999), with the exception of the cog-1, unc-37, and lin-49 results (our own results; the ASE defects are described in more detail in other figures). ASE(L/R) asymmetry was monitored using the lim-6 reporter transgene otIs6 or otIs114. A complete list of mutants tested for effect on asymmetric ASE(L/R) expression patterns is shown in Supplementary Table 1. References for the individual genes and alleles can be found in Troemel et al. (1999) and at http://www.wormbase.org. Notes: 1Ectopic lim-6::gfp expression is observed in a set of neurons other than ASE(L/R). 2The effect of the lin-49(ot74) null allele on str-2 expression (which in wild-type animals is 100% “one AWC on”; Troemel et al. 1999) is as follows: 38% “one AWC on,” 38% “no AWC on,” 14% “two AWC on,” 10% “more than two cells on” (n = 42). In contrast to these pleiotropic effects on AWC(L/R), the effect of lin-49 on lim-6 is qualitatively different; there is a stereotyped gain of ASER fate at the expense of the ASEL fate (see Fig. 5B). Given this qualitative difference and also given the molecular identity of LIN-49 as a broadly expressed transcriptional cofactor with roles in multiple tissue types (see text), we do not consider the effect of lin-49 on AWC and ASE as specific evidence that the AWC and ASE asymmetries are mechanistically related.
The unbiased asymmetry exemplified through str-2 expression in either AWCL or AWCR is analogous to the unbiased asymmetry of many animal morphological features and hence can be termed “antisymmetry” (Palmer 1996). The phenomenon of antisymmetry is contrasted with “directional asymmetry”, characterized by the sidedness of morphological features not being randomly distributed within a population, but strongly biased to one side of each individual (Palmer 1996). A clear case of directional asymmetry is displayed by the main class of C. elegans gustatory neurons called ASE. Like the AWC odorsensory class, the ASE neuron class consists of two bilaterally symmetric cells, called ASEL (ASE left) and ASER (ASE right), that are symmetric with regard to cell position, axonal and dendritic morphology, and patterns of synaptic connectivity (Fig. 1; White et al. 1986). However, three putative sensory receptors of the guanyl cyclase receptor family are asymmetrically expressed in ASEL (gcy-6 and gcy-7) or ASER (gcy-5; Fig. 1; Yu et al. 1997). Laser ablation of ASEL or ASER revealed that these asymmetric gene expression profiles correlate with functional asymmetry of the two neurons; each neuron is responsible for detecting a distinct class of water-soluble chemicals (Pierce-Shimomura et al. 2001). The functional significance of this L/R separation of chemosensory capacities was revealed through the analysis of lim-6 mutant animals in which the ASEL neuron, which normally senses sodium, but not chloride, now adopts the chloride-sensing feature of the ASER neuron (Hobert et al. 1999; Pierce-Shimomura et al. 2001). Hence, in lim-6 mutant animals, ASEL has the capacity to sense both sodium and chloride. These animals fail to effectively discriminate between the two chemicals; that is, they fail to sense one ion in the presence of the other (Pierce-Shimomura et al. 2001). The L/R separation of the chemosensory capacities of the ASE neurons in wild-type animals therefore increases the chemosensory capacities of the animals.
What are the cellular and molecular mechanisms that lead to functional diversification of ASEL and ASER? We have previously shown that the lim-6 LIM homeobox gene is required to repress expression of the ASER-specific guanyl-cyclase gene gcy-5 in ASEL. However, lim-6 is only one of presumably many factors that establishes asymmetry because first, lim-6 itself is already asymmetrically expressed in ASEL but not in ASER (Hobert et al. 1999) and second, ASEL-specific expression of the guanyl cyclase genes gcy-6 and gcy-7 is unaffected in lim-6 null mutants (Hobert et al. 1999). In order to elucidate the molecular mechanisms required to restrict expression of lim-6 and the guanyl cyclase genes in an asymmetric manner to just one of the two ASE neurons, we have undertaken a genetic screen to uncover mutants that show symmetrization of normally asymmetric ASE(L/R) features and report here the molecular identity of a subset of these mutants. Our study thus provides novel insights into the as yet poorly understood mechanisms of L/R diversification in the nervous system.
Results
L/R asymmetry in odorsensory and gustatory neurons is genetically separable
We first tested whether the determination of directional asymmetry in the ASE(L/R) gustatory neurons shares mechanistic similarities with the antisymmetric gene expression pattern of str-2 in the AWC(L/R) odorsensory neurons. Mutants that were previously shown to affect antisymmetry of str-2 gene expression in AWC(L/R) fall into three broad categories.
First, mutations that disrupt axon guidance cause str-2 asymmetry defects (Troemel et al. 1999). This defect has been attributed to a failure of the establishment of a direct cell–cell contact between the axons of AWCL and AWCR, which normally meet at the dorsal midline, run in close proximity to one another, and make reciprocal synaptic contacts with one another (White et al. 1986). Although making no reciprocal synaptic contact (White et al. 1986), the axons of ASEL and ASER also run in close proximity to one another after meeting at the dorsal midline (D. Hall and O. Hobert, unpubl.). Disruption of these contacts through the use of the same set of axon guidance mutants that disrupt asymmetric str-2 expression in AWCL/R has, however, no effect on asymmetric reporter gene expression in ASE(L/R) (Fig. 1; Supplementary Table 1).
Second, mutations that disrupt calcium signaling and mitogen-activated protein kinase (MAPK) signaling within AWC(L/R) cause str-2 asymmetry defects (Troemel et al. 1999; Sagasti et al. 2001). The same set of signaling mutants has no effect on asymmetric reporter gene expression in ASE(L/R) (Fig. 1; Supplementary Table 1).
Third, antisymmetric str-2 expression in AWC(L/R) was found to be affected in mutants that disrupt olfactory sensory processing (Troemel et al. 1999). We tested whether more than a dozen mutants that disrupt taste perception affect ASE(L/R) asymmetry. With the exception of che-1 (described following), we found this not to be the case (Fig. 1; Supplementary Table 1).
Last, as we will describe following, we have identified mutant alleles in which ASE(L/R) asymmetry is affected, but AWC(L/R) is not (Fig. 1). Consistent with their distinct appearance (antisymmetry vs. directional asymmetry), we conclude that the mechanisms of establishment of AWC(L/R) and ASE(L/R) asymmetry are genetically separable.
Identification of genes that affect ASE(L/R) asymmetry
Besides the mutant backgrounds mentioned earlier, we tested a variety of candidate genes for an effect on asymmetric expression of ASEL markers, including lin-12/Notch, receptor tyrosine kinase (RTK)-, transforming growth factor (TGFβ)-, and Wnt-signaling mutants and various transcription factors and other known patterning mutants and did not observe any defects (Supplementary Table 1). Given the previously reported impact of TGFβ-like signaling on the determination of L/R asymmetry in vertebrate organ and brain development (Ramsdell and Yost 1998; Mercola and Levin 2001), the absence of a defect in null mutants of the daf-4 gene, which codes for the sole type II TGFβ-receptor protein in the C. elegans genome (Estevez et al. 1993; Ruvkun and Hobert 1998), is of interest because it points to a different mechanism of regulation of L/R asymmetry.
Using transgenic reporter strains that express green fluorescent protein (gfp) exclusively in ASEL (lim-6::gfp and gcy-7::gfp), we then conducted unbiased genetic screens for mutants that display defects in asymmetric ASE marker gene expression (see Materials and Methods). Mutants derived from these screens were examined for defects in asymmetric expression of three ASE asymmetry markers, lim-6::gfp and gcy-7::gfp for ASEL and gcy-5::gfp for ASER. Because two ASEL markers, lim-6::gfp and gcy-7::gfp, show identical behaviors in all mutant backgrounds tested, we have not included a third ASEL marker, gcy-6::gfp, in our analysis but assume that it behaves similarly to lim-6::gfp and gcy-7::gfp. Consistent with this notion, lim-6, gcy-7 and gcy-6 contain significant patches of sequence similarity in their cis-regulatory regions (data not shown).
We retrieved three classes of mutants from our screen. In class I and class II mutants, the overall identity of the ASE neurons, as assessed by cell position, axon morphology, and bilaterally symmetric gene expression profiles is unaffected. In class I mutants, however, the ASEL markers lim-6::gfp and gcy-7::gfp are expressed in both ASEL and ASER, and gcy-5::gfp expression is concomitantly lost in ASER (“two ASEL”-phenotype; see Fig. 2 for examples). In class II mutants, lim-6::gfp and gcy-7::gfp fail to be expressed in ASEL, and there is concomitant ectopic expression of gcy-5::gfp in ASEL (“two ASER”-phenotype; see Fig. 5, below, for examples). Last, in class III mutants, the ASE neurons are generated but lackthe expression of several identity-determining markers, including all three ASEL- and ASER-specific asymmetry markers as well as bilaterally symmetric markers (see Fig. 8, below, for examples; Fig. 9A, below, shows a schematic summary of all mutant phenotypes in single and double mutants). We termed mutants from the first two classes “lsy” mutants (pronounced “lousy”), for lim-6 symmetry mutant. We also noted that lsy mutants do not disrupt the L/R asymmetric placement of a specific unilateral neuron (RIS) or the L/R asymmetric migration of the Q neuroblasts (data not shown).
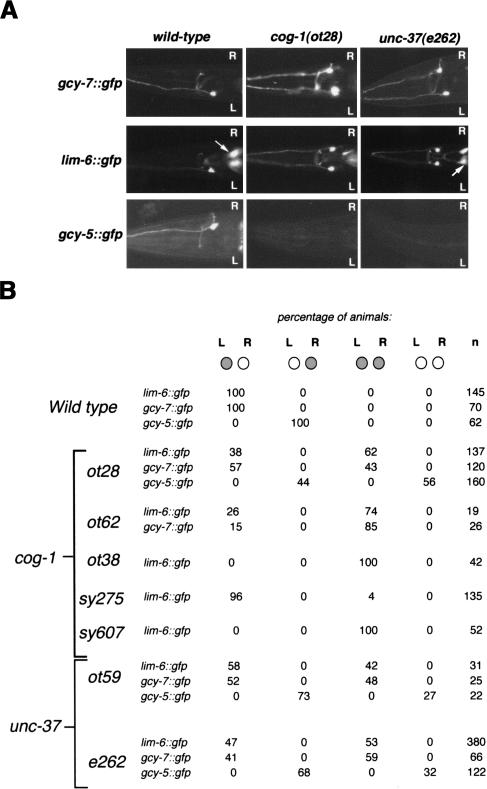
Figure 2.
Mutations resulting in a symmetric, “two ASEL” phenotype (class I phenotype). (A) Expression patterns of gcy-7::gfp, lim-6::gfp, and gcy-5::gfp in wild-type, cog-1(ot28), and unc-37(e262) mutant backgrounds. Adult animals are shown. Both gcy-7::gfp and lim-6::gfp are derepressed in ASER in unc-37 and cog-1 backgrounds, whereas gcy-5::gfp expression is lost in ASER. Arrows denote the excretory gland cell that expresses lim-6::gfp. (B) Quantification of the effects of cog-1 and unc-37 on ASE asymmetry. Animals were scored as adults (with the exception of ot59, which, because of their lethality, were scored as L1s). Note that derepression of lim-6 in ASER is always followed by a concomitant loss of gcy-5 expression.
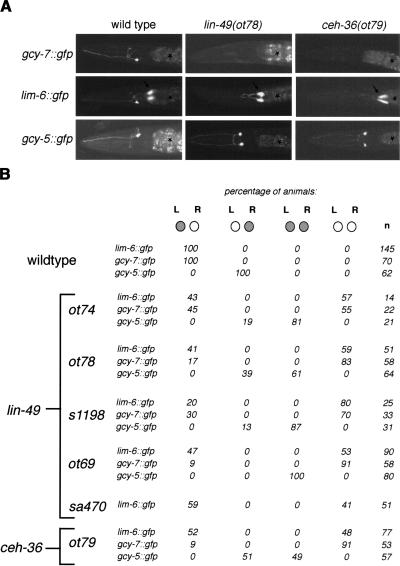
Figure 5.
Mutations resulting in a symmetric, “two ASER” phenotype (class II phenotype). (A) Expression patterns of gcy-7::gfp, lim-6::gfp, and gcy-5::gfp in wild-type, lin-49(ot78), and ceh-36(ot79) adult animals. Arrows denote the excretory gland cell that expresses lim-6::gfp. Asterisks denote gut autofluorescence. (B) Quantification of the effects of lin-49 and ceh-36 on ASE asymmetry. Animals were scored as adults (with the exception of s1198 and ot74 animals which, because of their late L1 lethality, were scored as viable L1s). The lackof a perfect correlation between loss of lim-6 expression and gain of gcy-5 expression [e.g., in lin-49(ot69), 100% of animals gain gcy-5 expression, but only 53% lose lim-6 expression] is likely a levels issue; that is, animals whose lim-6 expression levels appear unaffected by reporter gene analysis may have experienced a drop of lim-6 levels below a critical threshold required for gcy-5 repression.
Figure 8.
Mutations in the che-1 locus eliminate adoption of the ASE(L/R) fate (class I phenotype). (A) ASE(L/R) expression of gcy-7::gfp (otIs3), lim-6::gfp (otIs114), gcy-5::gfp (ntIs1), and flp-6::gfp (otIs125) in adult wild-type and adult che-1 (ot27) mutant animals. ASE-specific expression of all reporters is lost in che-1 mutants. Arrows in the “lim-6::gfp” panel denote the excretory gland cell that expresses lim-6::gfp and the ADF or AFD neuron class in the flp-6::gfp panel; note that gfp expression in neither of these cell types is affected in che-1 mutants. Asterisks denote gut autofluorescence. (B) Quantification of che-1 effects on ASE gene expression patterns. (C) Schematic CHE-1 protein structure (drawn to scale), denoting the position of mutant alleles retrieved from our screen. The deletion breakpoints in ot66 have not been sequenced but are inferred by PCR.
Figure 9.
Transcriptional cascade regulating ASE(L/R) asymmetry. (A) Summary of expression of transcription factors and their effectors in ASE(L/R). Circles indicate gfp expressing (green)/nonexpressing (not filled) ASEL and ASER cells. The ASE(L/R) patterns of expression of ubiquitously expressed genes (unc-37 and lin-49) are not shown. Notes: 1Low levels of cog-1 activity in ASEL are inferred from reporter gene assays as well as the observation that cog-1 is required in ASEL in a ceh-36 mutant to repress lim-6 expression. 2lim-6 is not required to initiate its own expression but to maintain it (data not shown); the empty circles refer to expression in the adult, after autoregulation has been established. 3flp-6::gfp was used to assess bilaterally symmetric ASE fate. 4See Figure 7C for an explanation of the reappearance of ASE asymmetry. (B) A molecular model for the establishment of asymmetric gcy-5 and gcy-7 expression. Arrows reflect genetic pathway interactions, which subsume either a direct interaction of a protein with the respective transcriptional regulatory elements or the presence of intermediary factors. Different sizes of the COG-1 protein in ASEL versus ASER are meant to reflect different protein levels brought about by differential activation or repression of cog-1 transcription (model #1 or model #2, which are not mutually exclusive). Note that low levels of cog-1 must be present in ASEL because, in a ceh-36 and lin-49 mutant background, a role for cog-1 is revealed in ASEL (Fig. 7). Differential expression of COG-1 in ASEL and ASER is either achieved through differential transcriptional repression in ASEL (model #1) or differential transcriptional activation in ASER (model #2). Because the loss of lim-6 results in activation of gcy-5 expression, we invoke CHE-1 as a potential direct positive regulator of gcy-5 expression, a notion supported by ectopically expressed CHE-1 being able to induce gcy-5 expression (data not shown; Uchida et al. 2003). We and others have also shown that CHE-1 regulates bilaterally symmetric features of ASE fate (Uchida et al. 2003; this paper). CEH-36 and LIN-49 do not regulate these features (data not shown).
We mapped most lsy alleles to specific chromosomal intervals. A combination of complementation testing and comparison of map position allowed us to conclude that we retrieved a total of 11 complementation groups, five displaying the class II phenotype (“two ASEL”), another five displaying the class I phenotype (“two ASER”), and one displaying the class III phenotype (“ASEL/R off”). One more class I mutant has not yet been ordered into a complementation group. Several of the complementation groups are represented only by single alleles, demonstrating that our screening efforts have not yet reached saturation. In this paper, we will describe the molecular characterization of two complementation groups that display the class I lsy phenotype (cog-1 and unc-37), two complementation groups that display the class II lsy phenotype (ceh-36 and lin-49), and the single complementation group that displays the class III phenotype (che-1).
Class I mutants (`two ASEL') affect the ASE(L/R)-expressed COG-1 and UNC-37 transcription factors
The four class I lsy mutants ot28, ot38, ot62, and ot59 define two complementation groups (Fig. 2). Through single-nucleotide polymorphism (SNP) mapping, complementation testing, transformation rescue, and allele sequencing, we demonstrated that ot28, ot38, and ot62 are allelic to cog-1 (Figs. 3A, 4). cog-1 was recently shown to code for a homeobox gene orthologous to the vertebrate Nkx6.1 and Nkx6.2 genes and to be involved in vulval patterning (Palmer et al. 2002). The previously described cog-1 alleles, sy607 and sy275, also display a lsy phenotype (Fig. 2B).
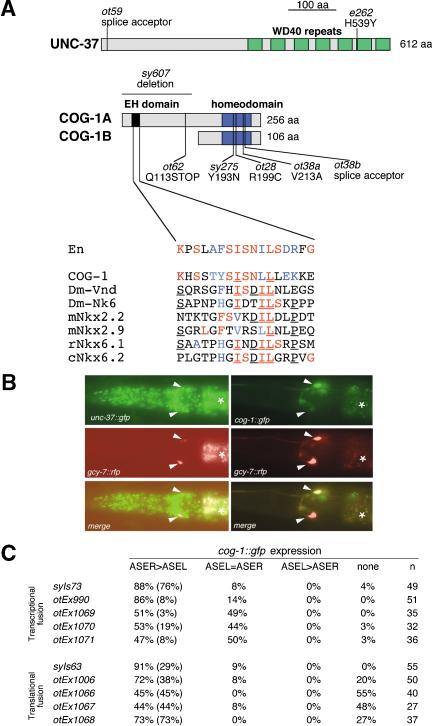
Figure 3.
Mutations in cog-1 and unc-37, two transcription factors expressed in ASE(L/R), cause ASE asymmetry defects. (A) Schematic depiction of the UNC-37 and COG-1 protein structure (drawn to scale) with mutant alleles noted. The blowup of the COG-1 sequence depicts a motif similar to the eh1 domain [alignment extended from Muhr et al. (2001)]. Colored residues indicate conservation to the Engrailed protein (red, identity; blue, conservative substitution), underlining indicates identity (in >50% of aligned sequences) among the NK-type protein that cannot be observed in the Engrailed protein. In contrast to the previously characterized cog-1 alleles sy607 and sy275, which still produce progeny, the ot38 and ot62 alleles, the latter of which only affects the cog-1a splice form (A), are completely sterile. ot28 animals are fertile. The lsy phenotype of ot62, but not any other cog-1 allele, is dominant (23% of ot62/+ animals are lsy; n = 22). cog-1(ot62) animals may produce a truncated COG-1A protein that contains only its transcriptional repressor domain, the eh1 domain, but not its DNA-binding domain. In heterozygous animals, this truncated protein may interfere in a dominant-negative manner with the activity of the wild-type copy of the COG-1A protein. (B) Expression of a translational UNC-37::GFP fusion protein (left panel; Kelly et al. 1997) and a translational COG-1::GFP fusion protein (right panel; transgenic line, syIs63; Palmer et al. 2002) in the ASE neurons in midlarval stage animals (white arrowheads). ASEL and ASER are marked with the ASE(L/R)-expressed reporter rfp transgene otIs131 (Is[gcy-7::rfp]) or otEx445 (Ex[gcy-7::rfp]). Asterisks denote gut autofluorescence. (C) Quantification of cog-1 reporter gene expression in different transgenic lines in ASEL and ASER. The top five lines carry transcriptional reporter fusions that contain the promoter of the cog-1 gene. syIs73 is a chromosomally integrated array (Palmer et al. 2002). The bottom five lines carry translational reporter fusions that contain the promoter as well as all coding sequences of cog-1. otEx1006 rescues the cog-1 mutant phenotype (Fig. 4A; other lines were not tested for rescue). ASER > ASEL indicates stronger gfp fluorescence in ASER compared with ASEL. Numbers in parentheses indicate complete absence of fluorescence in ASEL. In theory, the occasional, less consistent expression of cog-1 in ASEL could be a reporter gene artifact, explained through the titration of an ASEL-specific negative regulator of cog-1 expression. However, genetic evidence described in the text indicates that, in specific mutant backgrounds (ceh-36, lin-49), a function for cog-1 in ASEL is revealed, demonstrating that low levels of cog-1 activity are indeed present in ASEL. All animals contained gcy-7::rfp reporters in the background (otIs131 or otEx445), were photographed at midlarval stages in which gcy-7::rfp reporter is expressed in ASEL and ASER, and were scored for the gfp phenotype as gravid adults.
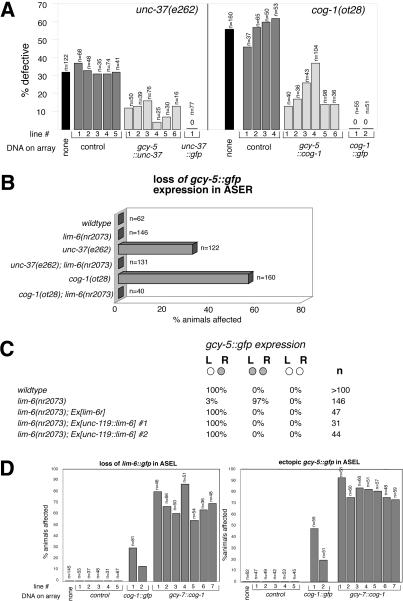
Figure 4.
cog-1 and unc-37 act autonomously to repress ASEL fate via repression of lim-6. (A) cog-1 and unc-37 act cell autonomously. Transformation rescue data of the unc-37 and cog-1 mutant defects are shown. Numbers below bars indicate independent transgenic strains. “% defective” refers to the absence of gcy-5::gfp expression in ASER, assessed with the ntIs1 (Is[gcy-5::gfp]) integrated transgene. “cog-1::gfp” (otEx1006, otEx1007; see Materials and Methods) and “unc-37::gfp” (Kelly et al. 1997) are translational gfp fusions in which the respective genomic locus is fused to gfp. cog-1::gfp was injected at 10 ng/μL and gcy-5::cog-1 at 2 ng/μL (lines 1 and 2), 5 ng/μL (lines 3 and 4), and 50 ng/μL (lines 5 and 6). gcy-5::unc-37 was injected at 50 ng/μL. rol-6 was the injection marker. Control lines have the cog-1 or unc-37 coding region replaced with gfp and were generated at 50 ng/μL injected DNA. We explain the ability to rescue gcy-5 expression through supplying cog-1 and unc-37 under control of the gcy-5 promoter by the gcy-5 promoter not being entirely shut off in the respective mutants. (B) cog-1 and unc-37 act through lim-6. Loss of gcy-5::gfp expression (monitored with ntIs1) in ASER in cog-1 and unc-37 is suppressed by removing lim-6 activity. (C) lim-6 is not sufficient to repress gcy-5 expression. gcy-5::gfp (ntIs1) is in the background of all strains. “lim-6r” is a rescuing lim-6 genomic fragment previously described to rescue other lim-6 mutant defects (Hobert et al. 1999) and was injected at 20 ng/μL. unc-119::lim-6 was injected at 20 ng/μL (#1) or 5 ng/μL (#2) and resulting F1s were scored. (D) Converting ASEL to ASER fate through raising the activity of cog-1. cog-1 activity was raised by generating multicopy arrays of a gfp-tagged cog-1 genomic clone (cog-1::gfp) or of a cog-1 cDNA driven by the gcy-7 promoter (gcy-7::cog-1). Arrays were expressed in wild-type animals, except in the right panel, where cog-1::gfp was expressed in a cog-1(ot28) mutant background to assess its rescuing capacity. The left panel shows the repression of lim-6 in ASEL and the right panel shows the concomitant gain of ASER features in ASEL (assessed with ntIs1). Note that cog-1::gfp is a rescuing gfp construct that, as shown in A, rescues the gcy-5 expression in ASER, but, as shown here, also causes ectopic gcy-5 expression in ASEL—both effects that can be attributed to the repression of the lim-6 repressor. Thus, gcy-5 expression is observed in ASEL + ASER in cog-1-overexpressing animals, only in ASER in wild-type animals, and neither in ASEL nor in ASER in cog-1 mutants. cog-1::gfp was injected at 5 ng/μL and gcy-7::cog-1 was injected at 50 ng/μL. As a control, gcy-7::rfp was injected at 50 ng/μL. Animals were scored as adults.
In vitro binding assays revealed that the vertebrate orthologs of COG-1 interact through the conserved engrailed homolog (eh1) domain with the transcriptional corepressor Groucho (Muhr et al. 2001). The C. elegans ortholog of Groucho, unc-37, maps to a chromosomal region on linkage group I (Pflugrad et al. 1997), to which we mapped another asymmetry mutant, ot59, with a lsy phenotype similar to our cog-1 alleles (Fig. 2). ot59 animals also displayed a characteristic unc-37-like locomotory defect. We sequenced the unc-37 gene in ot59 mutant animals and found an early splice site mutation (Fig. 3A). The canonical allele of unc-37, e262, also shows a class I lsy phenotype, which can be rescued through the introduction of the wild-type unc-37 locus (Fig. 4A).
cog-1 and unc-37 interact genetically
UNC-37 has recently been shown to interact with the engrailed homology (eh1) domain of the UNC-4 homeodomain protein (Winnier et al. 1999). The eh1 domain is also conserved in COG-1 (Fig. 2A). Moreover, the vertebrate orthologs of cog-1 and unc-37, Nkx6.1 and Grg4/Groucho, directly interact in vitro through the eh1 domain (Fig. 2A; Muhr et al. 2001), suggesting that COG-1 and UNC-37 may also directly interact to affect ASE asymmetry. To corroborate this notion, we examined a potential genetic interaction between cog-1 and unc-37. Lowering the dose of either cog-1 or unc-37 through placing a wild-type copy of the respective gene over a hypomorphic allele has no effect on asymmetry (0 out of 115 cog-1(ot28)/+ animals show ectopic gcy-7::gfp expression in ASER; 0/84 unc-37(e262)/+ animals show defects). If, however, the dosage of both genes is simultaneously reduced in a transheterozygous state, a significant asymmetry defect becomes obvious [26/95 unc-37(e262)/+; +/cog-1(ot28) animals show ectopic gcy-7::gfp expression in ASER]. Later we report that cog-1 and unc-37 act in a similar cell to affect asymmetry, thus leading us to conclude that, like their vertebrate orthologs, COG-1 and UNC-37 are likely to physically associate to regulate asymmetric patterns of gene expression.
unc-37 and cog-1 expression in the ASE neurons
Consistent with the ubiquitous expression of its vertebrate and fly orthologs, unc-37/Groucho was previously reported to be broadly expressed (Pflugrad et al. 1997); however, its expression was not specifically examined in individual head neurons. We therefore examined the expression of unc-37 in ASE(L/R) through the use of an UNC-37::GFP translational reporter (Kelly et al. 1997), which we found to rescue the asymmetry defects of unc-37 mutants (Figs. 3B, 4A). A red fluorescent protein rfp-expressing transgene, otIs131, which is exclusively expressed in larval ASE(L/R) neurons, shows cofluorescence with the unc-37::gfp reporter signal (Fig. 3B), thus demonstrating unc-37 expression in ASEL and ASER.
A cog-1::gfp reporter gene fusion was previously reported to be expressed in three classes of head sensory neurons, including ASEL and ASER (Palmer et al. 2002). Using previously described reporter lines as well as newly constructed reporter lines, we made the intriguing observation that all transgenic lines tested show significantly higher levels of cog-1 expression in ASER versus ASEL (Fig. 3B,C). Higher levels of expression of cog-1 can be observed with a rescuing cog-1::gfp reporter gene fusion, which contains the complete coding region of cog-1, as well as with reporter gene fusions in which only the promoter of cog-1 is fused to gfp (Fig. 3C). This observation indicates that differential expression of cog-1 in ASER versus ASEL is mediated via the transcriptional level.
cog-1 and unc-37 act in ASER and through lim-6 to affect gcy-5 expression
We first focused on the effects of cog-1 and unc-37 mutations in ASER, which loses gcy-5 expression (ASER marker) and gains lim-6 expression (ASEL marker) in the respective mutants. First, we tested whether cog-1 and unc-37 function is cell autonomous. We found that cDNAs of unc-37 and cog-1 driven by a postmitotic, ASER-specific transcriptional regulatory element derived from the gcy-5 locus rescue the respective mutant phenotypes (Fig. 4A), indicating that these two genes may act postmitotically in ASER to repress ASEL-specific features.
Second, we examined whether the effect of cog-1 and unc-37 on asymmetric gcy-5 expression is mediated through the regulation of the homeobox gene lim-6. lim-6, which is normally expressed exclusively in ASEL, is required to repress gcy-5 expression in ASEL (Hobert et al. 1999); in cog-1 and unc-37 mutants, lim-6 is ectopically expressed in ASER, and gcy-5 expression is concomitantly lost in ASER. To ask whether this loss of gcy-5 expression is due to ectopic lim-6 expression, we eliminated lim-6 in an unc-37 mutant and in a cog-1 mutant background. We find that in both cog-1; lim-6 and unc-37; lim-6 double mutants, gcy-5 expression in ASER reappears (Fig. 4B). Hence, in wild-type animals, cog-1 and unc-37 allow gcy-5 to be expressed through inhibiting the expression of the gcy-5 repressor lim-6. The lack of a perfect correlation between gain of lim-6 expression (e.g., 53% of unc-37 mutant animals gain lim-6 in ASER, but only 32% concomitantly lose gcy-5 expression; Fig. 2B) may be a reflection of a need for a threshold level of ectopic lim-6 expression to achieve gcy-5 repression; this level may not be reached in all animals.
After having shown that lim-6 is required to repress gcy-5 expression, we next tested whether lim-6 alone is sufficient to repress gcy-5 expression. To this end, we expressed lim-6 in both ASEL and ASER (and in all other neurons of the nervous system, using the unc-119 promoter; see Materials and Methods) in a lim-6 null mutant background. We found that, in these transgenic animals, the loss of gcy-5 repression in ASEL is rescued, confirming that this promoter produces sufficient levels of a functional LIM-6 protein (Fig. 4C). However, in none of the animals in which the repression of gcy-5 in ASEL is reestablished can we observe a concomitant repression of gcy-5 expression in ASER. The requirement but lack of sufficiency for lim-6 function suggests that the ectopic lim-6 expression observed in cog-1 and unc-37 mutants is necessary, but not sufficient, to repress gcy-5 expression. Other factors must similarly be derepressed in ASER in cog-1 and unc-37 mutants to allow lim-6 to repress gcy-5 expression.
Overexpression of cog-1 can convert ASEL to ASER
We next asked whether raising the levels of cog-1 in ASEL may be sufficient to repress ASEL features (lim-6) and allow ASER features (gcy-5) to appear. We used two approaches: First, we generated multicopy arrays of the cog-1 locus and second, we expressed a cog-1 cDNA under control of the gcy-7 promoter, which is active in ASEL and ASER embryonically and becomes restricted to ASEL postembryonically. We found that transgenic wild-type animals expressing either of these two constructs show repression of lim-6 expression in ASEL and a concomitant gain of the normally ASER-specific marker gcy-5 expression in ASEL (Fig. 4D).
In conclusion, cog-1 activity appears to be tightly regulated in ASER versus ASEL. In wild-type animals, cog-1 is present in ASEL and ASER, but effective as a repressor of lim-6 and gcy-7 only in ASER, possibly because of higher levels of expression. Raising the activity of cog-1 in ASEL through overexpression reveals that cog-1 can act as a repressor in ASEL as well. Following we describe a set of transcription factors that are intimately tied to the differential activity of COG-1/UNC-37.
Class II mutants (`two ASER') affect the LIN-49 and CEH-36 transcription factors
In order to better understand how cog-1 and unc-37 affect asymmetric lim-6 expression, we molecularly characterized class II asymmetry mutants derived from our screen. Those mutants display an opposite phenotype to cog-1 and unc-37; that is, they display a “two ASER” phenotype (Fig. 5A,B). Bilaterally symmetric features of ASE(L/R), such as the expression of the flp-6 gene, are unaffected in class II mutants (data not shown), suggesting that these mutants specifically disrupt the asymmetric gene expression programs. We will first describe two of these class II genes and in the next section we will describe their genetic interactions with cog-1 and unc-37.
lin-49: Through SNP mapping, complementation testing, allele sequencing, and transformation rescue, we found that the three recessive ot69, ot74, and ot78 alleles are mutations in the lin-49 gene, which codes for a predicted nuclear protein with two plant homeodomain (PHD)-finger domains and a bromodomain (Chamberlin and Thomas 2000). The PHD-finger domains, found in many chromatin-associated proteins (Aasland et al. 1995), and the bromodomain, an acetyllysine-binding domain (Dyson et al. 2001), indicate that the LIN-49 protein may be a general transcriptional cofactor involved in chromatin remodeling. Consistent with this notion, lin-49 was reported to be broadly expressed and shown to affect the development of several tissue types including the gut, the egg-laying system, and male mating structures (Chamberlin and Thomas 2000). We corroborated the broad and possibly ubiquitous expression of lin-49 using a gfp fusion construct that rescues the mutant phenotype (Fig. 6C).
Figure 6.
Mutations in lin-49 and ceh-36, two transcription factors expressed in ASE(L/R), cause ASE asymmetry defects. (A) The protein structure of LIN-49 and CEH-36 is schematically depicted with mutant alleles noted. The SMART domain search tool (http://smart.embl-heidelberg.de) was used to predict all domains shown here. ceh-36(ot79) is a likely hypomorph. Like the previously characterized s1198 allele (which also displays a lsy phenotype; Fig. 5B), the ot74 allele is an early stop codon in the lin-49 gene that also causes larval lethality; both alleles are likely null alleles (A). ot78 and ot69 are viable, yet ot69 is completely vulvaless and of low brood size. All alleles are recessive for their lsy phenotype. (B) Dendrogram showing the relationship of the homeodomain of CEH-36 to other OTX-type homeodomains (created with ClustalX and NJPlot). The homeodomain of the TTX-3 LIM-type homeodomain protein was used as an outlier. (C) Rescue of ceh-36(ot79) with a genomic ceh-36 fragment and an rfp-tagged ceh-36 construct and of lin-49(ot78) with a gfp-tagged lin-49 genomic fragment. “Control” indicates injection marker (rol-6) alone. Numbers refer to number of transgenic lines. Rescuing constructs were injected at 2.5 ng/μL. Animals were scored as adults. (D) Expression of a ceh-36 transcriptional gfp reporter fusion construct (promoter only) in ASEL and ASER in midlarval stage animals (transgenic line, otEx862). An additional pair of amphid sensory neurons is out of the plane of focus. ASEL and ASER were visualized with otIs131. A translational gfp reporter and a translational rfp reporter, which includes all exons and introns and rescues the mutant phenotype (C), show identical patterns of expression.
ceh-36: Through SNP mapping, allele sequencing, and transformation rescue, we found that ot79 is an allele of the previously uncharacterized ceh-36 gene, one of three orthodenticle-type homeobox genes predicted in the C. elegans genome (Fig. 6A,B; Ruvkun and Hobert 1998). The ot79 allele is completely recessive but unlikely to be a null allele because the truncation introduced by the premature stop codon still leaves the homeodomain intact (Fig. 6A). A ceh-36::rfp fusion construct that is capable of rescuing the mutant phenotype (Fig. 6C), as well as a fusion of the ceh-36 upstream regulatory region to gfp, showed exclusive expression in two pairs of head sensory neurons in postembryonic animals, one being ASE(L/R) (Fig. 6D; A. Lanjuin and P. Sengupta, pers. comm.).
Because both lin-49 and ceh-36 mutants have similar effects on lim-6 and gcy gene expression, we considered whether they may do so in a sequential manner through activating each other's expression. Since we consider it highly unlikely that a broadly expressed transcriptional cofactor is under control of the 2-neuron-specific transcription factor ceh-36, we only tested the possibility that ceh-36 expression may be regulated by lin-49. We found that a ceh-36::gfp reporter construct is normally expressed in lin-49 null mutants; it is also unaffected in cog-1 and unc-37 mutants (data not shown). Following we present genetic epistasis data that indicate that ceh-36 and lin-49 show very similar patterns of interaction with other transcription factors, suggesting that CEH-36 and LIN-49 may act together, possibly in a complex analogous to COG-1 and UNC-37.
The cog-1/unc-37 repressor complex antagonizes the ceh-36/lin-49-mediated activation of lim-6 expression
We have shown that lin-49 and ceh-36 are required for lim-6 and gcy-7 expression in ASEL. Yet ceh-36 (and the ubiquitously expressed lin-49 gene) is expressed in both ASEL and ASER. What prevents ceh-36 and lin-49 from activating lim-6 expression in ASER? One possibility is that their activity is antagonized in ASER by the transcriptional repressors cog-1 and unc-37. We tested this hypothesis by asking whether lowering the activity of cog-1 and unc-37 now allows lin-49 and ceh-36 to activate lim-6 expression in ASER. To this end, we constructed a variety of double mutant animals. We find that ectopic expression of lim-6 in ASER in cog-1 and unc-37 mutants requires the activity of both lin-49 and ceh-36 because, in either double mutant combination, ectopic lim-6 expression in ASER is diminished (Fig. 7A). The model of an antagonism between cog-1/unc-37 and ceh-36/lin-49 is further corroborated by two experiments. First, as described earlier, raising the levels of cog-1 in ASEL (which normally expresses lower levels of cog-1 than does ASER) counteracts the normal activation of lim-6 by ceh-36 and lin-49 in ASEL and hence leads to a loss of lim-6 expression (Fig. 4D; see also model in Fig. 7C). Second, we find that reduction of ceh-36 or lin-49 activity does not merely lead to a loss of transcriptional activation of lim-6 in ASEL but, to the contrary, leads to a cog-1-dependent repression of lim-6. This is because, if we lower cog-1 activity in a ceh-36 or lin-49 hypomorphic background, lim-6 expression in ASEL is at least partially, if not completely, restored (Fig. 7B,C; a concomitant repression of gcy-5 in ASER that correlates with restored lim-6 expression can also be observed; data not shown). This experiment corroborates our observation of low levels of cog-1 expression in ASEL, whose repressive activity in wild-type animals seems to be antagonized by ceh-36 and lin-49 activity; lowering of ceh-36 and lin-49 activity hence allows cog-1 to repress lim-6.
Figure 7.
Genetic interactions between cog-1, unc-37, ceh-36, and lin-49 and lim-6. Animals contained either the integrated lim-6 or gcy-5 reporter arrays (otIs114 or ntIs1) and were scored as adults, except for ot74 animals, which were scored as larvae because of their lethality. The single mutant data is also shown in previous figures and is shown here for comparison. (A) cog-1 and unc-37 antagonize lin-49 and ceh-36 to affect asymmetric ASER properties. Ectopic lim-6::gfp expression in ASER in cog-1 and unc-37 mutants is significantly reduced on reduction of lin-49 or ceh-36 activity (left panel). Consequently, the loss of gcy-5::gfp expression in ASER in cog-1 and unc-37 mutants, presumably caused by ectopic expression of lim-6, is suppressed by lowering ceh-36 or lin-49 activity (right panel). (B) cog-1 function in ASEL. Lowering of ceh-36 or lin-49 activity leads to a reduction of lim-6 expression in ASEL. This reduction requires cog-1 because, in the double mutant, the loss of lim-6 expression is suppressed. (C) Reasoning for the restoration of asymmetric lim-6 expression in cog-1; ceh-36 doubles. The size of the circles correlates with the level of gene activity. Note that the tested ceh-36 and cog-1 alleles are hypomorphic alleles that do not completely eliminate gene function, but merely reduce it (arrow pointing down). Overexpression of cog-1 in ASEL (see Fig. 4D) is reflected with an arrow pointing up.
Also consistent with the notion of cog-1/unc-37 and ceh-36/lin-49 acting in parallel rather than sequentially, we find that L/R asymmetric expression of a cog-1 reporter gene construct is unaffected in ceh-36 and lin-49 mutants and that ceh-36 reporter gene expression in a cog-1 and unc-37 mutant background is unaffected (data not shown).
One way to fit this genetic data into a molecular model is to suppose that a COG-1/UNC-37 repressor complex competes with CEH-36 and LIN-49 for control over the activity of the lim-6 and gcy-7 promoters (or the promoter of an intermediary factor that positively regulates these factors). In ASER, the COG-1/UNC-37 complex prevails over CEH-36 and LIN-49; loss of the COG-1/UNC-37 complex in ASER hence reveals the activity of CEH-36 and LIN-49. In ASEL, however, CEH-36 and LIN-49 prevail over the COG-1/UNC-37 complex, likely because of the lower expression of COG-1 in ASEL; the loss of CEH-36 and LIN-49 hence reveals the activity of COG-1/UNC-37 (Figs. 7C, 9).
Although ceh-36 and lin-49 show no mutant phenotype in the transheterozygous state (data not shown), we hypothesize that both proteins act in a common transcriptional activation complex because both genes behave indistinguishably in genetic interaction tests, loss of either causes the same effects on the expression of downstream genes, they are both required for ASEL-marker expression in ASER in the absence of cog-1 or unc-37, and their reduction of activity in ASER is compensated by removal of cog-1. The domain structures of the two proteins are also supportive of a role for these proteins in a single transcriptional activation complex in which CEH-36 may provide the DNA binding specificity and LIN-49 the connection to the chromatin remodeling activities required for gene activation.
che-1 is a positive regulator of several ASEL and ASER features
We have described earlier a set of genes that act as positive regulators of gcy-7 and lim-6 in ASEL, with the latter factor being a repressor of the ASER marker gcy-5. But what factor is required for the activation of gcy-5 expression in ASER? In cog-1 and unc-37/groucho mutants, gcy-5 expression in ASER is lost. Groucho has previously been shown to be able to convert transcriptional activators, such as Runt or Dorsal, into repressors (Fisher and Caudy 1998). It could have thus been envisioned that cog-1 is an unc-37/groucho-independent activator of gcy-5 expression and an unc-37/groucho-dependent repressor of lim-6. However, our genetic interaction data show that this is not the case, because we can restore gcy-5 expression in cog-1 mutants if we remove the lim-6 gene. cog-1 thus “activates” gcy-5 expression through repressing the gcy-5 repressor lim-6.
Activation of gene expression through counteracting repression automatically invokes the existence of a basal transcriptional activation mechanism. We considered it possible that class III mutants that we have retrieved from our screen may shed light on the issue of transcriptional activation of ASER features. In all class III mutants gcy-5 expression is lost (Fig. 8B). Because lim-6 expression is also lost in these mutants (Fig. 8B), the loss of gcy-5 expression cannot be due to ectopic expression of lim-6 in ASER. We hence decided to undertake a molecular characterization of class III mutants, which may define direct or indirect activators of gcy-5 expression. We found that all class III mutants fall into a single complementation group that is allelic to the che-1 locus (Fig. 8A). che-1 was recently shown to be a Zn-fingercontaining transcription factor related to the Drosophila Glass transcriptional activator (Uchida et al. 2003). We found that in che-1 mutants not only are ASER markers lost, but also the ASEL markers lim-6 and gcy-7 (Fig. 8B). We furthermore examined expression of the ASE(L/R)-expressed cog-1 and ceh-36 genes in che-1 mutants and found a complete loss of their expression as well (data not shown). Another symmetric ASE(L/R) cell fate marker, flp-6, also fails to be expressed in che-1 mutants (Fig. 8A,B). The loss of expression of all of these genes is not a reflection of a failure of the neuron to be generated or to adopt a neuronal fate, because a pan-neuronal gfp marker is still expressed in ASE(L/R) and because the anatomy of ASE is only partially affected in che-1 mutants (Lewis and Hodgkin 1977; Uchida et al. 2003). We propose that che-1 is a permissive transcriptional regulator, located at the top and at the lower levels of a transcriptional regulatory cascade required to initiate asymmetric profiles of gene expression in ASEL and ASER.
Discussion
Cellular diversification along the L/R axis is a common feature in nervous systems as diverse as nematodes and humans, yet its molecular basis is still poorly understood (Hobert et al. 2002). The genetic amenability of C. elegans enabled us to identify a set of proteins involved in determining neuronal L/R patterning, most of which are highly conserved across phylogeny. In C. elegans, these factors serve to eventually segregate the expression of putative sensory receptors encoded by the gcy genes into otherwise largely bilaterally symmetric types of cells. The separation of distinct sensory capacities into two separate, yet bilaterally symmetric cells, each of which has a similar set of downstream synaptic partners (White et al. 1986), is a prerequisite for the capacity of worms to appropriately discriminate between distinct sensory inputs and hence to navigate in a complex sensory environment (Pierce-Shimomura et al. 2001). The increase in the discriminatory sensory potential through asymmetric chemoreceptor gene expression is not only a feature of the gustatory ASEL and ASER neurons, but also of the olfactory AWCL and AWCR neurons (Wes and Bargmann 2001). Despite their similar purpose, our genetic analysis points to the distinctiveness of the molecular mechanisms of establishment of AWC and ASE asymmetries. Although in the former case, cell–cell contact, calcium, and MAPK signaling are required to establish antisymmetric gene expression pattern (Troemel et al. 1999; Sagasti et al. 2001), none of these events is required to establish directional ASE asymmetry.
A molecular model that sheds some light onto the L/R asymmetric segregation of chemosensory receptor expression has emerged from our genetic studies and can be summarized as follows (Fig. 9). The Zn finger transcription factor CHE-1 acts at the top of the regulatory hierarchy to determine subtype-specific and bilateral-symmetric features of ASE, including the expression of terminal differentiation markers such as the neuropeptide gene flp-6, cyclic nucleotide ion channels, and orphan serpentine receptors (Uchida et al. 2003; this paper). CHE-1 also triggers either directly or indirectly the expression of factors that are required to determine the L/R asymmetric state of ASEL and ASER, including cog-1, ceh-36, and lim-6. These factors serve to modulate what we consider an “ASER default state,” which is defined by the expression of gcy-5. This default state is also revealed through laser ablation studies, demonstrating that the removal of putative signaling cells causes ASEL to convert to the ASER state (R.J. Johnson Jr. and O. Hobert, unpubl.); features of this default state, such as gcy-5 expression, may be directly induced by CHE-1. After CHE-1 has induced CEH-36 and COG-1 expression, a putative CEH-36/LIN-49 complex activates expression of at least two factors, a gcy gene (gcy-7) that defines ASEL properties and a transcription factor (lim-6) that prevents the ASER-specific gcy-5 gene from being expressed (Fig. 9). Although the bilaterally symmetrically expressed CEH-36/LIN-49 proteins (which may in analogy to COG-1 and UNC-37 be acting in a complex) are capable of inducing lim-6 and gcy-7 expression in both ASEL and ASER, they are prevented from doing so in ASER through the activity of the COG-1/UNC-37 repressor complex, which thus helps preserve the ASER default state (Fig. 9). The activity of COG-1/UNC-37 antagonizes CEH-36/LIN-49 activity only in ASER because of higher levels of COG-1 protein in ASER. Consequently, raising the levels of COG-1 in ASEL can overcome the CEH-36/LIN-49-mediated induction of ASEL cell fate, causing the adoption of ASER fate.
Our studies have revealed two basic transcriptional regulatory principles in the novel context of L/R asymmetry determination. First, we have shown that the expression of ASER fate (gcy-5 expression) is mediated through a series of repressive transcriptional interactions; that is, COG-1 and UNC-37 repress lim-6 expression, which represses gcy-5 expression. This transcriptional regulatory principle is reminiscent of the sequential repressor scheme in the creation of cellular diversity in the vertebrate spinal cord, which, intriguingly, involves the human orthologs of COG-1, the Nkx6 proteins, and of unc-37, the Grg proteins (Muhr et al. 2001). We have provided further insight into the sequential repressor model by demonstrating that a second transcriptional regulatory mechanism is at workin ASEL and ASER, namely, a tightly balanced antagonistic relationship between a repressor (COG-1/UNC-37) and a putative activator (CEH-36/LIN-49) complex. We do not know at this point whether these two protein complexes functionally interact on the lim-6 and gcy-7 promoter level, or whether they act via intermediary factors. Under the assumption that they workon a common target promoter, we can envision two molecular models for the mechanistic basis of the antagonism. COG-1/UNC-37 and CEH-36/LIN-49 may directly compete for a single binding site on their target gene promoter, or, alternatively, they may occupy distinct sites on the promoter; different levels of COG-1 may be read out as a differential binding site occupancy. We favor the latter model because a preliminary mutational analysis of the gcy-7 promoter identified two sites required for transcriptional activation in ASEL and a separate site required for repression in ASER (S. Chang and O. Hobert, unpubl.).
Our studies suggest that it is the differential levels of cog-1 activity in ASER versus ASEL in wild-type animals that are responsible for the cellular diversification of ASEL and ASER fate. Moreover, our reporter gene studies indicate that L/R differential activity of cog-1 is conferred by differential transcription of cog-1 in ASER versus ASEL. Either a repressor acts in ASEL to lower the levels of cog-1 transcription in ASEL (e.g., through antagonizing CHE-1 mediated activation of cog-1 transcription) or, alternatively, an activator acts in ASER to increase cog-1 levels (e.g., through assisting CHE-1-depending activation of cog-1). The identification of the upstream regulator of cog-1 expression will represent a further step toward understanding the mechanism that is responsible for setting up asymmetric patterns of gene expression. This mechanism may rely on intrinsically programmed lineage cues and hence be a consequence of the initially highly asymmetric embryo (Schnabel and Priess 1997). Alternatively, asymmetry may be nonautonomously determined through an “asymmetry-inducing” signal that acts past the stage of early embryonic inductions, a notion that we find supported by preliminary cell ablation data (R.J. Johnston Jr. and O. Hobert, unpubl.). Our candidate gene approach has ruled out several canonical signaling molecules as determinants in this event, including TGFβ-type signaling, found to be involved in vertebrate L/R signaling (Mercola and Levin 2001). We expect that the further molecular characterization of as yet uncloned lsy mutants will provide us with a better understanding of the development of L/R asymmetry in the nervous system.
Materials and methods
Transgenic reporter lines
All strains were grown at 20°C and scored at room temperature as gravid adults if not otherwise indicated.
otIs6 and otIs114: Is[lim-6prom::gfp; rol-6(d)]. Both integrants derive from a previously described extrachromosomal line (Hobert et al. 1999). In contrast to previous nomenclature (Hobert et al. 1999), we will for the sake of simplicity from here on refer to “lim-6prom::gfp” as “lim-6::gfp.”
otIs3: Is[gcy-7::gfp; lin-15(+)]. In mid-embryogenesis, otIs3 is initially expressed in both ASEL and ASER; expression becomes restricted to ASEL in late embryogenesis. This integrant derives from an extrachromosomal line described in Yu et al. (1997).
otIs125: Is[flp-6::gfp]. The extrachromosomal line from which this integrant was derived was a gift from C. Li.
otIs131: Is[gcy-7::rfp; rol-6(d)], derived from otEx445 (see following).
ntIs1: Is[gcy-5::gfp; lin-15(+)] [a gift from S. Lockery; derived from an extrachromosomal line described in Yu et al. (1997)]. Expression of gfp is turned on exclusively in ASER in embryogenesis after the generation of ASER.
kyIs140: Is[str-2::gfp; lin-15(+)] (Troemel et al. 1999), a 5′ fusion (i.e., containing only regulatory sequences upstream of the start codon).
otEx445: Ex[gcy-7::rfp; rol-6(d)], a 5′ fusion. Because of the delayed maturation of the RFP protein (compared with the GFP protein expressed from the otIs3 transgene mentioned earlier), GCY-7::RFP expression from this array can be observed in both ASEL and ASER until midlarval stages and becomes restricted to ASEL thereafter.
otEx862: Ex[ceh-36prom::gfp; rol-6(d)], a 5′ fusion.
otEx1030, otEx1031: Ex[ceh-36::rfp; rol-6(d)], a translational fusion (i.e., containing upstream regulatory sequences as well as all exons and introns of the locus).
otEx863: Ex[lin-49prom::gfp; rol-6(d)], a 5′ fusion.
otEx1025–1029: Ex[lin-49::gfp; rol-6(d)], a translational fusion.
syIs73: Is[pBP164 {cog-1prom::gfp; dpy-20(+)}], a 5′ fusion (Palmer et al. 2002).
syIs63: Is[cog-1::gfp; dpy-20(+)] (Palmer et al. 2002), a translational fusion.
otEx990: Ex[cog-1prom::gfp; rol-6(d)], a 5′ fusion.
otEx1069–1071: Ex[cog-1prom::gfp], a 5′ fusion.
otEx1006–1007: Ex[pBP159 (cog-1::gfp); rol-6(d)], a translational fusion.
otEx1066–1068: Ex[pBP159 (cog-1::gfp); unc-122::gfp], a translational fusion.
PD8100: Ex[unc-37::gfp; rol-6] (Kelly et al. 1997).
Mutant screen for lsy genes
otIs6 or otIs3 animals were mutagenized with ethyl methanesulfonate (EMS) using standard procedures (Brenner 1974). The progeny of individually picked F1 animals derived from the mutagenized P0 population were analyzed under a dissecting scope equipped with a fluorescent light source. Using otIs6 as a marker, we screened through 3200 haploid genomes, and using otIs3, we screened through 12,200 haploid genomes. A total of 28 mutants were retrieved. Through chromosomal linkage and complementation testing, we found these mutants to define 11 complementation groups; one mutant has not yet been assigned to a group. Representatives from each complementation group were backcrossed and transferred into several different gfp transgenic backgrounds that assess gcy-5, gcy-7, and lim-6 expression (otIs6, otIs3, otIs114, ntIs1).
SNP-based mapping
We made use of SNPs present in the Hawaiian C. elegans isolate CB4856 identified by the Washington University Genome Sequencing Center and by Ronald Plasterkand colleagues (Wicks et al. 2001). The respective mutant strain was crossed with CB4856 and resultant mutant F2 progeny were singled out. Their progeny were lysed using standard procedures, genomic fragments that included the SNP were amplified by PCR, and the SNP was analyzed by RFLP. In order to facilitate the identification of recombinants, mutant animals were in some cases marked with a visible marker and mutant recombinants that lost the visible marker were chosen for SNP analysis. che-1, cog-1, and lin-49 were identified as lsy genes after initial SNP mapping of the lsy alleles to chromosomal intervals that contained these genes (cog-1, LGII, between K10H10 and Y53F4B; lin-49, LGIV, between cosmid D2096 and ZK596) and subsequent complementation testing and allele sequencing.
Positional cloning of ceh-36(ot79)
Of all transcription factors described in this paper, ceh-36 is the only one for which no mutant allele was previously reported. On mapping of the lsy mutants retrieved from our mutant screen, we noted that ot79 was linked tightly to a canonical linkage SNP on the right arm of chromosome X at 16.01 m.u. (R03E1; 0/92 recombinants from a cross with CB4856). Left- and right-hand boundaries were established at 15.42 m.u. (C33G3; 1/92 recombinants) and 17.28 m.u. (F23D12; 1/92 recombinants), respectively. This region includes 60 predicted protein-coding genes. Because at that point we had shown that numerous transcription factors play a role in asymmetry, we sequenced all four predicted transcription factors in this region (F28H6.2, C37E2.4, C37E2.5, K04C1.3) and found a mutation in the coding region of one of them, ceh-36/C37E2.4.
DNA constructs
Reporter gene constructs: gcy-7::rfp was constructed by replacing gfp in the gcy-7::gfp construct with dsRed2 (Clontech). gcy-7::gfp contained 1.3 kb of the 5′ upstream regulatory region of the gcy-7 gene (Yu et al. 1997). A cog-1 transcriptional reporter was constructed by PCR fusion (Hobert 2002) using 4.6 kb of genomic region upstream of the ATG. A cog-1-rescuing gfp plasmid (pBP159) was kindly provided by Takao Inoue and Paul Sternberg [described by Palmer et al. (2002)]. A lin-49 translational gfp reporter was constructed by PCR fusion (Hobert 2002), including 2 kb of 5′ region (up to the preceding gene) and all exons and introns of lin-49. A ceh-36 translational rfp reporter was constructed by including 5.1 kb of 5′ region and all exons and introns of ceh-36; the rfp coding region derived from the dsRed2 (Clontech, Inc.) vector. The marker for all injections was pRF4/rol-6(d) at either 50 or 100 ng/μL. The unc-37 translational gfp reporter was reported by Kelly et al. (1997).
Heterologous expression constructs: Three promoters were used for heterologous expression, unc-119 (Maduro and Pilgrim 1995), gcy-5, and gcy-7 (Yu et al. 1997). The latter two promoters had to be recloned; for gcy-5, 3.1 kb of the 5′ upstream regulatory region of the gcy-5 gene were amplified from N2 genomic DNA using primers containing BamHI sites at either end and subcloned into the gfp vector pPD95.75 (a gift from A. Fire) to create gcy-5::gfp; for gcy-7, 1.3 kb of the 5′ upstream regulatory region of the gcy-7 gene were amplified from N2 genomic DNA using primers with PstI/BamHI sites at either end. The amplicon was subcloned into the gfp vector pPD95.75 to yield gcy-7::gfp. A shorter deletion derivative, gcy-7Hind::gfp, was constructed by releasing a HindIII fragment from gcy-7::gfp, which left 188 bp of the gcy-7 promoter, which still yielded completely penetrant ASEL expression in adults. The gcy-5::cog-1 and gcy-5::unc-37 rescuing plasmids were constructed by releasing gfp from gcy-5::gfp and replacing it with the cog-1a and unc-37 cDNAs. The unc-37 cDNA was provided by David Miller (Pflugrad et al. 1997); the cog-1a cDNA was obtained by PCR from an incomplete EST clone, providing the missing 5′ sequence in the primer sequence. The gcy-7::cog-1 expression plasmid was constructed by replacing gfp from gcy-7Hind::gfp with a cog-1a cDNA. unc-119::lim-6 was constructed by amplifying the genomic lim-6 region from the start to the stop codon and subcloning it into the pBY103 vector (Maduro and Pilgrim 1995).
Acknowledgments
We thank David Miller for unc-37 reagents, Shawn Lockery for ntIs1, Takao Inoue and Paul Sternberg for cog-1 reagents, Chris Li for providing flp-6::gfp, Qi Chen for expert technical assistance, several rotating and visiting students (Liat Hopper, Kristin Klemenhagen, Sera Konur, and John Etchberger) for their involvement in the isolation and mapping of lsy mutants and other parts of the project, David Hall for assistance in tracing ASEL and ASER axons in EM sections, the NIH-funded CGC for providing strains, and Piali Sengupta, Iva Greenwald, Sophie Jarriault-Reina, Richard Mann, and David Miller for comments on the manuscript. We are grateful to Anne Lanjuin and Piali Sengupta for sharing their results on ceh-36 function and expression prior to publication. S.C. was supported by an NIH predoctoral fellowship and R.J.J. by an NSF predoctoral fellowship. O.H. is a Searle, Irma T. Hirschl, Klingenstein, Rita Allen, and Sloan Scholar.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1117903.
Supplemental material is available at http://www.genesdev.org.
References
- Aasland R., Gibson, T.J., and Stewart, A.F. 1995. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20: 56–59. [DOI] [PubMed] [Google Scholar]
- Bargmann C.I., Hartwieg, E., and Horvitz, H.R. 1993. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74: 515–527. [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin H.M. and Thomas, J.H. 2000. The bromodomain protein LIN-49 and trithorax-related protein LIN-59 affect development and gene expression in Caenorhabditis elegans. Development 127: 713–723. [DOI] [PubMed] [Google Scholar]
- Concha M.L., Burdine, R.D., Russell, C., Schier, A.F., and Wilson, S.W. 2000. A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron 28: 399–409. [DOI] [PubMed] [Google Scholar]
- Davidson R.J. and Hugdahl, K. 1994. Brain asymmetry. MIT Press, Cambridge, MA.
- Dyson M.H., Rose, S., and Mahadevan, L.C. 2001. Acetyllysinebinding and function of bromodomain-containing proteins in chromatin. Front. Biosci. 6: D853–D865. [DOI] [PubMed] [Google Scholar]
- Essner J.J., Branford, W.W., Zhang, J., and Yost, H.J. 2000. Mesendoderm and left–right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development 127: 1081–1093. [DOI] [PubMed] [Google Scholar]
- Estevez M., Attisano, L., Wrana, J.L., Albert, P.S., Massague, J., and Riddle, D.L. 1993. The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature 365: 644–649. [DOI] [PubMed] [Google Scholar]
- Fisher A.L. and Caudy, M. 1998. Groucho proteins: Transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes & Dev. 12: 1931–1940. [DOI] [PubMed] [Google Scholar]
- Galaburda A.M. 1991. Asymmetries of cerebral neuroanatomy. In Biological asymmetry and handedness (ed. L. Wolpert), pp. 219–226. Wiley, West Sussex, UK. [DOI] [PubMed]
- Glick S.D. and Ross, D.A. 1981. Lateralization of function in the rat brain. Trends Neurosci. 4: 196–199. [Google Scholar]
- Hobert O. 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32: 728–730. [DOI] [PubMed] [Google Scholar]
- Hobert O., Tessmar, K., and Ruvkun, G. 1999. The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development 126: 1547–1562. [DOI] [PubMed] [Google Scholar]
- Hobert O., Johnston Jr., R.J., and Chang, S. 2002. Left–right asymmetry in the nervous system: The Caenorhabditis elegans model. Nat. Rev. Neurosci. 3: 629–640. [DOI] [PubMed] [Google Scholar]
- Kelly W.G., Xu, S., Montgomery, M.K., and Fire, A. 1997. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146: 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.A. and Hodgkin, J.A. 1977. Specific neuroanatomical changes in chemosensory mutants of the nematode Caenorhabditis elegans. J. Comp. Neurol. 172: 489–510. [DOI] [PubMed] [Google Scholar]
- Liang J.O., Etheridge, A., Hantsoo, L., Rubinstein, A.L., Nowak, S.J., Izpisua Belmonte, J.C., and Halpern, M.E. 2000. Asymmetric nodal signaling in the zebrafish diencephalon positions the pineal organ. Development 127: 5101–5112. [DOI] [PubMed] [Google Scholar]
- Ludwig W. 1932. Das Rechts-Links Problem im Tierreich und beim Menschen. Springer, Berlin.
- Maduro M. and Pilgrim, D. 1995. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141: 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercola M. and Levin, M. 2001. Left–right asymmetry determination in vertebrates. Annu. Rev. Cell Dev. Biol. 17: 779–805. [DOI] [PubMed] [Google Scholar]
- Miklosi A., Andrew, R.J., and Savage, H. 1997. Behavioural lateralisation of the tetrapod type in the zebrafish (Brachydanio rerio). Physiol. Behav. 63: 127–135. [DOI] [PubMed] [Google Scholar]
- Muhr J., Andersson, E., Persson, M., Jessell, T.M., and Ericson, J. 2001. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell 104: 861–873. [DOI] [PubMed] [Google Scholar]
- Palmer A.R. 1996. From symmetry to asymmetry: Phylogenetic patterns of asymmetry variation in animals and their evolutionary significance. Proc. Natl. Acad. Sci. 93: 14279–14286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer R.E., Inoue, T., Sherwood, D.R., Jiang, L.I., and Sternberg, P.W. 2002. Caenorhabditis elegans cog-1 locus encodes GTX/Nkx6.1 homeodomain proteins and regulates multiple aspects of reproductive system development. Dev. Biol. 252: 202–213. [DOI] [PubMed] [Google Scholar]
- Pflugrad A., Meir, J.Y., Barnes, T.M., and Miller III, D.M. 1997. The Groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4 to govern motor neuron identity in C. elegans. Development 124: 1699–1709. [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura J.T., Faumont, S., Gaston, M.R., Pearson, B.J., and Lockery, S.R. 2001. The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature 410: 694–698. [DOI] [PubMed] [Google Scholar]
- Ramsdell A.F. and Yost, H.J. 1998. Molecular mechanisms of vertebrate left–right development. Trends Genet. 14: 459–465. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. and Hobert, O. 1998. The taxonomy of developmental control in Caenorhabditis elegans. Science 282: 2033–2041. [DOI] [PubMed] [Google Scholar]
- Sagasti A., Hisamoto, N., Hyodo, J., Tanaka-Hino, M., Matsumoto, K., and Bargmann, C.I. 2001. The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell 105: 221–232. [DOI] [PubMed] [Google Scholar]
- Schnabel R. and Priess, J.R. 1997. Specification of cell fates in the early embryo. In C. elegans II (eds. D.L. Riddle, T. Blumenthal, B.J. Meyer, and J.R. Priess), pp. 361–382. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Troemel E.R., Sagasti, A., and Bargmann, C.I. 1999. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99: 387–398. [DOI] [PubMed] [Google Scholar]
- Uchida O., Nakano, H., Koga, M., and Ohshima, Y. 2003. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development 130: 1215–1224. [DOI] [PubMed] [Google Scholar]
- Wes P.D. and Bargmann, C.I. 2001. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature 410: 698–701. [DOI] [PubMed] [Google Scholar]
- White J.G., Southgate, E., Thomson, J.N., and Brenner, S. 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314: 1–340. [DOI] [PubMed] [Google Scholar]
- Wicks S.R., Yeh, R.T., Gish, W.R., Waterston, R.H., and Plasterk, R.H. 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- Winnier A.R., Meir, J.Y., Ross, J.M., Tavernarakis, N., Driscoll, M., Ishihara, T., Katsura, I., and Miller III, D.M. 1999. UNC-4/UNC-37-dependent repression of motor neuron-specific genes controls synaptic choice in Caenorhabditis elegans. Genes & Dev. 13: 2774–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Avery, L., Baude, E., and Garbers, D.L. 1997. Guanylyl cyclase expression in specific sensory neurons: A new family of chemosensory receptors. Proc. Natl. Acad. Sci. 94: 3384–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]