Abstract
We used several of the genetic lesions commonly associated with human liver tumors to reconstruct genetic progression to hepatocellular carcinoma and adenoma in mouse models. We initiated tumorigenesis with a transgene of the protooncogene MET or by hydrodynamic transfection of MET in combination with other genes into the livers of adult animals. Hepatocellular carcinoma in both instances arose from cooperation between MET and constitutively active versions of β-catenin. In contrast, adenomas were produced by cooperation between MET and defective signaling through the transcription factor HNF1α. Prompted by these findings, we uncovered a coincidence between activation of the protein-tyrosine kinase encoded by MET and activating mutations of β-catenin in a subset of human hepatocellular carcinomas. Inactivation of MET transgenes led to regression of hepatocellular carcinomas despite the persistence of activated β-catenin. The tumors eventually recurred in the absence of MET expression, however, presumably after the occurrence of one or more events that cooperated with activated β-catenin in lieu of MET. These results offer insight into hepatic tumorigenesis, provide mouse models that should be useful in the further study of hepatic tumorigenesis and for preclinical testing, and identify a subset of human hepatocellular carcinomas that may be susceptible to combination therapy directed against Met and the Wnt signaling pathway.
Keywords: β-catenin, hepatocyte nuclear factor 1α, liver cancer, MET, mouse, hepatocellular carcinoma
Liver cancer is among the deadliest of human malignancies, with an annual worldwide incidence of 600,000 cases and a mean survival time of 6 months from time of diagnosis (1). The principal causes of liver cancer are infection with hepatitis B or C virus, chronic alcoholism, aflatoxin exposure, or other circumstances that predispose to cirrhosis. These causes are believed to produce liver cancer by inducing repeated rounds of hepatocyte death and proliferation (2), creating a permissive environment in which genetic or epigenetic changes could occur that confer gain of function on protooncogenes or loss of function on tumor suppressor genes (3). Many such changes have been reported, but the combinations of these changes that operate to produce human hepatocellular carcinoma (HCC) remain largely uncharacterized. Frequent among the changes known to occur in human HCC are overexpression, amplification, or mutation of the protooncogene MET, which encodes the receptor protein tyrosine kinase Met (4–6) and activation of the Wnt signaling pathway by mutation of the genes encoding either β-catenin (CTNNB1), axin (AXIN1), or axin 2 (AXIN2) (7). These changes provided points of departure for the present study.
Humans also develop hepatocellular adenomas (HCAs), a relatively rare benign tumor of the liver found most frequently in women with a history of oral contraceptive use (8). In contrast to HCC, the most frequent genetic change that has been clearly implicated in HCA is mutation that inactivates the TCF1 gene, which encodes the transcription factor hepatocyte nuclear factor 1α (HNF1α). Biallelic inactivating mutations of TCF1 are found in 50% of sporadic HCAs, and some families with heterozygous germ-line mutations in TCF1 display an adenomatosis syndrome, in which individuals develop 10 or more HCAs that exhibit a loss of heterozygosity for TCF1. Although tcf1−/− mice develop hepatomegaly and die around the time of weaning, they have not been reported to show evidence of neoplasia (9). Hence, other events in addition to inactivation of TCF1 are likely to be necessary for HCA genesis.
We previously reported that overexpression of wild-type MET, as observed in a substantial fraction of human HCCs, can initiate HCC genesis in mice (10). We have now used those mice to reconstruct prospectively the genomic progression to both HCA and HCC. The results appear to replicate events that occur during the genesis of both benign and malignant tumors in the human liver. The mouse models described here should prove useful for the further study of tumorigenesis in the liver and for preclinical testing of new therapeutics.
Results
Benign and Malignant Tumors in MET Transgenic Mice.
We previously generated four independent lines of mice that overexpress a wild-type allele of human MET specifically in hepatocytes under the control of doxycycline (10). Use of a human allele of MET allowed discrimination between the transgenic and endogenous Met proteins by immunoanalysis. Two of these lines (TRE-MET lines 3 and 4) developed HCC and HCA often in the same liver (Fig. 1) (10). Typically there were between one and five separate tumor nodules in any given liver. HCCs predominated in line 3, whereas HCAs were dominant in line 4 (data not shown).
Fig. 1.
Gross pathology of the liver in a MET transgenic mouse. Shown is a liver removed from an 8-month-old line 4 LAP-tTA/TRE-MET transgenic mouse. Doxycycline was withheld for the life of the animal. The liver contained nodules of both HCA (a) and HCC (c), which were verified by histological analysis.
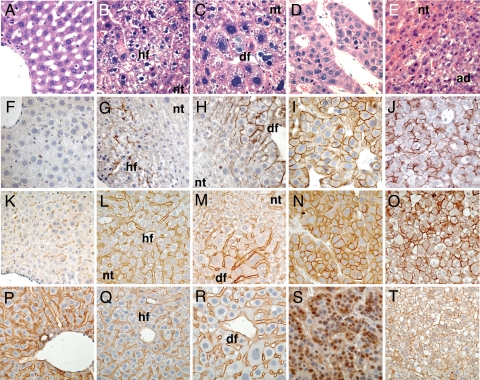
Histologically, the livers sequentially developed hyperplastic foci, dysplastic foci, and, by 3 months of age, overt tumors (Fig. 2 A–E) (10). The HCCs were composed predominantly of hepatocytic cells, had cell plates greater than three cells thick, a paucity of bile duct cells, and lacked lobular architecture (Fig. 2D) (10). In addition, the HCCs expressed the fetal marker alpha-fetoprotein (AFP) [supporting information (SI) Fig. 5N] as found in human HCC. The HCAs also were composed of hepatocytic cells, had a paucity of bile duct cells, and lacked lobular architecture (Fig. 2E). Unlike the HCCs, however, the HCAs had cell plates that were only one to two cells thick (Fig. 2E) and did not express AFP (SI Fig. 5O). We did not observe HCCs developing within HCAs or vice versa. We conclude that the HCCs and HCAs probably arose independently of one another, as is validated further later.
Fig. 2.
Morphological and molecular progression during tumorigenesis in the livers of MET transgenic mice. Doxycyline was withheld for the life of all animals. Sections of livers from LAP-tTA transgenic animals (A, F, K, and P) or line 3 LAP-tTA/TRE-MET animals at 1 month of age (B, G, L, and Q), 2 months of age (C, H, M, and R), or >3 months of age (D, E, I, J, N, O, S, and T) are shown. Livers were sectioned and analyzed by microscopy after H&E staining (A–E), immunohistochemistry with an antibody against phosphorylated Met as a surrogate for kinase activity (F–J), immunohistochemistry with an antibody against human Met (K–O), or immunohistochemistry with an antibody against β-catenin (P–T). Images represent control livers (A, F, K, and P), hyperplastic foci (B, G, L, and Q), dysplastic foci (C, H, M, and R), HCCs (D, I, N, and S), or HCAs (E, J, O, and T). hf, hyperplastic focus; df, dysplastic focus; nt, nontumor tissue; ad, HCA.
Distinct Pathways of Tumorigenesis in MET Transgenic Mice.
We used immunohistochemistry to monitor the expression and activity of Met in various tissues. Phosphorylation of Met served as a surrogate for direct assay of enzymatic activity, which is not presently possible for analyses in situ. There is a well established correlation between enzymatic activity and autophosphorylation of human Met on tyrosine at residues 1234 and 1235 in the activation loop of Met (11, 12). The autophosphorylation can be detected either by use of antibodies specific for the phosphorylated tyrosine residues or immunoprecipitation with an antibody against generic phosphotyrosine and detection with an antibody against Met. In our experience, the two assays have given identical results when performed in parallel on the same samples, so we have used them interchangeably.
Although Met was expressed in all hepatocytes in the livers of transgenic mice (Figs. 1M and 2L), phosphorylated Met was detected only in hyperplastic foci (Fig. 2G), dysplastic foci (Fig. 2H), HCCs (Fig. 2I), and HCAs (Fig. 2J). Furthermore, continued expression of the MET transgene was apparently necessary for maintenance of hyperplastic and dysplastic foci because these lesions were not observed in transgenic animals maintained in the absence of doxycycline for 6 months and then placed on doxycycline for 6 months (data not shown). We conclude that activation of Met coincided spatially and temporally with the onset of preneoplastic lesions in the liver, and that continued expression of the MET transgene was necessary for the maintenance of those lesions. Although it is possible that the presence of phosphorylated Met is simply a marker of proliferating hepatocytes and not the cause of the proliferation, we favor the latter explanation because silencing of the MET transgene expression halts proliferation in both preneoplastic lesions and the subsequent HCC that develop (data not shown) (10).
Taking a clue from previous findings with human HCC, we next examined whether activation of β-catenin was involved in the progression from hyperplastic or dysplastic foci to HCC in the transgenic mice. We tested for activation of β-catenin by two separate assays: nuclear accumulation of β-catenin (7) and expression of a β-catenin target, the gene glul, which encodes the protein glutamine synthetase (GS) (13). Nuclear β-catenin was detected in sections from HCC nodules (Fig. 2S), whereas only membrane staining was present in sections from normal liver, hyperplastic foci, dysplastic foci, and HCAs (Fig. 2 P–R and T). In agreement with the nuclear β-catenin staining, GS was expressed in HCC nodules, but absent from HCA nodules (SI Fig. 5 D and E). As in the normal liver (13), expression of GS in hyperplastic and dysplastic foci was limited to a zone immediately adjacent to the central vein (SI Fig. 5 A–C).
To determine the mechanism of β-catenin activation in HCCs of MET transgenic mice, we sequenced the ctnnb1 in tumors. Twenty of the 21 HCC nodules that we analyzed harbored a heterozygous activating mutation in ctnnb1 (SI Fig. 6 and SI Table 2). The mutations bear a strong resemblance to those found in the ctnnb1 of human HCC (14). In particular, by eliminating crucial sites of phosphorylation, they stabilize β-catenin, causing it to accumulate and acquire constitutive activity in the Wnt signaling pathway (7). In contrast, no HCA nodule that we analyzed (0 of 13) had a mutation in ctnnb1 (data not shown). We conclude that β-catenin was activated in the progression to HCC, but not to HCA.
In the seven instances where we analyzed multiple nodules of HCC from the same liver, separate nodules harbored different ctnnb1 mutations, indicating that each nodule represented an independent clone (SI Table 2). It is possible that these independent nodules arose from a single neoplastic clone and later acquired different mutations in ctnnb1. We do not favor this explanation because these independent clones were frequently in separate lobes of the liver (data not shown), making origin from a single clone less likely.
Motivated again by the example of data from human tumors, we explored the possibility that loss of HNF1α function was involved in HCA genesis. The HNF1α target gene, pah, encoding the protein phenylalanine hydroxylase (PAH) (15) was not expressed in HCA nodules (SI Fig. 5J), but was expressed in normal liver, hyperplastic and dysplastic foci, and HCC nodules (SI Fig. 5 F–I). Despite these findings, we were unable to find any defect in either the structure or expression of HNF1α in the HCAs (data not shown), so we cannot presently explain the absence of PAH in the HCAs. By using hydrodynamic transfection, however, we were able to confirm that the expression of PAH appears to be dependent on signaling by way of HNF1α, thus authenticating PAH as a surrogate marker for such signaling, and that a deficiency in the signaling cooperates with MET to elicit HCAs. Thus, we suspect that HCAs found in the MET transgenic mice arose from a similar collaboration. The absence of β-catenin mutations in the HCAs and the evidence for functional signaling through HNF1α in the HCCs, as opposed to HCAs, support the view that the two types of tumors had independent origins.
Induction of Tumorigenesis in Mice by Hydrodynamic Transfection.
Our results with MET transgenic mice suggested a forked pathway in which activation of the Met kinase initiates tumorigenesis, and progression to HCC or HCA is favored by activation of β-catenin or loss of HNF1α function, respectively. To test this model directly, we used hydrodynamic transfection with a transposable vector to stably express exogenous genes in the liver (16–20).
In an effort to reconstruct the pathogenesis of HCCs and HCAs by using hydrodynamic transfection, we generated vectors containing wild-type human MET, a constitutively active version of CTNNB1 (ΔN90-CTNNB1) (21) and a dominant-negative version of TCF1 containing only the N-terminal 290 aa (DNHNF1α) (22), similar to mutants that are responsible for familial adenomatosis (23). We were able to detect expression of each of the transfected genes by immunohistochemistry with antibodies against epitope tags (SI Fig. 7 A–D). Stable expression occurred in 1–20% of hepatocytes (data not shown) in accord with previous experience (19). We were not able to detect phosphorylated Met in sections of liver from animals that were transfected with MET alone (data not shown). Instead, we inferred that the protein product of transfected MET was functionally active from three phenotypic responses in the liver: induction of widespread and numerous foci of dysplasia (SI Fig. 8B and data not shown), up-regulation of E-cadherin (data not shown), and cooperation with activated β-catenin in tumorigenesis.
Hydrodynamic transfection of ΔN90-CTNNB1 induced patchy expression of the β-catenin target glul throughout the liver (SI Fig. 8I). Hydrodynamic transfection of DNHNF1α gave rise to scattered hepatocytes with a foamy appearance (SI Fig. 8D) and inhibited expression of the HNF1α target pah (SI Fig. 8P). This inhibition sustains the view that expression of pah depends on HNF1α in hepatocytes, in turn validating the use of pah expression as a surrogate for HNF1α activity in liver tissue.
Over the course of 1 year, none of the animals transfected with MET, ΔN90-CTNNB1, or DNHNF1α alone showed any evidence of liver tumors (Fig. 3, SI Fig. 8 B–D, and data not shown). One year after transfection with MET, three animals died of unknown causes, but the remaining six animals remained healthy and without liver tumors (Fig. 3 and data not shown). We conclude that the alleles we generated were functional, but were not sufficient individually to induce tumors over the time period we analyzed. Given that a MET transgene can initiate tumorigenesis, we cannot explain the absence of tumors in mice transfected with MET. The discrepancy may be due either to differences in the levels of expression of Met or the relative number of cells expressing the gene (which is much greater in the transgenic animals).
Fig. 3.
Survival of mice after hydrodynamic transfection. Six- to 8-week-old FVB/N mice were hydrodynamically transfected with the indicated constructs and observed. The number of animals in each group is indicated to the right of each listed group. The black arrowhead indicates the point of hydrodynamic transfection.
None of the genes produced immediate responses of any sort when transfected individually. In contrast, 21% (7 of 34) of animals hydrodynamically transfected with MET in combination with ΔN90-CTNNB1 died within 3 days thereafter (Fig. 3). We cannot explain these deaths decisively, but suggest that the combined activities of β-catenin and Met interfered with recovery from the liver damage that is known to follow hydrodynamic transfection (19). Hydrodynamic transfection of the combination of MET and ΔN90-CTNNB1 gave rise to HCC in 74% (20 of 27) of surviving animals within 1 month and death within 3 months (Fig. 3, SI Fig. 8E, and data not shown). In the seven animals that did not develop HCC, we were unable to detect the protein products of the transfected genes; the transfections had apparently failed. The HCCs were multifocal, with >50 nodules per liver. The nodules contained both phosphorylated Met and ΔN90-β-catenin (SI Fig. 7 E, F, and H). Like the HCCs in MET transgenic mice, the tumors induced by hydrodynamic transfection of MET and ΔN90-CTNNB1 expressed GS, PAH, and AFP (SI Fig. 8 K, Q, and W). Livers from these animals showed no evidence of HCA. Hydrodynamic transfection of MET followed by ΔN90-CTNNB1 3 weeks later, or in reverse order at the same interval, yielded similar results to simultaneous transfection of the two genes (data not shown). We conclude that cooperation between MET and ΔN90-CTNNB1 rapidly induced multifocal HCC irrespective of the order in which the genes were delivered.
Hydrodynamic transfection with a combination of MET and DNHNF1α gave rise to HCA (SI Fig. 8F and data not shown), which occurred as multiple nodules in 50% (5 of 10) of animals within 1 month (data not shown). These animals were still alive after 10 months of observation (Fig. 3), reflecting the indolent nature of HCA. In the five animals that did not develop tumors, we were unable to detect the protein products of the transfected genes. Like the HCAs from MET transgenic mice, the tumors induced by hydrodynamic transfection of MET and DNHNF1α did not express GS, PAH, or AFP (SI Fig. 8 L, R, and X). Immunohistochemistry confirmed that DNHNF1α was expressed in the nuclei of all hepatocytic cells within each HCA nodule (SI Fig. 7G). Western Blotting confirmed that phosphorylated Met was present in HCA nodules, albeit at lower levels than in HCC nodules (SI Fig. 7H). We conclude that cooperation between transfected MET and DNHNF1α rapidly induced multifocal HCA, in accord with our previous inference that a deficiency of signaling by way of HNF1α might be involved in the genesis of HCA in the MET transgenic mice.
Activation of Met and β-Catenin in Human HCC.
To explore whether the genetic pathway to HCC that we delineated in our mouse model also operates in humans, we analyzed samples of human HCC for activation of Met and β-catenin (Table 1 and SI Fig. 9). Among tumors with easily detectable activation of Met, 60% had an activating mutation in CTNNB1 (Table 1). In contrast, only 12% of those tumors with a low or undetectable amount of activated Met harbored an activating mutation in CTNNB1 (Table 1). Among the subset of HCCs with low or undetectable amounts of activated Met, high amounts of total Met were detected in 43% (18 of 41) (SI Fig. 9, lanes 9 and 10, and data not shown), compared with 100% (15 of 15) of tumors with readily detectable activated Met (SI Fig. 9, lanes 1–6, and data not shown). We cannot explain the absence of Met kinase activity in some of the tumors that overexpressed the protein, but the finding raises a caution against using protein levels as the sole assessment of Met in human tumors. In summary, the majority of human tumors (60%) containing activated Met carried mutant alleles of CTNNB1, and the majority of tumors (64%) carrying mutant alleles of CTNNB1 contained activated Met (Table 1). The association of activated Met and mutant alleles of CTNNB1 was statistically significant, with a P value of <0.001 by the χ2 test.
Table 1.
Association of phosphorylated Met with mutant CTNNB1 in human tumors
| Variable | Low or undetectable phosphorylated Met (N = 41) | High phosphorylated Met (N = 15) |
|---|---|---|
| CTNNB1 mutant (N = 14) | 5 (12) | 9 (60) |
| CTNNB1 wild type (N = 42) | 36 (88) | 6 (40)* |
Met activation was detected by immunoprecipitation with an anti-phosphotyrosine antibody, followed by Western blotting with an anti-human Met antibody. Samples were considered to have high phosphorylated Met if they produced a distinct band at intermediate exposures of Western blots. Samples were considered to have low or undetectable phosphorylated Met if they produced an indistinct or absent band by Western blot at the same duration of exposure. Representative data are illustrated in SI Fig. 9. DNA was extracted from human HCCs samples and then subjected to PCR analysis with primers specific for exon three of the CTNNB1 gened. The number of tumors in each group is listed. Numbers in parentheses represent the percentage of tumors with the indicated level of phosphorylated Met. The P value was <0.001 by χ2 for the association of phosphorylated Met with mutant CTNNB1.
*One of the six tumors with wild-type CTNNB1 had high levels of GS, indicating that β-catenin may have been activated by another mechanism in that tumor.
Recurrence After Tumor Regression.
We previously reported regression of HCC in the MET transgenic animals after inactivation of the transgene by administration of doxycycline (10). In an effort to detect residual tumor cells after regression, we exploited the nuclear localization of β-catenin in the cells of HCC. We maintained MET transgenic animals in the absence of doxycycline for 7 months to permit the development of HCC. We then selected animals with enlarged abdomens, indicating the presence of tumor, and placed these animals on doxycycline for 6 months. The tumors regressed, but microscopic foci of small eosinophilic hepatocytic cells containing nuclear β-catenin were detectable within scar tissue (Fig. 4 A and B). Immunohistochemical analysis demonstrated the absence of transgenic Met and phosphorylated Met in these cells (data not shown). In contrast, we never observed hepatocytes with nuclear accumulation of β-catenin outside of scar tissue (Fig. 4B and data not shown), the presence of which would be expected if normal liver tissue had been reconstituted by differentiation of tumor cells.
Fig. 4.
Recurrence of HCC after regression. Eight-month-old LAP-tTA/TRE-MET line 3 mice with tumors were either placed on doxycycline (E, dashed line; n = 18) or continued on a regular diet (E, solid line; n = 15) and then followed for the remainder of the experiment or killed after 6 months (A–D and F). (A–D) Livers from killed mice were sectioned and analyzed by microscopy after H&E staining (A and C) or immunohistochemistry with an antibody against β-catenin (B and D). (A and B) Putative residual tumor cells (arrowheads) embedded in scar tissue adjacent to normal parenchyma. (C and D) Cells from a recurrent tumor nodule. (E) Survival of tumor-bearing mice was analyzed after administration of doxycycline (black arrow). (F) Western Blot analysis was performed on lysates from liver with antibodies against the indicated antigens. Lane 1, nontumor tissue from a LAP-tTA/TRE-MET line 3 mouse with recurrent tumor; lanes 2–4, recurrent tumors from LAP-tTA/TRE-MET line 3 mice maintained in the presence of doxycycline for 1 year; lane 5, wild-type FVB/N control; lane 6, nontissue from a LAP-tTA/TRE-MET line 3 mouse maintained in the absence of doxycycline; lane 7, HCC from a LAP-tTA/TRE-MET line 3 mouse maintained in the absence of doxycycline.
Despite being maintained on doxycycline after regression of HCCs, the MET transgenic mice began to succumb to recurrent tumors within a few months (Fig. 4E). The recurrent tumors displayed the histological appearance of HCC and had nuclear accumulation of β-catenin (Fig. 4 C and D). Western Blot analysis of lysates from the recurrent tumors demonstrated the absence of both phosphorylated and unphosphorylated Met (Fig. 4F). We conclude that inactivation of the MET transgene was sufficient to induce tumor regression even in the presence of mutant ctnnb1. The eventual recurrence of tumors was presumably because of an event that somehow complemented the absence of Met.
We have not ascertained whether HCAs regress when transgenic animals are placed on doxycycline.
Discussion
Distinct Pathways of Tumorigenesis in the Liver.
Our study of transgenic mice provided correlative evidence that tumorigenesis initiated by MET could take different routes depending on which of two spontaneously occurring genetic events arose first: activation of β-catenin or inactivation of the HNF1α pathway. We were able to substantiate this conclusion by introducing exogenous genes into the adult liver with hydrodynamic transfection. Hydrodynamic transfection with suitable combinations of oncogenes produced a large number of tumors rapidly, suggesting that few, if any, additional cooperating events were required to generate hepatic tumors in this experimental setting.
The consistency with which spontaneous activating mutations of ctnnb1 were found in HCCs initiated by the MET transgene presumably reflects a powerful selection for the Wnt signaling pathway as a collaborator with MET during genesis of the tumors and suggests that activation of β-catenin is for some reason favored in that selection. We encountered a similar, albeit not inevitable, pairing of activated Met and β-catenin in human HCCs as well, a correspondence that argues for some measure of authenticity in the mouse model.
Why are mutations in the gene for β-catenin particularly favored in the tumorigenic collaboration with Met? One possibility is that these mutations augment a direct biochemical interaction between MET and β-catenin. Indeed, MET has been reported to directly phosphorylate β-catenin, thereby facilitating its activation (24, 25). However, the mutations described here are known to independently activate β-catenin, so it seems just as likely that signaling from Met and β-catenin are independent variables that cooperate in tumorigenesis for reasons as yet unknown.
Histopathological changes initiated by a MET transgene developed sequentially: hyperplasia first, followed by dysplasia and then HCC. We cannot conclude definitively that each type of morphological lesion was derived from the previous type in this sequence. However, the temporal sequence of morphological lesions paralleled nicely the sequential activation of Met and β-catenin, both of which were present in the eventual HCC. This morphological sequence also is reminiscent of that seen with experimental chemical carcinogenesis in the liver (26). Furthermore, prospective studies have shown the development of HCC within dysplastic nodules in humans with liver disease (27). Nonetheless, definitive proof of the potential for each of these preneoplastic lesions to progress to cancer awaits proper lineage-tracing experiments. HCC and HCA might arise from bipotential liver stem cells, committed hepatocyte progenitors, or from the mature hepatocytes, but it is presently impossible to discern whether the same type of cell can give rise to both types of tumors.
Pathways of Tumorigenesis in the Human Liver.
In our analysis, the majority of human HCCs with activated β-catenin also contained activated Met. However, there was a subset of human HCCs in which β-catenin was activated in the absence of activated Met. Apparently, one or more events other than activation of Met also can cooperate with β-catenin in hepatic tumorigenesis, although such an event might simply affect an element in one of the signaling pathways commanded by Met and, thus, create a phenocopy of Met activity. Analysis of the recurrent tumors may provide access to one or more of those events. Our findings suggest that ≈20% of human HCC may arise through the cooperation of Met and β-catenin, a subset that may correspond to the subset of HCC recently described as expressing a Met signature (28). The implication is that such cooperation is just one of several pathways that engender human HCC. The transgenic model studied here has fortuitously imitated the particular pathway that employs Met and β-catenin. The model could prove useful in the preclinical testing of therapeutics directed at either the Met or Wnt signaling pathway.
A recent report describes a subset of HCAs that is particularly prone to malignant progression and also harbors a mutation in the gene-encoding β-catenin (29). Our transgenic model of HCA apparently represents another subset of human HCAs that does not tend to progress and does not have mutations in the gene-encoding β-catenin. The newly described, premalignant subset of human HCAs may be analogous to premalignant dysplastic foci, which possess some, but not all, of the mutations necessary to drive malignant behavior.
Therapeutic Implications of Tumor Regression in the Mouse Models.
As reported previously (10), tumors initiated by transgenic MET regressed when the transgene was inactivated despite the sustained presence of mutant ctnnb1. Small foci of abnormal cells harboring activated β-catenin remained in scars at the sites of regression. We presume that these were residual tumor cells placed into dormancy by the absence of Met activity. It has been reported previously that regression of various tumors can occur in the presence of presumptive cooperating mutations, but the existence and identity of such mutations was not established (30). However, some tumors do not regress in the presence of a cooperating oncogene (31). It appears that the potential for regression may vary depending on the tissue type and individual oncogenes involved in tumorigenesis.
The regression of HCC after inactivation of a MET transgene helps validate Met as a potential target in the therapy of human HCC. Moreover, our results define a subset of human HCCs that might be susceptible to combination therapy directed against the Met and Wnt signaling pathways. The eventual recurrence of HCC in the mice without reactivation of the MET transgene dramatizes two points. First, resistance to a targeted therapy may arise from an event that bypasses the tumor's original dependence on the therapeutic target, as opposed to a mutational change in the target. Second, any effort to use Met as a target in the treatment of human HCC should anticipate the need for combination therapy to reduce the likelihood of relapse.
Methods
Mice.
Transgenic mice that express the tet transactivator in a liver-specific fashion (LAP-tTA) were mated with transgenic mice that express wild-type human MET under the control of the tetracycline response element (TRE-MET) to generate double-transgenic mice (LAP-tTA/TRE-MET) (10). All mice were on the FVB/N background. LAP-tTA littermates or FVB/N mice were used as controls. Doxycycline was administered in the food (200 mg/kg) for suppression of transgene expression. Genotyping was performed by PCR as described (10).
Hydrodynamic Transfection.
Procedures were as described previously (16–19). Ten to 50 micrograms of the plasmids encoding the Sleeping Beauty transposase and transposons with oncogenes of interest in a ratio of 1:25 were diluted in 2.5 ml of filtered 0.9% NaCl and then injected into the lateral tail veins of 6- to 8-week-old FVB/N mice (Charles River Breeding Laboratories, Portage, MI).
Histology.
Animals were killed, and their livers were removed and rinsed in PBS. One piece was snap-frozen in liquid nitrogen for preparation of lysates, and other pieces were fixed overnight in freshly prepared, cold 4% paraformaldehyde. Fixed tissue samples were then washed three times in PBS and stored in 70% ethanol until they were embedded in paraffin. Five-micrometer sections were placed on slides and stained with H&E.
Immunohistochemistry.
Paraffin was removed from unstained slides with xylenes. The slides were then rehydrated through a series of washes with incrementally decreasing percentages of ethanol. Antigen retrieval was done in 10 mM sodium citrate buffer (pH 6.0) by placement in a microwave on high for 10 min, followed by a 20-min cool down at room temperature. Samples were then subjected to 3% hydrogen peroxide for 10 min to quench endogenous peroxidase activity. Blocking was done with the Avidin-Biotin blocking kit (Vector Laboratories, Burlingame, CA) in combination with either goat serum or the mouse-on-mouse peroxidase kit (Vector Laboratories). Primary antibody binding was done for either 30 min at room temperature or overnight at 4°C. Detection was performed with the ABC-Elite peroxidase kit (Vector Laboratories) by using the DAB substrate kit (Vector Laboratories). Counterstaining was done by a 5-sec dip in hematoxylin Gill 3 (Sigma–Aldrich, St. Louis, MO). Antibodies and dilutions were as follows: anti-phospho-Met (Tyr-1234/1235) antibody (1:25; Cell Signaling Technology, Danvers, MA), anti-human Met (1:500; Zymed Laboratories, South San Francisco, CA), anti-AFP (1:1,000; Dako North America, Carpenteria, CA), anti-glutamine synthetase (1:500; BD Biosciences, San Jose, CA), and anti-phenylalanine hydroxylase (1:500; BD Biosciences).
Preparation of Lysates.
Lysates were made by taking a sample of frozen liver tissue and placing it in a tissue grinder with lysis buffer, which was composed of 1% Nonidet P-40, 50 mM Hepes (pH 7.5), 150 mM NaCl, 10% glycerol, 1.5 mM MgCl2, 1 mM EDTA, 183 mg/ml NaVO4, 100 mM NaF, and a mixture of protease inhibitors consisting of leupeptin, aprotinin, and Pefabloc (Roche Diagnostics, Indianapolis, IN). After homogenization, insoluble debris was removed by centrifugation. Samples from the lysates were then subjected to a BCA protein assay (Pierce Chemical, Rockford, IL).
Immunoprecipitation.
For each sample, 10 μl of anti-phosphotyrosine antibody (4G10) was added to 1 mg of protein in 200 μl of lysis buffer and placed on a rocker overnight at 4°C. Twelve microliters of protein G beads were added to each sample, which was placed on a rocker at 4°C for 1 h. The beads were washed three times with 1 ml of lysis buffer and then boiled in 50 μl of SDS sample buffer; 20 μl was then loaded per lane and subjected to Western Blotting as described next.
Western Blot Analysis.
Fifty-microgram protein samples were subjected to SDS/PAGE. Proteins were transferred to PVDF membranes and blocked with 5% milk. Primary antibody binding was done for either 1 h at room temperature or overnight at 4°C. Detection was performed by ECL (Amersham Biosciences, Piscataway, NJ). Antibodies and dilutions were as follows: anti-phospho-Met (Tyr-1234/1235) antibody (1:1,000; Cell Signaling Technology), anti-human Met (1:2,000; Santa Cruz Biotechnology, Santa Cruz, CA), anti-glutamine synthetase (1:5,000; BD Biosciences), and anti-tubulin (1:250; Abcam, Cambridge, MA).
DNA Sequence Analysis.
Tumor DNA was extracted with a QIAamp Tissue Kit (Qiagen, Valencia, CA) and then subjected to PCR under the following conditions: 94° for 5 min; 35 cycles each of 94° for 30 sec, 56° for 30 sec, and 68° for 1 min; and then a final extension step of 68° for 7 min. Platinum Pfx polymerase was used for all PCR for sequencing. The sequences of the PCR primers, which also were used as sequencing primers, were: mouse BCAT ex2 F (ctgcccgtcaatatctgaaaa), mouse BCAT ex2 R (tcccatggagctcatactgac), human BCAT ex3 F (caatgggtcatatcacagat), and human BCAT ex3 R (agtgacattgctattactctc).
Supplementary Material
Acknowledgments
We thank Cliff Lowell, Kevin Shannon, and Julie Sneddon for critical reading of the manuscript; Linda Prentice for assistance with histology; Luda Urisman for assistance with animal husbandry; and Hans Clevers, Osamu Tetsu, James Nelson, and Frank McCormick for providing plasmids. This work was supported by the George W. Hooper Research Foundation (J.M.B.); National Institutes of Health Grants CA009043 (to J.M.B.), DK49022 (to M.A.K.), and K01 096774 (to X.C.); and National Institute of General Medical Science Grant 5T32GMO7618 (to A.D.T.).
Abbreviations
- AFP
alpha-fetoprotein
- GS
glutamine synthetase
- HCA
hepatocellular adenoma
- HCC
hepatocellular carcinoma
- HNF1α
hepatocyte nuclear factor 1-α
- PAH
phenylalanine hydroxylase.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnasorg/cgi/content/full/0706578104/DC1.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Bruix J, Boix L, Sala M, Llovet JM. Cancer Cell. 2004;5:215–219. doi: 10.1016/s1535-6108(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 3.Thorgeirsson SS, Grisham JW. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 4.Park WS, Dong SM, Kim SY, Na EY, Shin MS, Pi JH, Kim BJ, Bae JH, Hong YK, Lee KS, et al. Cancer Res. 1999;59:307–310. [PubMed] [Google Scholar]
- 5.Tavian D, De Petro G, Benetti A, Portolani N, Giulini SM, Barlati S. Int J Cancer. 2000;87:644–649. [PubMed] [Google Scholar]
- 6.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giles RH, van Es JH, Clevers H. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 8.Zucman-Rossi J. J Hepatol. 2004;40:1036–1039. doi: 10.1016/j.jhep.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Pontoglio M, Barra J, Hadchouel M, Doyen A, Kress C, Bach JP, Babinet C, Yaniv M. Cell. 1996;84:575–585. doi: 10.1016/s0092-8674(00)81033-8. [DOI] [PubMed] [Google Scholar]
- 10.Wang R, Ferrell LD, Faouzi S, Maher JJ, Bishop JM. J Cell Biol. 2001;153:1023–1034. doi: 10.1083/jcb.153.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naldini L, Vigna E, Ferracini R, Longati P, Gandino L, Prat M, Comoglio PM. Mol Cell Biol. 1991;11:1793–1803. doi: 10.1128/mcb.11.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longati P, Bardelli A, Ponzetto C, Naldini L, Comoglio PM. Oncogene. 1994;9:49–57. [PubMed] [Google Scholar]
- 13.Cadoret A, Ovejero C, Terris B, Souil E, Levy L, Lamers WH, Kitajewski J, Kahn A, Perret C. Oncogene. 2002;21:8293–8301. doi: 10.1038/sj.onc.1206118. [DOI] [PubMed] [Google Scholar]
- 14.de La Coste A, Romagnolo B, Billuart P, Renard CA, Buendia MA, Soubrane O, Fabre M, Chelly J, Beldjord C, Kahn A, Perret C. Proc Natl Acad Sci USA. 1998;95:8847–8851. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pontoglio M, Faust DM, Doyen A, Yaniv M, Weiss MC. Mol Cell Biol. 1997;17:4948–4956. doi: 10.1128/mcb.17.9.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 17.Liu F, Song Y, Liu D. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 18.Zhang G, Budker V, Wolff JA. Hum Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 19.Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay MA. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 20.Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mol Cell Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barth AI, Stewart DB, Nelson WJ. Proc Natl Acad Sci USA. 1999;96:4947–4952. doi: 10.1073/pnas.96.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaxillaire M, Abderrahmani A, Boutin P, Bailleul B, Froguel P, Yaniv M, Pontoglio M. J Biol Chem. 1999;274:35639–35646. doi: 10.1074/jbc.274.50.35639. [DOI] [PubMed] [Google Scholar]
- 23.Reznik Y, Dao T, Coutant R, Chiche L, Jeannot E, Clauin S, Rousselot P, Fabre M, Oberti F, Fatome A, et al. J Clin Endocrinol Metab. 2004;89:1476–1480. doi: 10.1210/jc.2003-031552. [DOI] [PubMed] [Google Scholar]
- 24.Monga SP, Mars WM, Pediaditakis P, Bell A, Mule K, Bowen WC, Wang X, Zarnegar R, Michalopoulos GK. Cancer Res. 2002;62:2064–2071. [PubMed] [Google Scholar]
- 25.Zeng G, Apte U, Micsenyi A, Bell A, Monga SP. Exp Cell Res. 2006;312:3620–3630. doi: 10.1016/j.yexcr.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitot HC. Lancet. 2000;358:859–860. doi: 10.1016/S0140-6736(01)06038-X. [DOI] [PubMed] [Google Scholar]
- 27.Libbrecht L, Desmet V, Roskams T. Liver Int. 2005;25:16–27. doi: 10.1111/j.1478-3231.2005.01016.x. [DOI] [PubMed] [Google Scholar]
- 28.Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. J Clin Invest. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zucman-Rossi J, Jeannot E, Nhieu JT, Scoazec JY, Guettier C, Rebouissou S, Bacq Y, Leteurtre E, Paradis V, Michalak S, et al. Hepatology. 2006;43:515–524. doi: 10.1002/hep.21068. [DOI] [PubMed] [Google Scholar]
- 30.Felsher DW. Curr Opin Genet Dev. 2004;14:37–42. doi: 10.1016/j.gde.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 31.D'Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE, Cox JD, Ha SI, Belka GK, Golant A, et al. Nat Med. 2001;7:235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.