Abstract
The multiple steps of DNA transposition take place within a large complex called the transpososome, in which a pair of transposon DNA ends are synapsed by a multimer of the transposase protein. The final step, a DNA strand transfer reaction that joins the transposon ends to the target DNA strands, entails no net change in the number of high-energy chemical bonds. Physiology demands that, despite remaining stably associated with the transpososome, the strand transfer products undergo neither the reverse reaction nor any further cleavage reactions. Accordingly, when the Mu or Tn10 strand transfer complex was produced in vitro through transposase-catalyzed reaction steps, reverse reactions were undetectable. In contrast, when the Mu or Tn10 strand transfer complexes were assembled from DNA already having the structure of the strand transfer product, we detected a reaction that resembled reversal of target DNA strand transfer. The stereoselectivity of phosphorothioate-containing substrates indicated that this reaction proceeds as the pseudoreversal of the normal target DNA strand transfer step. Comparison of the reactivity of closely related Mu substrate DNA structures indicated that the configuration of the flanking DNA outside of the transposon sequence plays a key role in preventing the transposon end cleavage reaction after the strand transfer step.
Keywords: DNA transposition, phage Mu, recombination
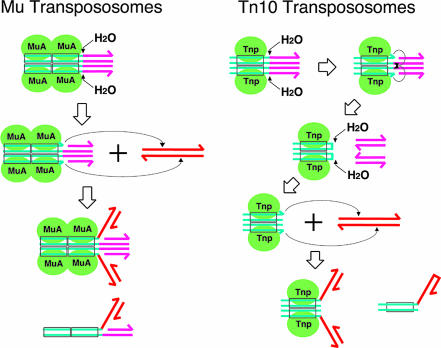
Mobile genetic elements are common throughout nature. Many transposons, including retrotransposons and retroviruses, are mobilized by closely related mechanisms (1, 2). Transposition entails a series of DNA cleavage and joining events. First, the transposon–host junctions are hydrolytically nicked to expose the 3′-OH ends of the transposons. These ends are later joined to the two strands of a target DNA at the site of insertion by one-step transesterification (DNA strand transfer; see Fig. 1) (3). In the case of phage Mu, related replicative transposons, and retroviruses, only one strand of the initial transposon end-containing duplex is cleaved. Other transposons, through a variety of mechanisms, also cut the transposon sequence at or near the 5′ ends, freeing the transposon from its previous surrounding DNA before proceeding with strand transfer. Tn10 uses the 3′ hydroxyls created in the initial step to cut the opposite strand, generating hairpinned transposon ends that are subsequently opened hydrolytically to produce blunt ends (Fig. 1) (4). Thus, Tn10 and its relatives use a four-step process to join the transposon end to a target DNA strand, in contrast to the two-step process of Mu.
Fig. 1.
Transpososomes and transposition reaction steps. Blue-green double lines represent transposon end DNA, and pink double lines represent transposon-flanking DNA. Red double lines are the new target DNA. Rectangles indicate the transposase binding sequences. Transposase molecules are shown as green ovals. Small arrows indicate the nucleophilic attack either by water molecules or by 3′-OH with the arrowheads pointing to the scissile phosphate. Next to the STC, the DNA substrates used for the disintegration reaction are depicted.
These steps are catalyzed by a transposon-encoded transposase in the context of a higher-order protein–DNA complex called a transpososome. The core of the Mu transpososome is composed of two Mu-end DNA segments synapsed by a stably bound tetramer of the transposase, MuA (5–7). Tn5 and Tn10 contain a dimer of transposase (8–10). In the case of Mu and Tn10, a single transposase active site within a transpososome has been shown to catalyze all of the successive chemical reaction steps that take place at each of the transposon ends (9, 11).
Whereas the hydrolytic DNA cleavage steps of transposition are practically irreversible, the strand transfer steps are in principle reversible. This raises the question as to why neither reversal of strand transfer nor the cutting of the new junction between the target and transposon DNA takes place? These reactions, which would be counterproductive for transposons, are considered to be rare, if they occur at all, under physiological conditions. However, efficient reversal of target DNA strand transfer has been observed in cell-free reactions for HIV-1 integrase starting with substrate DNA that mimics the strand transfer product, and this reaction has been termed “disintegration” (12). Determination of the stereochemical course of this reaction indicated that it is mechanistically equivalent to the true reversal of the target DNA strand transfer, rather than mimicry of the viral 3′ end processing reaction with mistaken identity of the nucleophile (13). Whereas this reaction is rather efficient with certain substrate DNAs in vitro, it is suspected to be rare in vivo. An efficient disintegration reaction has not been noticed after transposon end cutting and target strand transfer for the Tn10 or Mu transposition reactions under standard reaction conditions, although heat treatment of transpososomes relaxes this prohibition (ref. 14 and this work). Presumably the architecture of the normal product transpososome prevents such deleterious reactions.
We are interested in finding out what structural features of the transpososome, present after the target strand transfer [strand transfer complex (STC)], prevent cutting of the new transposon–target junction or reversal of the strand transfer reaction. To elucidate how the activity of the transpososome is controlled, we studied the requirements for the disintegration reaction for the Mu and Tn10 transposition, systems that do not normally undo the strand transfer step. We found that the configuration of the flanking DNA that is connected to the transposon end is critical in controlling the enzymatic activity of a Mu transpososome. We also found that the disintegration reaction can be detected for Mu and Tn10 transposition when transpososomes are assembled from DNA substrates that mimic strand transfer products, thus bypassing the earlier reaction steps. Stereoselectivity for chiral phosphorothioate-containing substrates indicated that the mechanism of the Mu and Tn10 disintegration reactions resembles the reverse of the target strand transfer reaction. However, the disintegration reaction is undetectable when the STC is formed through the normal course of the reaction steps, namely transposase-mediated transposon end cutting and target strand transfer, indicating a pathway-dependent conformational difference within the STC.
Results
The Composition of the Flanking DNA Regulates the Transpososome's Activity.
Functional Mu transpososomes can be assembled starting either with intact Mu end DNA segments or with precut Mu end DNA (15, 16). Mu transpososomes can also be assembled from Mu end DNA that has noncomplementary flanking strands (frayed-end DNA) (17). Frayed-end DNA assembles into transpososomes even more readily than the standard Mu end DNA, presumably because it requires less energy to deform the segment adjacent to the transposon end, a necessary step for transpososome assembly. In all cases, the resulting transpososomes efficiently undergo Mu end cleavage and subsequent strand transfer.
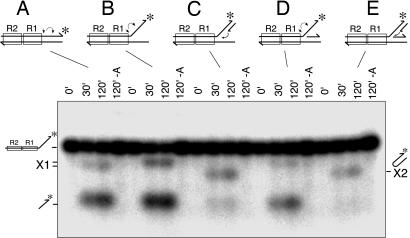
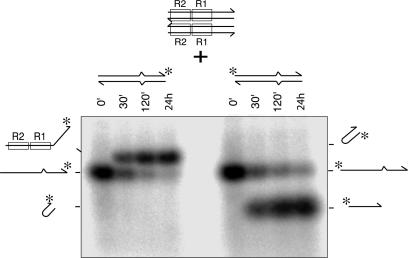
The frayed-end DNA resembles the branched structure of the strand transfer product but differs in that its flanking strands are not associated with respective complementary strands. Therefore, we examined the impact on Mu end cleavage activity of adding the complementary strand to one or both of the flanking DNA strands (Fig. 2). Compared with the reaction with the standard Mu end DNA (Fig. 2A), the 3′ end cleavage reaction is more efficient with frayed-end DNA (Fig. 2B). Addition of the complementary strand to the 3′ flanking strand (Fig. 2C) or to the 5′ flanking strand (Fig. 2D) suppressed the 3′ end cleavage reaction 5- and 3-fold, respectively, and addition of both complementary strands suppressed the cleavage efficiency ≈20-fold (Fig. 2E). Thus, having a duplex DNA structure on the two branches of the flanking DNA segment strongly suppresses the 3′ end cleavage reaction. This inhibition is not due to poor transpososome assembly; analysis of protein–DNA complexes by native agarose gel electrophoresis indicated that these substrates assembled into transpososomes with efficiencies similar to that of the standard substrate (70–80% of input DNA within 30 min; data not shown). Thus, the flanking DNA structure present in the normal STC inhibits Mu 3′ end cleavage.
Fig. 2.
Flanking DNA configuration controls Mu transpososome activity. The substrate DNA used for each set of reactions is depicted above the lanes with asterisks showing the position of the label. Small arrows indicate the reaction that produced the products marked X1 or X2. Each substrate was reacted with MuA for 0 min, 30 min, and 120 min, and products were analyzed by alkaline agarose gel electrophoresis. The fourth lane in each set was incubated in the absence of MuA for 120 min.
In this set of reactions, each substrate yielded other products in addition to the simple 3′ end cleavage product. The gel mobility of the products, labeled “X” in Fig. 2, depended on the substrate. Those (X1) produced from the standard Mu end DNA and substrates with a single strand 3′-flanking segment (Fig. 2 B and D) are, we believe, the result of a preferential strand transfer reaction (after the 3′ end cleavage) into the cleaved flanking DNA. The same products could be detected regardless of whether the top strand was labeled at the 3′ or 5′ end, indicating the labeled strand experienced an internal deletion (data not shown). These products were not analyzed further. The second type of product (X2), made from the substrates containing a duplex 3′-flanking branch, had a hairpin structure as described below.
The extent of hairpin formation was typically limited to 5–10% even after prolonged reaction (data not shown), although a large fraction of the substrate assembled into transpososomes. In contrast, the donor 3′ end cleavage with normal Mu donor DNA with duplex flanking segment can proceed well beyond 50% (Fig. 2A). The majority of MuA molecules in the transpososome remain active during an extended reaction at least up to 48 h [see supporting information (SI) Fig. 8]; thus, inactivation of the transposase during the reaction is not responsible for the limited extent of the reaction.
The 3′ Branch Can Be Removed as a Hairpin as Well as by Hydrolysis.
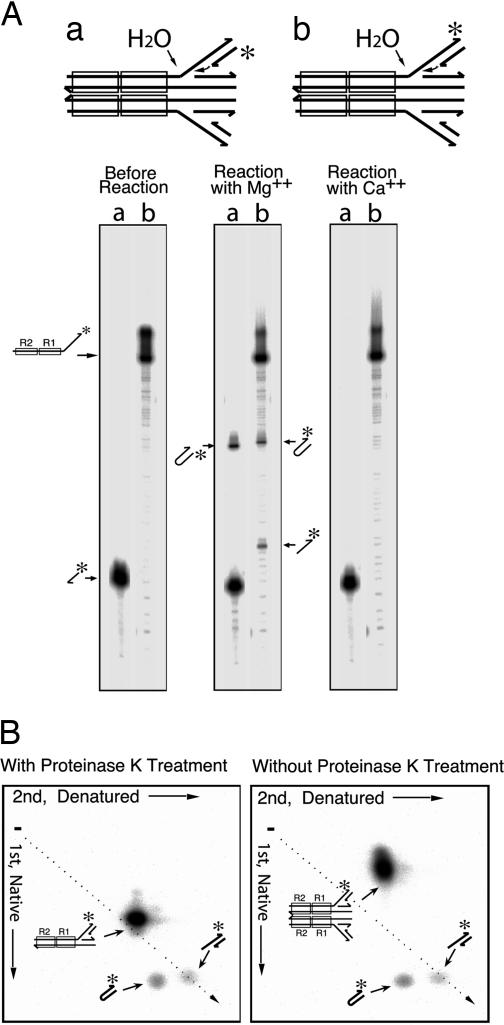
Several lines of evidence verified the hairpin structure of the product yielded by substrates containing duplex 3′-flanking branches. The total length of this product is consistent with this conclusion as judged by its electrophoretic mobility in both alkaline agarose and urea-polyacrylamide gels (Figs. 2 and 3A). The same product was detected when the label was on either strand of this branch (Fig. 3A). Furthermore, the mobility of this product in two-dimensional gel electrophoresis, the first dimension in neutral and the second in alkaline buffers, is consistent with a DNA hairpin (Fig. 3B). The DNA hairpin runs at approximately the same place as the simple cleavage product in the neutral buffer and more slowly in the alkaline buffer, as expected for the approximately double length of the denatured hairpin. Therefore, we conclude that this product was generated when the free 3′-OH end of the complementary strand attacked the opposite strand at the Mu–host junction, yielding a DNA hairpin. Analysis of the products by two-dimensional electrophoresis without prior proteinase treatment, to preserve the structural integrity of the transpososome, demonstrated that neither the hairpin nor the simple cleavage product was stably retained within the transpososome (Fig. 3B Right).
Fig. 3.
MuA generates hairpin flanking DNA upon incubation with the strand transfer product DNA. (A) Urea acrylamide gel analysis. The two substrates, a and b, which differ by the position of the label indicated by asterisks, are depicted on the top and indicated above the lanes. (B) Two-dimensional agarose gel analysis of the products. The sample in Left was deproteinized before electrophoresis, and the sample in Right was electrophoresed as a protein–DNA complex in the first dimension.
It should be noted here that, whereas the substrate for the experiments of Fig. 2 had one unique target branch sequence, experiments in Fig. 3 were carried out by using equimolar mixtures of two substrates whose 3′ (target) branches differed in length and sequence, and only one of which was radiolabeled. Half of the transpososomes assembled in these reactions would have contained one short and one long target branch. Intermolecular splicing of the two target branches, indicative of true reverse strand transfer, would have yielded a mixed-length, non-hairpin product: this was not detected. Each of these substrates contained a 5-nt single-stranded gap, reflecting the 5-bp staggered target DNA cuts during normal target strand transfers. The single-stranded gaps of the two versions of the substrate DNA were complementary to one another, but not self-complementary. We did not detect any significant difference in the efficiency of hairpin formation based on the presence or absence of this complementarity. We conclude that cross-splicing of the two halves of the target DNA branch, a reaction equivalent to the true reversal of the normal target DNA strand transfer, does not take place efficiently for Mu under the conditions studied. The difference from true reversal of the normal strand transfer reaction is also reflected in the metal cofactor specificity. The strand transfer reaction can take place in the presence of Ca2+, instead of Mg2+ (18), but hairpin formation cannot (Fig. 3A).
Branch Removal Shows the Stereoselectivity Expected for Disintegration.
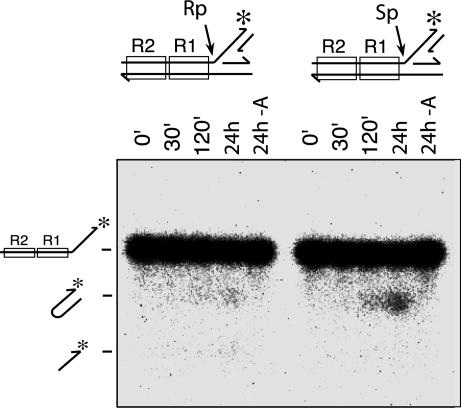
Cutting of the target DNA branch by Mu transposase to produce hairpin flanking DNA ends is chemically similar to reversal of the target DNA strand transfer step of the normal Mu transposition. From the catalytic point of view, is it the reverse of strand transfer, or an aberrant version of the initial 3′ end cleavage reaction using the wrong nucleophile, the nearby 3′-OH of the complementary strand rather than a water molecule? To answer this question, we investigated the chiral phosphorothioate stereoselectivity of this reaction. Every step catalyzed by the DNA transposase/integrase family of proteins so far studied proceeds through inversion of the chirality at the phosphorus reaction center (13, 19–23), indicating an in-line nucleophilic substitution mechanism (24, 25). When the substrate contains a chiral phosphorothioate at the scissile position, transposases exhibit clear preferences for one stereoisomer over the other (21, 19). This stereoselectivity presumably reflects the orientation of the substrate and the contacts made to it within the active site. Both the Mu donor DNA cleavage and target DNA strand transfer steps prefer the Rp diastereomer (19, 21). Because there is inversion of the chirality at each step, the reverse of the target DNA strand transfer step should prefer the Sp diastereomer, as has been shown for the HIV-1 DNA disintegration reaction (13). We prepared two versions of the branched DNA substrate, one containing an Rp and the other an Sp phosphorothioate at the scissile position, and looked at the impact of these substitutions on the Mu disintegration reaction (Fig. 4). The Sp diastereomer was clearly preferred over the Rp form. Therefore, we conclude that this reaction mechanistically mimics the reverse of the target DNA strand transfer reaction, rather than the donor 3′ end cleavage reaction.
Fig. 4.
Phosphorothioate stereoselectivity of the Mu flanking hairpin formation reaction. Each substrate had Rp or Sp phosphorothioate at the scissile position as indicated. The time course of the reaction was monitored by alkaline agarose gel electrophoresis. The last lane in each set was incubated for 24 h without MuA.
No Disintegration Takes Place After Target Strand Transfer.
Does removal of the target DNA branch from the strand transfer product take place during the normal transposition reaction? We investigated this question by using mismatch-specific targeting of Mu transposition. Although normally nonspecific, the Mu strand transfer reaction strongly prefers mismatch-containing target sites, placing the mismatch at the center of the 5-nt target core (26). This avoids the technical difficulty of detecting potential products with size heterogeneity.
A mismatch-containing target DNA was prepared with one strand labeled either at the 5′ or the 3′ end. A limiting quantity of this target DNA was reacted with an excess of the Mu end donor DNA and MuA (Fig. 5). Under the reaction conditions, 50% or more of the target DNA was converted to the strand transfer product with the expected size within 30 min, and after 24 h ≈90% was converted to the product. Strand transfer joined the 3′ portion of the target to the cleaved Mu DNA strand, yielding a slower-migrating species on the alkaline agarose gel. The displaced 5′ end of the target was not joined to the donor DNA and migrated faster (Fig. 5). However, we could not detect any unique band at the position expected for the hairpin product. Even when the products were analyzed by two-dimensional gel electrophoresis (similar to Fig. 3B), which displays a lower background at the expected position of the hairpin, no such product was detected (data not shown). Therefore, we conclude that, when the transpososome is assembled from uncleaved Mu donor DNA substrate and the reaction proceeds through Mu 3′ end cleavage, target capture, and then strand transfer, the target DNA branch within the final complex does not undergo cleavage to form the hairpin product.
Fig. 5.
Hairpin formation does not take place with the authentic STC. Unlabeled duplex Mu end DNA (100 nM) was reacted with MuA (300 nM) in the presence of a limiting concentration of end-labeled mismatch containing target DNA (20 nM). MuB and ATP were present in the reaction to enhance strand transfer efficiency. The target DNA had a 3′ or a 5′ end label as indicated. Products were analyzed by alkaline agarose gel electrophoresis. Positions of the substrate and primary products, as well as the expected positions of the hairpin products, are indicated.
Tn10 Disintegration Also Shows the Stereoselectivity Expected for the Reverse of the Strand Transfer Reaction.
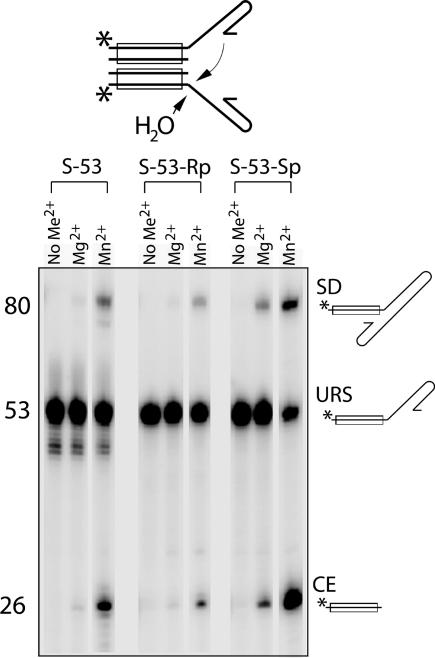
Tn10 transposase also has been shown to efficiently assemble a DNA substrate that mimics the strand transfer product into transpososomes and then catalyze disintegration at a low efficiency (27). As with Mu, this product-mimicking DNA is a poor substrate for the transposon 3′ end cleavage reaction compared with the normal uncleaved transposon end DNA. The two products result from hydrolysis at the 3′ end of the Tn10 sequence and intermolecular single-strand disintegration. A limited amount of double strand disintegration was also detected in the presence of Mn2+ (see SI Fig. 9). Here we investigated the phosphorothioate stereoselectivity of the Tn10 disintegration reaction. We have previously shown that the forward reaction steps of Tn10 display the same phosphorothioate stereoselectivity (Rp) as the corresponding steps of the Mu and HIV-1 reactions (13, 19). Fig. 6 shows that Tn10 disintegration prefers the Sp diastereomer over the Rp form, consistent with its occurring as the reverse of the target DNA strand transfer reaction, as shown above for Mu.
Fig. 6.
Phosphorothioate stereoselectivity of the Tn10 disintegration reaction. The Tn10 strand transfer product DNA used as the substrate with the 5′ end label position indicated by an asterisk is depicted above. Arrows indicate the two types of reaction detected, hydrolytic cleavage and disintegration by transesterification. At the position of the scissile phosphate, each substrate had normal phosphate (S-53), Rp-phosphorothioate (S-53-Rp), or Sp-phosphorothioate (S-53-Sp) as indicated. Reactions were carried out without divalent metal ions, with Mg2+ (5 mM), or with Mn2+ (0.8 mM). Products were analyzed by urea-acrylamide gel electrophoresis. URS, unreacted substrate; CE, cleaved end; SD, single-end disintegration product.
Note that the hydrolysis reaction and disintegration by transesterification both contribute to the product labeled “CE.” The fact that CE production also clearly favors the Sp diastereomer over the Rp form indicates that the hydrolysis reaction also prefers the Sp form. This is the opposite stereoselectivity from that of the other two hydrolysis reactions catalyzed by Tn10 transposase (first-strand cleavage and hairpin opening) (19). Therefore, the major pathway for target branch hydrolysis is mechanistically distinct from the normal strand cleavage reaction steps and appears to be akin to the reversal of strand transfer with a water molecule being used as the nucleophile instead of the cut target 3′ end.
Although the Tn10 disintegration reaction and the Mu target hairpinning reaction exhibit the same phosphorothioate stereoselectivity, there is a critical difference: The Mu reaction is intramolecular, and the corresponding hairpin products have not been detected for Tn10 disintegration. Instead, Tn10 joins the two target DNA branches by intermolecular splicing, more akin to the true reversal of the target strand transfer, which we did not detect for the Mu system. In addition, the stereoselectivity of the hydrolytic cleavage of Mu and Tn10 target branch differ (data for Mu, based on overexposed gel of Fig. 4, which is not shown). Tn10 disintegration can apparently allow a water molecule to act as the nucleophile in place of the cut target 3′-OH, whereas in the Mu case target branch hydrolysis appears mechanistically similar to the first step of normal Mu transposition.
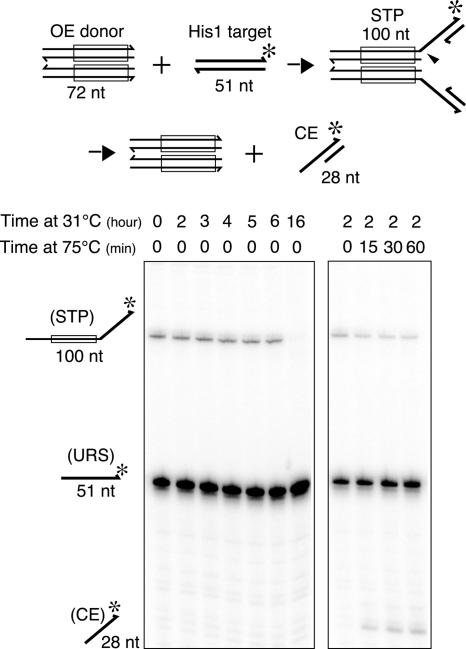
Although the above results as a whole indicate that the disintegration reactions reported here mechanistically mimic the reverse reaction of target strand transfer, they also suggest that the transpososome conformation is not exactly the same as that after the normal target strand transfer step. Like the authentic Mu STC, which does not generate the hairpin product, the authentic Tn10 STC does not cut off the target DNA joined to the Tn10 ends by strand transfer even after a 16-h reaction. However, when the STC was incubated at 75°C the hydrolysis product was readily observed (Fig. 7). An additional observation further supports this notion. A mutant Tn10 transposase carrying an amino acid substitution near the active site (P167S) is defective for the target strand transfer reaction but is as efficient as the wild-type protein for the disintegration reaction (see SI Fig 9). Harshey and colleagues (14) have similarly shown that authentic Mu STCs will also catalyze disintegration when heated to 75°C.
Fig. 7.
Target DNA hydrolysis in the context of the authentic Tn10 STC. STC was assembled by using precleaved OE (72 bp) and a 50-bp target DNA containing the HisG1 hotspot for Tn10 insertion. The latter was labeled with 32P at the 3′ terminus of one strand. Aliquots were removed at the indicated time points and analyzed on an 8% urea-polyacrylamide gel (Left). A portion of the 2-h reaction was saved and subjected to further incubation at 75°C for the indicated amounts of time (Right) and analyzed as above. The appearance of the 28-nt species is indicative of hydrolysis precisely at the target–transposon junction.
Discussion
Control of a transposase's activity is important considering the fact that the same active site must catalyze multiple distinct reaction steps in a defined order to produce a physiologically sensible transposition product. In this article, using phage Mu transposition as an example, we demonstrated that the configuration of the DNA flanking the transposon ends exerts control over the catalytic activities of the transposase. Although the transposon end DNA within the complex is believed to retain its basic structure throughout the reaction, the connectivity to, and the configuration of, the adjacent flanking DNA are significantly altered by the reaction steps. Also, two (Mu) or three (Tn10) different phosphate groups are engaged in the same active site during different reaction steps. Therefore, it would be sensible for the catalytic core of the transpososome to be responsive to the flanking DNA configuration and to use this structural information in dictating whether to catalyze another reaction step, and, if so, which one. Footprinting experiments have demonstrated extended protection of the flanking DNA segment within transpososomes (6, 28, 29), indicating that changes in interactions within the transpososome could be used in sensing changes in the DNA configuration. However, further understanding of how this control is accomplished would require high-resolution structures of the transpososome trapped at each step of the reaction. Control mechanisms similar to those discussed here probably exist for many other enzymes that mediate multiple reaction steps within higher-order protein–nucleic acid complexes, such as the RNA splicing machinery.
Our data imply that, when Mu transpososomes are assembled from DNA segments mimicking the strand transfer product, up to three distinct, very slowly interconverting species of transpososome resulted: minor ones that allow target hairpin formation or hydrolytic cleavage, and a major one that is inactive. We propose this because, although <10% of the target branches were removed even after prolonged reaction, the efficiency of transpososome assembly was often >80%. Although this could reflect the equilibrium of this reaction, branch removal is likely to be energetically downhill because the cleaved fragment is released from the transpososome. We therefore favor the alternative interpretation that only a small percentage of the transpososomes were assembled in a configuration that allows branch removal. This interpretation is attractive because target hairpin formation is not the true reverse of strand transfer, which should have regenerated intact target duplex. The complex that generates this product is thus not the authentic STC. Indeed, the authentic STC that forms after the normal course of reaction steps is inactive for target cleavage under the same reaction conditions. Therefore, we presume that the chemically inactive majority species corresponds to the authentic STC and that the target hairpin is produced by a “misfolded” STC. The hydrolytic cleavage of the target branch may reflect yet another type of complex akin to the synaptic complex that normally contains the duplex donor DNA, in which the “target branch” in the substrate was mistaken as a strand of the flanking host DNA in the duplex donor DNA. Target branch hairpin formation is not an artifact caused by the use of the short DNA fragments as the substrate because a similar reaction has been reported using purified Mu strand transfer product DNA that was made from plasmid DNA substrates (14).
How does the structure of this misfolded STC differ from the authentic one, which bars any further catalysis? Perhaps the 5-nt single-strand gap at the junction loops back and the positions of the right and left target branches are swapped compared with the authentic STC. This possibility has been independently suggested by Au et al. (14). This would allow the scissile phosphate to be positioned in a similar configuration as in the STC, yet somehow, in the authentic STC, the active site is locked into an inactive configuration.
Unlike the Mu disintegration reaction, the Tn10 disintegration reaction appears to be chemically the true reverse reaction of target strand transfer: it is intermolecular rather than intramolecular. Significant hydrolytic target branch cleavage was also observed, and, also unlike Mu, this reaction appeared to proceed by a mechanism distinct from the normal 3′ end cleavage reaction. Because the stereoselectivity of these two Tn10 reactions is the same, they may reflect the use of two different nucleophiles (water and a 3′-OH) within a structurally similar disintegration complex.
Like the Mu reaction, only a small fraction of the Tn10 substrate target branches were removed (either by disintegration or hydrolysis), and no target DNA segment removal was observed with the authentic STC that was generated through the normal course of reaction steps. However, when the authentic Tn10 or Mu STC was heated at 75°C to encourage conformational changes that normally do not take place, removal of the target branch was readily observed (this work and ref. 14). Thus, although Tn10 disintegration is closer to a true reversal of strand transfer than Mu disintegration, in both cases the conformation of the authentic STC prevents this reaction.
Reversal of the strand transfer reaction, although unproductive for the transposon, would be thermodynamically favorable in the absence of protein: the net number of phosphodiester bonds is unchanged, but the system entropy is expected to increase because of the release of the cleaved branch. In the context of the transpososome, the strand transfer products must be stabilized by the protein's preferential interaction with them (product binding energy). Although true catalysts cannot change the overall equilibrium of a reaction, transposases, which generally do not turn over by themselves, can accomplish this apparent violation of thermodynamics by refusing to dissociate from tightly bound products. In the case of Mu transposition, the authentic STC is disassembled in an ATP-dependent manner by ClpX (30, 31).
An efficient disintegration reaction has been reported for RAG1/2-mediated transposition of the V(D)J recombination signal sequence (32). It is possible that, like the disintegration reactions studied here, the RAG1/2-mediated “V(D)J disintegration reaction” is inefficient under physiological circumstances. However, an alternative possibility is also attractive. The RAG1/2 reaction is widely believed to have evolved from a transposon, most likely a member of the Transib family (33), because of the closely related reaction mechanism to HAT family transposition (34). But in its modern-day domesticated role in vertebrate cells, which is to recombinationally assemble antigen receptor gene segments, the transposition reaction is not only nonproductive, but dangerous for the organism (35). Whereas transposition activity has been detected for the RAG system in a cell-free reaction (36, 37), in vivo RAG-mediated transposition appears to be very rare (38). An efficient disintegration reaction may be a contributing factor to help avoid accidental transposition, which could cause cell death or oncogenesis in lymphoid tissues.
Experimental Procedures
Purification of the MuA protein and Mu transposition reaction conditions were as previously described (19, 39). Preparation of the Tn10 transposase and Tn10 reaction conditions have been published (40, 41). Oligonucleotides were purchased from Operon Biotechnologies (Huntsville, AL) or from Sigma–Genosys (Oakville, ON, Canada), and appropriate ends were labeled with 32P by using either [γ-32P]ATP (NEN, Waltham, MA) and T4 polynucleotide kinase or [α-32P]ddATP (Amersham, Piscataway, NJ) and terminal deoxynucleotidyl transferase. The details of the oligonucleotide sequences used are described in SI Text, and the substrates assembled from them are depicted in SI Fig. 10. Methods for the purification of the diastereomers of chiral phosphorothioate containing oligonucleotide and assembly of the substrates were essentially as described (19) and are summarized in SI Text.
Alkaline agarose gel electrophoresis was carried out in 4% MetaPhor agarose (Cambrex Bio Science, Rockland, ME) using 50 mM NaOH/0.1 mM EDTA as the buffer. Two-dimensional gels were run by using Tris acetate buffer in the first dimension and the alkaline buffer in the second. Gels were neutralized and dried, and the autoradiography was done by using a Fuji BAS2000 or Fuji BAS2500.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health, the National Institutes of Health Intramural AIDS Targeted Antiviral Program (M.M., P.A.R., and K.M.), and the Canadian Institutes of Health Research (S.J.W. and D.B.H.).
Abbreviation
- STC
strand transfer complex.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706556104/DC1.
References
- 1.Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: Am Soc Microbiol; 2002. [Google Scholar]
- 2.Curcio MJ, Derbyshire KM. Nat Rev Mol Cell Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 3.Mizuuchi K. Genes Cells. 1997;2:1–12. doi: 10.1046/j.1365-2443.1997.970297.x. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy AK, Guhathakurta A, Kleckner N, Haniford DB. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 5.Baker TA, Mizuuchi K. Genes Dev. 1992;6:2221–2232. doi: 10.1101/gad.6.11.2221. [DOI] [PubMed] [Google Scholar]
- 6.Lavoie BD, Chan BS, Allison RG, Chaconas G. EMBO J. 1991;10:3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizuuchi M, Baker TA, Mizuuchi K. Cell. 1992;70:303–311. doi: 10.1016/0092-8674(92)90104-k. [DOI] [PubMed] [Google Scholar]
- 8.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 9.Bolland S, Kleckner N. Cell. 1996;84:223–233. doi: 10.1016/s0092-8674(00)80977-0. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy AK. London, ON, Canada: Univ of Western Ontario; 1999. Ph.D. thesis. [Google Scholar]
- 11.Williams TL, Jackson EL, Carritte A, Baker TA. Genes Dev. 1999;13:2725–2737. doi: 10.1101/gad.13.20.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow SA, Vincent KA, Ellison V, Brown PO. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 13.Gerton JL, Herschlag D, Brown PO. J Biol Chem. 1999;274:33480–33487. doi: 10.1074/jbc.274.47.33480. [DOI] [PubMed] [Google Scholar]
- 14.Au TK, Pathania S, Harshey RM. EMBO J. 2004;23:3408–3420. doi: 10.1038/sj.emboj.7600344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craigie R, Mizuuchi K. Cell. 1987;51:493–501. doi: 10.1016/0092-8674(87)90645-3. [DOI] [PubMed] [Google Scholar]
- 16.Mizuuchi M, Mizuuchi K. Cell. 1989;58:399–408. doi: 10.1016/0092-8674(89)90854-4. [DOI] [PubMed] [Google Scholar]
- 17.Savilahti H, Rice PA, Mizuuchi K. EMBO J. 1995;14:4893–4903. doi: 10.1002/j.1460-2075.1995.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savilahti H, Mizuuchi K. Cell. 1996;85:271–280. doi: 10.1016/s0092-8674(00)81103-4. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy AK, Haniford DB, Mizuuchi K. Cell. 2000;101:295–305. doi: 10.1016/s0092-8674(00)80839-9. [DOI] [PubMed] [Google Scholar]
- 20.Engelman A, Mizuuchi K, Craigie R. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 21.Mizuuchi K, Nobbs TJ, Halford SE, Adzuma K, Qin J. Biochemistry. 1999;38:4640–4648. doi: 10.1021/bi990054p. [DOI] [PubMed] [Google Scholar]
- 22.vanGent DC, Mizuuchi K, Gellert M. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 23.Mizuuchi K, Adzuma K. Cell. 1991;66:129–140. doi: 10.1016/0092-8674(91)90145-o. [DOI] [PubMed] [Google Scholar]
- 24.Knowles JR. Annu Rev Biochem. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- 25.Eckstein F. Annu Rev Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- 26.Yanagihara K, Mizuuchi K. Proc Natl Acad Sci USA. 2002;99:11317–11321. doi: 10.1073/pnas.132403399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart BJ, Wardle SJ, Haniford DB. EMBO J. 2002;21:4380–4390. doi: 10.1093/emboj/cdf425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuuchi M, Baker TA, Mizuuchi K. Proc Natl Acad Sci USA. 1991;88:9031–9035. doi: 10.1073/pnas.88.20.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crellin P, Chalmers R. EMBO J. 2001;20:3882–3891. doi: 10.1093/emboj/20.14.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levchenko I, Luo L, Baker TA. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 31.Kruklitis R, Welty DJ, Nakai H. EMBO J. 1996;15:935–944. [PMC free article] [PubMed] [Google Scholar]
- 32.Melek M, Gellert M. Cell. 2000;101:625–633. doi: 10.1016/s0092-8674(00)80874-0. [DOI] [PubMed] [Google Scholar]
- 33.Kapitonov VV, Jurka J. PloS Biol. 2005;3:998–1011. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou LQ, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 35.Jones JM, Gellert M. Immunol Rev. 2004;200:233–248. doi: 10.1111/j.0105-2896.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 36.Agrawal A, Eastman QM, Schatz DG. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 37.Hiom K, Melek M, Gellert M. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 38.Reddy YVR, Perkins EJ, Ramsden DA. Genes Dev. 2006;20:1575–1582. doi: 10.1101/gad.1432706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizuuchi M, Mizuuchi K. EMBO J. 2001;20:6927–6935. doi: 10.1093/emboj/20.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chalmers RM, Kleckner N. J Biol Chem. 1994;269:8029–8035. [PubMed] [Google Scholar]
- 41.Sakai J, Chalmers RM, Kleckner N. EMBO J. 1995;14:4374–4383. doi: 10.1002/j.1460-2075.1995.tb00112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.