Abstract
The regulated accumulation of the contact-dependent extracellular C-signal morphogen in the bacterium Myxococcus xanthus ensures the temporal and spatial coordination of multicellular morphogenesis and cellular differentiation during fruiting bodyformation. Synthesis of the C-signal depends on the csgA gene. The CsgA protein exists in two forms, the full-length 25-kD protein (p25), which is homologous to short-chain alcohol dehydrogenases, and a 17-kD protein (p17). The molecular nature of the C-signal has remained elusive. Here we show that p25 and p17 are associated with the outer membrane and that p17 copurifies with C-signal activityfrom M. xanthus cells. p17 corresponds to the C-terminal part of p25. A recombinant p17 protein, which lacks the N-terminal coenzyme binding pocket and which fails to bind NAD+ in vitro, has C-signal activity. These data provide evidence that p17 is the active species in C-signaling and that p17 does not act as a short-chain alcohol dehydrogenase to generate the C-signal. We further provide evidence that p17 is synthesized by N-terminal proteolytic processing of p25 by a serine protease. Compared to other bacterial signaling molecules, p17 is unusual with respect to size and cell-surface association. In these regards, C-signal is functionally analogous to eukaryotic signaling proteins.
Keywords: Extracellular signaling, proteolysis, morphogen, development, protein secretion
A recurring theme in developmental biology is the induction of different responses by the same extracellular signaling protein. These responses may be context-dependent or in the case of morphogens they may be concentration-dependent. Examples of extracellular signaling proteins acting as morphogens in eukaryotic systems are Wingless, Hedgehog, and Dpp in the Drosophila wing and leg imaginal discs (Teleman et al. 2001). In the Gram-negative bacterium Myxococcus xanthus, the extracellular C-signal acts as morphogen and induces distinct responses at discrete thresholds during the developmental process that leads to formation of multicellular, spore-filled fruiting bodies. The molecular nature of the C-signal has remained elusive. To understand a signaling pathway, it is essential to define the biochemical nature of the signaling molecule and the mechanisms by which cells produce this signal. Here, we describe the identification of the C-signal and analyze how it is produced.
Fruiting body formation in M. xanthus is initiated by starvation of cells at a high cell density on a solid surface. Controlled changes in organized cell movements from swarming to aggregation constitute the basis for the formation of fruiting bodies (Dworkin 1996). The first signs of morphogenesis are evident after 6 h with the formation of small aggregation foci. These foci enlarge into mounds as the result of continued aggregation of cells, and at 24 h immature fruiting bodies have formed, each containing 105 densely packed cells. Within the immature fruiting bodies, the rod-shaped cells differentiate into spores, resulting in the formation of mature fruiting bodies. Fruiting body morphogenesis is accompanied by temporal changes in gene expression (Inouye et al. 1979; Kroos et al. 1986). Developmental gene expression, in turn, is also important for progression of the developmental program (Kroos et al. 1990).
Fruiting body formation is dependent on five extracellular signals (A to E) that act in a specific sequential order (Shimkets 1999). The C-signal is the latest acting of these signals and becomes important for development after 6 h (Kroos and Kaiser 1987). Mutants unable to synthesize the C-signal carry mutations in the csgA gene (Shimkets et al. 1983). csgA cells are deficient in aggregation and sporulation, and display reduced or abolished expression of genes normally induced after 6 h (Shimkets et al. 1983; Kroos and Kaiser 1987). The C-signal is not only required for these responses but directly induces aggregation, sporulation, and full expression of these genes (Kim and Kaiser 1991; Li et al. 1992; Kruse et al. 2001). C-signal elicits these responses at specific thresholds: At intermediate levels aggregation and early C-signal dependent genes are induced, and at high levels sporulation and the latest C-signal dependent genes are induced (Kim and Kaiser 1991; Li et al. 1992; Kruse et al. 2001). A regulated increase in the level of C-signaling during development ensures the correct temporal order of aggregation and sporulation (Kim and Kaiser 1991; Li et al. 1992; Kruse et al. 2001). Furthermore, the contact-dependent C-signal transmission mechanism (see below) ensures the spatial coupling of aggregation and sporulation (Kruse et al. 2001).
The CsgA protein exists in two forms, one with an approximate size of 25 kD (designated p25), which corresponds to full-length CsgA protein, and one with an approximate size of 17 kD (designated p17; Kruse et al. 2001), which is similar in size to the C-factor protein (Kim and Kaiser 1990a,b; see below). Only p25 is detected in vegetative cells, and both proteins accumulate during development (Kruse et al. 2001). C-signal transmission occurs by a contact-dependent mechanism (Kim and Kaiser 1990a). Consistently, antibodies against the C-terminal 166 residues of the 229-residue p25 protein block development of wild-type cells and recognize epitopes, which are located on the surface of developing wild-type cells (Shimkets and Rafiee 1990). The 17-kD C-factor protein was purified from starving wild-type cells by detergent extraction and biochemical fractionation on the basis of its ability to restore development to csgA cells (Kim and Kaiser 1990a). The understanding that the C-factor protein is the C-signal was questioned by several observations, which suggested that p25 might act as an enzyme to produce the C-signal. First, p25 is homologous to members of the short-chain alcohol dehydrogenase (SCAD) family of intracellular proteins (Baker 1994; Lee et al. 1995). SCAD enzymes contain two conserved sequence motifs, both of which are present in p25: an N-terminal motif corresponding to the NAD(P)-coenzyme binding pocket and a more C-terminal motif involved in the catalytic mechanism (see Fig. 3, below; Oppermann et al. 2003). Consistently, p25 binds NAD+ in vitro (Lee et al. 1995). Secondly, exogenous p25 purified from Escherichia coli restores development of csgA cells; however, exogenous p25 proteins carrying substitutions in either the coenzyme binding pocket or in the catalytic site fail to restore development of csgA cells (Lee et al. 1995). Moreover, a p25 protein which carries a substitution in the coenzyme binding pocket has reduced capacity to bind NAD+ in vitro (Lee et al. 1995). Finally, overproduction of the SocA protein, which is homologous to SCAD enzymes, restores development in a csgA mutant in vivo (Lee and Shimkets 1994, 1996). Enzymatic activity of p25, however, still remains to be shown. Likewise, the molecular difference between p25 and the C-factor protein has not been elucidated. Thus, it remains to be resolved whether one of the csgA proteins acts as a SCAD to produce the C-signal or whether one of the csgA proteins is the actual C-signal.
Figure 3.
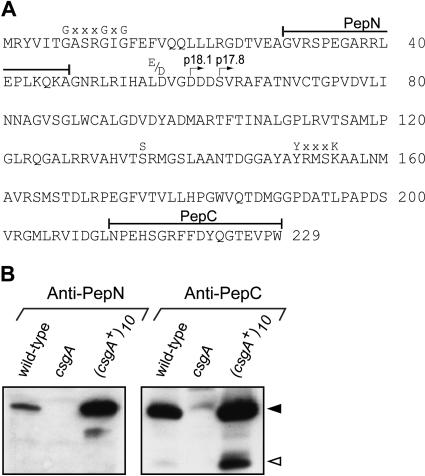
p17 corresponds to the C-terminal part of p25. (A) Primary amino acid sequence of p25. The PepN and PepC peptides are indicated. The csgA part of the recombinant MalE–p18.1 and MalE–p17.8 are marked with right-pointing arrows. Conserved residues in the coenzyme binding pocket [G(x)2–3 GxG(x)33–55(D/E)], and in the catalytic site [S(x)12–14Y(x)3K] of SCADs are shown. (B) Immunoblot assay of M. xanthus total cell lysates. Wild type (DK1622), csgA (DK5208), and (csgA+)10 (DK1622/pTK98-10) were starved for 12 h in submerged culture, and proteins from 2.5 × 108 input cells were loaded per lane. The immunoblots were probed with anti-PepN and anti-PepC antibodies as indicated. Closed and open arrowheads indicate p25 and p17, respectively.
Here, we examined the nature of the C-signal. We present evidence that p25 as well as p17 are associated with the outer membrane. Our data are consistent with the notion that p17 corresponds approximately to the C-terminal 17 kD of p25. Recombinant csgA proteins, which correspond to the C-terminal 18.1 and 17.8 kD of p25, have C-signal activity. These recombinant csgA proteins lack the NAD(P)+ binding pocket. Consistently, the recombinant protein with a size of 18.1 kD is unable to bind NAD+ in vitro. From these observations, we conclude that p17 does not act as a SCAD to produce the C-signal. Rather, these data provide evidence that p17 is the C-signal. Furthermore, we provide biochemical evidence that p17 is produced by N-terminal proteolytic processing of p25 by a serine protease.
Results
The csgA proteins are associated with the outer membrane
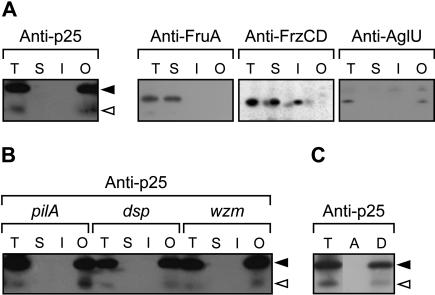
C-signal transmission occurs by a contact-dependent mechanism (Kim and Kaiser 1990a), suggesting that the active species in C-signaling is exposed on the cell surface. To determine the subcellular localization of p17 and p25, we carried out biochemical fractionation experiments on whole-cell lysates of wild-type cells (DK1622) that had been starved for 24 h (Materials and Methods). Proteins were separated into three fractions enriched for soluble proteins, inner membrane proteins, or outer membrane proteins. Next, proteins in these fractions were subjected to immunoblot analysis using antibodies against p25. Both p17 and p25 were detected in the outer membrane fraction and were absent from the inner membrane fraction and the fraction containing soluble proteins (Fig. 1A). As controls, the cytoplasmic FruA protein (Ogawa et al. 1996; Ellehauge et al. 1998) was found in the fraction containing soluble proteins; the FrzCD protein, a cytoplasmic protein which is associated with the inner membrane (McBride et al. 1992) was found in the soluble fraction as well as in the inner membrane fraction; and the outer membrane protein AglU (Thomasson et al. 2002) was found in the outer membrane fraction (Fig. 1A). To determine whether the association of p17 and p25 with the outer membrane fraction was due to ionic interactions, the fractionation was repeated in the presence of 1 M NaCl in all buffers. Following this fractionation, p17 and p25 were still detected only in the outer membrane fraction (data not shown). Thus, the association of p17 and p25 with the outer membrane is not simply due to ionic interactions.
Figure 1.
Biochemical fractionation of starved M. xanthus cells. (A) Wild-type cells (DK1622) were starved for 24 h on TPM-agar. The total cell lysate (T) was separated into fractions enriched for soluble proteins (S), inner membrane proteins (I), and outer membrane proteins (O). The fractions were subjected to immunoblot analyses and probed with anti-p25 antibodies. As controls, the association of FruA, FrzCD, and AglU with the different fractions was determined using anti-FruA, anti-FrzCD, and anti-AglU antibodies, respectively. (B) pilA cells (DK10407), which are unable to synthesize type IV pili; dsp cells (DK3470), which are unable to synthesize extracellular fibrils; and wzm cells (HK1321), which are deficient in lipopolysaccharide O-antigen synthesis, were starved and fractionated as in A and probed with anti-p25 antibodies. (C) DK1622/pTK98-10 cells were starved for 24 h. The lysate was phase separated with Triton X-114 into an aqueous (A) and a detergent (D) phase. The immunoblots were probed with anti-p25 antibody. Closed and open arrowheads indicate p25 and p17, respectively.
To analyze whether the association of p17 and p25 with the outer membrane depends on type IV pili, extracellular fibrils, or lipopolysaccharide O-antigen, the subcellular localization of p17 and p25 during development was examined in cells lacking these structures. In all mutant backgrounds, the localization of p17 and p25 was similar to that observed in wild-type cells (Fig. 1B).
As a further test of the outer membrane association of p17 and p25, a whole-cell lysate of starved DK1622/pTK98-10 cells, which overproduce the csgA proteins, was subjected to a Triton X-114 extraction followed by phase separation (Materials and Methods). In a Triton X-114 phase separation, amphiphilic membrane proteins and lipoproteins partition to the detergent phase, and hydrophilic proteins partition to the aqueous phase. As shown in Figure 1C, both p25 and p17 partition to the detergent phase after the Triton X-114 phase separation. As a control, the Tgl lipoprotein (Rodriguez and Kaiser 1997) was found associated with the detergent phase (data not shown). These data indicate that p17 and p25 are either amphiphilic membrane proteins or subject to a hydrophobic modification, which anchors them in the outer membrane.
p17 copurifies with the C-signal
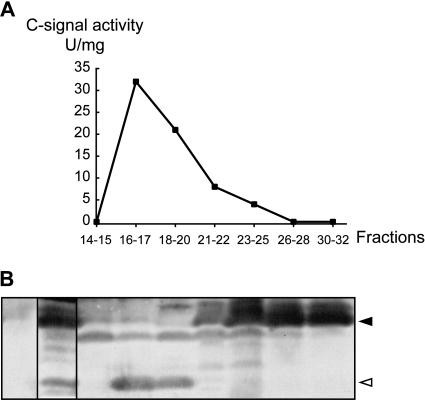
To determine which of the csgA proteins copurifies with the C-signal, the C-signal was partially purified from starving DK1622/pTK98-10 cells using the procedure of Kim and Kaiser (1990a,b; Materials and Methods). After elution of adsorbed proteins from a Mono Q anion-exchange column, fractions were analyzed for the presence of C-signal activity using the C-signal bioassay. In this assay, fractions to be tested for C-signal activity are added to starving csgA cells. Fractions containing C-signal restore aggregation and sporulation to the csgA cells. The C-signal eluted in a broad peak with C-signal activity being detected in the pooled samples of fractions 16–17, 18–20, 21–22, and 23–25 (Fig. 2A). This elution profile is similar to that previously reported for the C-factor protein (Kim and Kaiser 1990b).
Figure 2.
The C-signal copurifies with the p17 protein. (A) C-signal was purified from starved DK1622/pTK98-10 cells. Fractions eluted from a Mono Q column were pooled as indicated and tested for C-signal-specific activity. (B) Pooled fractions were subjected to immunoblot analysis using anti-p25 antibodies. The leftmost lane contains a total lysate of the csgA strain, DK5208, starved for 12 h; the second lane is the sample prior to Mono Q chromatography. Closed and open arrowheads indicate p25 and p17, respectively.
Next, the pooled samples were analyzed for the presence of p17 and p25 using the anti-p25 antibodies. p17 was detected in the samples consisting of the pooled fractions 16–17 and 18–20 (Fig. 2B). These samples correspond to the samples containing the highest specific activity of C-signal (Fig. 2A). High levels of p25 were detected in samples 23–25, 26–28, and 30–32. Among these samples, only sample 23–25 contained a low level of C-signal activity. Hypothetically, high protein content in the fractions containing p25 could have interfered with the C-signal assay. However, the protein content in the pooled samples varies by less than twofold (data not shown). These data provide evidence that the specific C-signal activity of p25 is significantly lower than that of p17. Moreover, they demonstrate that p17 copurifies with the C-signal. This observation is in agreement with that of Kim and Kaiser (1990a,b), who reported that the 17-kD C-factor protein copurified with the C-signal. We propose that p17 is identical to the C-factor protein.
p17 corresponds to the C-terminal part of p25
To determine the difference between p17 and p25, polyclonal antibodies were raised against peptides from the N and C termini of p25. These peptides correspond to residues 30–47 (PepN) and 212–229 (PepC) in p25, respectively (Fig. 3A). To analyze whether these antibodies recognize p17 and p25, immunoblots were carried out on total protein isolated from wild-type cells, cells that overproduce the csgA proteins (DK1622/pTK98-10) and csgA cells (DK5208) that had been starved for 12 h. The anti-PepN antibodies only detected p25 in wild-type cells and in the strain overproducing the csgA proteins (Fig. 3B). In contrast, the anti-PepC antibodies recognized p25 and p17 (Fig. 3B). These data provide evidence that p17 and p25 have similar C termini and different N termini. Moreover, these data indicate that p17 corresponds to an N-terminal truncated form of p25.
Recombinant p17 has C-signal activity
The estimated molecular mass of p17 from SDS-PAGE is 17.7 ± 0.4 kD (data not shown). The findings in the previous section in combination with the observations that p25 neither contains a signal peptide (Pugsley 1993) nor a signal peptide for export through the twin-arginine targeting protein export system (Yen et al. 2002) led us to investigate whether the expression in vivo of a csgA protein corresponding to the C-terminal 18.1 kD of p25 is sufficient to correct the developmental defects in a csgA mutant in the absence of p25. Accordingly, the plasmid pSL628, which contains a truncated csgA gene encoding residues 60–229 of p25 (the calculated molecular mass of this protein is 18.1 kD), was constructed (Materials and Methods) and integrated at the phage Mx8 attachment site in the csgA mutant DK5208. As a control, the plasmid pSL627, which contains the intact csgA gene, was also integrated at the Mx8 attachment site in DK5208. When exposed to starvation, DK5208/pSL627 displayed a wild-type developmental phenotype, whereas DK5208/pSL628 displayed a developmental phenotype characteristic of the csgA mutant (data not shown). Immunoblots of total protein from starving cells showed that DK5208/pSL627 synthesized p17 as well as p25 and DK5208/pSL628 synthesized a protein with a size of 18.1 kD (data not shown). However, this 18.1-kD protein remained in the soluble fraction and did not localize to the outer membrane (data not shown). Thus, the inability of the 18.1-kD protein to rescue development may be due to lack of proper localization of this protein. Similar results were obtained with csgA alleles encoding csgA proteins corresponding to amino acid residues 17–229 and 31–229 of p25 (data not shown). These observations suggest that p25 is required for proper localization of p17 to the outer membrane.
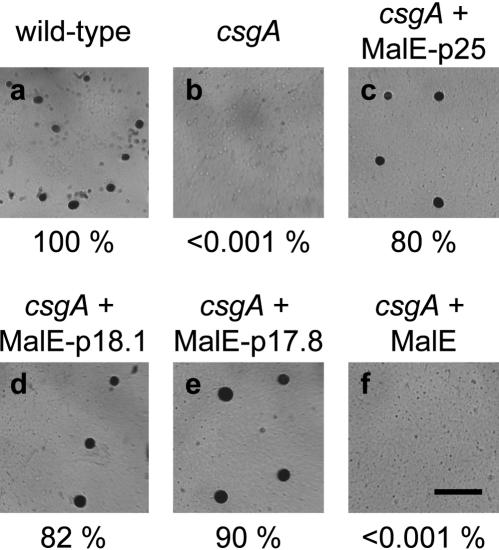
Therefore, to test whether a protein corresponding to the C-terminal 18.1 kD of p25 can correct the developmental defects in a csgA mutant in the absence of p25 by extracellular rescue, we purified a recombinant MalE–CsgA protein (designated MalE–p18.1) from E. coli which contains residues 60–229 of p25 and then tested it for biological activity in the C-signal bioassay. As a control, a MalE–CsgA fusion protein (designated MalE–p25), which contains amino acid residues 1–229 of p25, was also purified. Exogenous MalE–p18.1 protein corrected the developmental defects in csgA cells and restored aggregation as well as sporulation (Fig. 4d). Sporulation in the csgA cells was restored to the same level as that observed in wild-type cells (Fig. 4a,d). In agreement with previous observations (Lee et al. 1995), exogenous MalE–p25 protein also rescued the developmental defects in the csgA cells and restored aggregation and sporulation (Fig. 4c). A MalE–CsgA protein which contains residues 63–229 of p25 (designated MalE–p17.8) also corrects the developmental defects in csgA cells (Fig. 4e). As a control, exogenous MalE protein was tested in the bioassay. MalE protein did not rescue the developmental defects in csgA cells (Fig. 4f). The specific activity of the three fusion proteins is similar, 20 U/mg. A similar specific activity was reported for a slightly different MalE–p25 fusion protein (Lee et al. 1995; Søgaard-Andersen and Kaiser 1996). This activity is 2000-fold lower than that of C-factor protein purified to homogeneity from wild-type cells (Kim and Kaiser 1990b). We speculate that the low specific activities are due to missing posttranslational modification(s) in the csgA part of the fusion proteins and/or the presence of the large MalE moiety (42.5 kD) in the fusion proteins. From these experiments, we conclude that a recombinant p17 protein corresponding to the C-terminal 167 amino acid residues of p25 has C-signal activity. Even though the precise N terminus of p17 synthesized in vivo remains to be identified, these data argue that p17 synthesized in vivo corresponds to the C-terminal 17.7 ± 0.4 kD of p25.
Figure 4.
C-signal activity of MalE–p25, MalE–p18.1, and MalE–p17.8. Wild-type (DK1622, a) and csgA cells (DK5253, b) developed in submerged culture in A50-starvation buffer are shown together with csgA cells (DK5253) developed in the presence of one unit (corresponding to a final concentration of 2 μM) of MalE–p25 (c), MalE–p18.1 (d), or MalE–p17.8 (e). MalE protein was added to a final concentration of 2 μM (f). Fruiting bodies and spores were scored after 72 h of development. Sporulation frequencies are shown below each panel and are presented relative to the level of sporulation in DK1622, which had an absolute sporulation frequency of 0.96%. Bar, 0.5 mm.
Recombinant p17 does not bind NAD+ in vitro
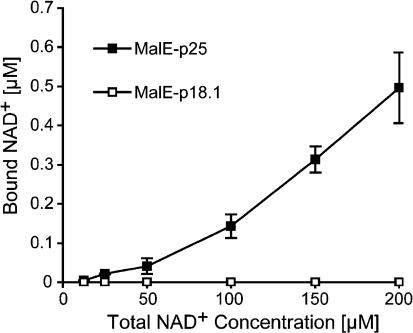
MalE–p25 protein binds NAD+ in vitro (Lee et al. 1995). The MalE–p18.1 and MalE–p17.8 proteins lack the coenzyme binding pocket (Fig. 3A). To test NAD+ binding by MalE–p18.1 in vitro, we used a filter binding assay. MalE–p25 binds NAD+ (Fig. 5). Importantly, binding of NAD+ by MalE–p18.1 is not detectable (Fig. 5). These data provide evidence that binding of a NAD-coenzyme by MalE–p18.1 is not a prerequisite for this protein to display C-signal activity and, thus, they argue that this protein does not act as a SCAD to generate the C-signal.
Figure 5.
Binding of NAD+ by MalE–p25 and MalE–p18.1. MalE–p25 or MalE–p18.1 was incubated with various concentrations of 32P-labeled NAD+, and NAD+ binding was analyzed in a filter binding assay. Closed and open squares indicate data for MalE–p25; and MalE–p18.1, respectively.
p17 is produced by proteolytic processing of p25
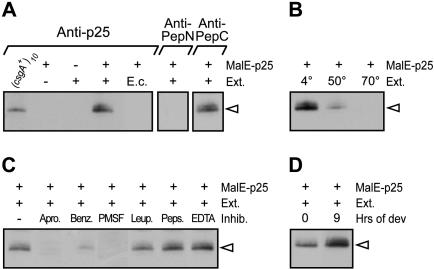
The csgA gene is transcribed from the same promoter during vegetative growth and development (Li et al. 1992), and the translation start codon for p25 synthesis is essential for synthesis of the C-signal (Lee et al. 1995). These observations argue against the hypothesis that p17 is synthesized from an alternative downstream start codon. Synthesis of p17 involving translational frame shifts appears unlikely, as this mechanism would involve at least two frameshifts. We therefore tested biochemically whether p17 is synthesized by proteolytic processing of p25 (Fig. 6). To accomplish this, a total cell extract was prepared from wild-type M. xanthus cells that had been starved for 9 h. Next, this extract was incubated with MalE–p25 protein purified from E. coli, and then subjected to immunoblot analysis with the anti-p25 antibodies. A protein with a size similar to that of p17 (from here on denoted p17vitro) was produced in samples in which the MalE–p25 protein was added to the cell extract. In the control reaction in which MalE–p25 was absent, p17vitro did not accumulate. Likewise, a control reaction including MalE–p25, but lacking cell extract, did not give rise to p17vitro. To verify that p17vitro is similar to p17 made in vivo, we tested whether the anti-PepN and anti-PepC antibodies recognized p17vitro. As shown in Figure 6A, the anti-PepC antibodies recognize p17vitro, whereas p17vitro is not recognized by the anti-PepN antibodies. These data suggest that p17vitro is similar to p17 produced in vivo and support the notion that p17 is produced by N-terminal proteolytic processing of p25.
Figure 6.
Proteolytic cleavage of MalE–p25 in vitro. (A) MalE–p25 was incubated as indicated with crude cell extracts (Ext.) of either wild-type M. xanthus cells developed on TPM-agar for 9 h (+) or E. coli cells (E.c.) and analyzed in immunoblot assays probed with anti-p25, anti-PepN, or anti-PepC antibodies. The first lane shows p17 from (csgA+)10 (DK1622/pTK98-10). (B) Cell extracts prepared from wild-type cells developed on TPM-agar for 9 h were incubated at the indicated temperature for 10 min prior to incubation with MalE–p25. (C) MalE–p25 was incubated in the presence of the protease inhibitors aprotinin (Apro), benzamidine (Benz), PMSF, leupeptin (Leup), pepstatin (Peps), or EDTA with crude cell extracts (Ext.) of wild-type cells developed on TPM-agar for 9 h. (D) Extracts of vegetative wild-type cells (0 h) or wild-type cells starved for 9 h on TPM agar (9 h) were used in the protease assay. The immunoblots in B, C, and D were probed with anti-p25 antibodies. The open arrowhead indicates p17vitro.
To analyze whether p17vitro synthesis is due to an autoproteolytic activity in MalE–p25, which is activated by the addition of a nonproteinaceous substance present in the cell extract, the cell extract was heated prior to the addition to MalE–p25. Incubation of the cell extract at 50°C for 10 min strongly reduced p17vitro synthesis (Fig. 6B). Incubation of the cell extract at 70°C for 10 min completely abolished p17vitro synthesis (Fig. 6B). These data in combination with the finding that MalE–p25 protein is stable at 4°C for several weeks (data not shown) indicate that p17vitro synthesis depends on a protease present in the cell extract rather than on autoprocessing of p25.
To investigate the specificity of the protease, we first tested whether addition of cell extract prepared from E. coli to MalE–p25 resulted in formation of p17vitro. As shown in Figure 6A, p17vitro did not accumulate after the addition of E. coli cell extract to MalE–p25. Next, protease inhibitors were included in reactions in which cell extract prepared from starved M. xanthus cells was added to MalE–p25 (Fig. 6C). Inhibitors of serine proteases [aprotinin, benzamidine, and phenylmethylsulfonyl fluoride (PMSF)] specifically blocked synthesis of p17vitro, whereas no effect was observed with inhibitors of cysteine proteases (leupeptin), aspartyl proteases (pepstatin), and metalloproteases (EDTA). These data suggest that the protease involved in N-terminal proteolytic processing of p25 is specific to M. xanthus and has the specificity of a serine protease.
To analyze whether the activity of this serine protease is developmentally regulated, cell extracts were prepared from vegetative and developing wild-type M. xanthus cells. Addition of cell extract from vegetative cells to MalE–p25 resulted in the accumulation of p17vitro (Fig. 6D), suggesting that the protease is present in vegetative cells. However, more p17vitro accumulated when extracts from starving cells were added to MalE–p25 (Fig. 6D). Taken together these results support the idea that p17 is produced by N-terminal processing of p25 by a serine protease, which displays increased activity during development.
Homology model of p25
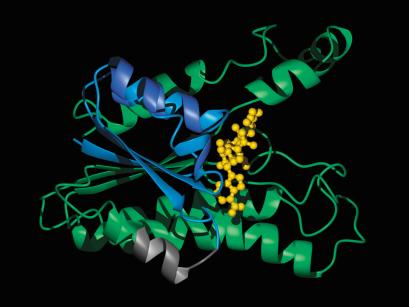
To examine the position of the potential cleavage site in the three-dimensional structure of p25, a homology model of p25 was constructed. Although the sequence identity between p25 and SCAD enzymes is only ∼30%, the three-dimensional structure of p25 can be modeled using solved structures of these proteins (Fig. 7). In the homology model, the tertiary structure of p25 is compatible with binding of NAD+. From the estimated size of 17.7 ± 0.4 kD of p17 synthesized in vivo, the cleavage site is predicted to be located between residues 60 and 68 in p25. In the homology model, the potential cleavage site is located partly in a surface-exposed loop and partly in a surface-exposed α-helix. This implies that the cleavage site is accessible to a protease even in a folded state of p25.
Figure 7.
Homology model of p25. Residues 1–59 are colored blue, the predicted cleavage site of p25 located between residues 60–68 is colored gray, residues 69–229 are colored green. The NAD+ molecule is shown in yellow. The illustration was prepared with the program MOLMOL, version 2k.2 (Koradi et al. 1996) and the POV-Ray rendering program (http://www.povray.org).
Discussion
The C-signal acts as a morphogen during fruiting body formation in M. xanthus and induces aggregation and sporulation at distinct thresholds. Moreover, the C-signal coordinates aggregation and sporulation temporally and spatially. The principal finding in this study is that the C-signal is a protein with an approximate size of 17 kD (p17). p17 is associated with the outer membrane. Moreover, evidence was obtained that the C-signal protein is synthesized by N-terminal proteolytic processing of the full-length CsgA protein (p25) by a serine protease, which displays increased activity during development.
Several lines of evidence support the conclusion that p17 is the genuine C-signal. Firstly, p17 copurifies with the C-signal from starving M. xanthus cells. Secondly, p25 produced by starving M. xanthus cells displays significantly lower C-signal activity than p17. Thirdly, p17 has a C terminus similar to that of p25, whereas the two proteins have different N termini. A MalE–CsgA protein, which contains the C-terminal 18.1 kD of p25, lacks the NAD(P)-coenzyme binding pocket present in p25. Consistently, this protein is unable to bind NAD+ in vitro. Nevertheless, this protein as well as a MalE–CsgA protein containing the C-terminal 17.8 kD of p25 rescues the developmental defects in csgA cells when added exogenously. Even though the precise N terminus of p17 has yet to be determined, these data argue against p17 acting as a SCAD to synthesize the C-signal. Likewise, they argue against p25 being the C-signal or acting as a SCAD to synthesize the C-signal. The data presented here are in agreement with those of Kim and Kaiser (1990a,b), who purified the C-signal from starving M. xanthus cells as a 17-kD protein referred to as the C-factor protein. However, in those studies the difference between C-factor protein and the full-length CsgA protein was not determined. Likewise, they did not rule out the possibility that the C-factor protein acted as a SCAD to produce the C-signal. We suggest that the C-factor protein purified by Kim and Kaiser (1990a,b) and p17 are identical.
In light of the data presented here, it is interesting that in an extracellular rescue assay a MalE–p25 protein purified from E. coli has C-signal activity (Lee et al. 1995), whereas p25 purified from M. xanthus cells does not display C-signal activity. We speculate that the C-signal activity displayed by the fusion protein in the extracellular rescue assay is either due to processing of the protein to p17 or to the ability of the protein to mimic p17. The inability of p25 purified from M. xanthus cells to display C-signal activity may be due to inefficient processing of this protein or an inability of this protein to mimic p17. Furthermore, we speculate that the inability of MalE–p25 proteins, which carry substitutions in the coenzyme binding pocket or in the catalytic site, to rescue development of csgA cells in the extracellular rescue assay (Lee et al. 1995) is due to a failure of these proteins to either become processed or mimic p17. The observation that overproduction of the SocA protein, which is homologous to SCAD's, rescues development of csgA cells in vivo (Lee and Shimkets 1994, 1996) could be explained by the SocA protein mimicking p17. It should be emphasized that the results presented here do not exclude the possibility that p25 may possess SCAD activity. This function of p25 is, however, not essential for fruiting body formation, as development of csgA cells is rescued by exogenous p17.
It was previously shown that the start codon for p25 synthesis is required for synthesis of the C-signal (Lee et al. 1995). Synthesis of p17 by proteolytic cleavage of p25 would explain this observation. To test whether p17 is produced by proteolytic cleavage of p25, we used an assay in which total M. xanthus cell extracts were added to a MalE–p25 protein purified from E. coli. Addition of this extract to MalE–p25 protein results in the synthesis of a protein (p17vitro) with a size similar to that of p17. This protein shares properties with p17: The anti-PepC antibodies recognize it whereas the anti-PepN antibodies do not. Four observations provide evidence that the proteolytic cleavage of MalE–p25 is the result of the action of a specific M. xanthus encoded protease: (1) The proteolytic synthesis of p17vitro from MalE–p25 is only observed after addition of cell extracts of M. xanthus cells, whereas the addition of cell extract of E. coli cells does not result in p17vitro synthesis. (2) The proteolytic cleavage is specifically inhibited by the addition of inhibitors of serine proteases. (3) Heating of the M. xanthus extract results in inactivation of the proteolytic activity, suggesting that a separate and specific serine protease catalyzes the proteolytic processing of MalE–p25 rather than MalE–p25 undergoing auto-proteolysis to produce p17vitro. (4) The serine protease is present in cell extracts from vegetative M. xanthus cells. However, higher levels of activity of the protease are detected in extracts from starving M. xanthus cells, suggesting that the activity or accumulation of the protease is developmentally regulated. This activity pattern is consistent with the observation that in wild-type cells p17 is only detected during starvation, whereas in a strain that overexpresses the csgA gene in vegetative cells, p17 is also detected during vegetative growth (Kruse et al. 2001).
The precise location of the proteolytic processing site in p25 remains to be identified. However, from the estimated molecular mass of p17 synthesized in vivo, we speculate that the cleavage site is located between amino acid residues 60 and 68 in p25. These residues are exposed on the surface in a homology model of p25, implying that the cleavage site would be accessible to the protease even in a folded state of p25. Experiments are in progress to determine the exact position of the cleavage site and to identify the serine protease. Proteases have previously been shown to have critical functions during fruiting body morphogenesis. Thus, synthesis of the A-signal involves at least two extracellular proteases (Plamann et al. 1992), and the Lon protease is important for the generation of the B-signal (Gill et al. 1993).
C-signal transmission involves a contact-dependent mechanism. Consistently, we found that p17 and p25 are associated with the outer membrane. This association is independent of the presence of type IV pili, extracellular fibrils, and the lipopolysaccharide O-antigen. From the homology of p25 to SCAD enzymes, which have an α/β-folding pattern (Fig. 7), it is apparent that p25 does not adopt a β-barrel structure typical of outer membrane proteins. Likewise, the homology model does not reveal surface-exposed hydrophobic patches, which could explain the membrane association. In addition, analysis of the primary sequence of p25 does not reveal putative membrane-spanning regions. Yet, p25 and p17 are associated with the outer membrane and partition to the detergent phase during a Triton X-114 extraction. From these observations, we suggest that p25 and p17 are anchored in the outer membrane by means of a hydrophobic modification. We do not know the nature of this modification or the position of the modification. However, some inferences can be made. Typically, secreted lipoproteins are modified on the N-terminal Cys residue (Pugsley 1993). p25 contains neither a signal peptide nor a lipoprotein signal peptide, suggesting that p25 and p17 may contain a different type of hydrophobic modification.
The association of p25 with the outer membrane argues that the p25 export mechanism does not involve proteolytic processing, which is consistent with the absent signal peptide. Likewise, p25 does not contain a signal peptide for export through the twin-arginine targeting protein export system (Yen et al. 2002). csgA proteins consisting of residues 17–229, 31–229, or 60–229 of p25 fail to localize to the outer membrane when synthesized in M. xanthus. These data may indicate that signals important for the proper export of p25 are located in the N-terminal region of p25. Alternatively, the intact three-dimensional structure of p25 is important for proper export. The failure of N-terminally truncated p25 derivatives to localize correctly to the outer membrane when synthesized in vivo has implications for the understanding of the proteolytic processing of p25 to p17. This observation suggests that in order for native p17 to localize to the outer membrane, it depends on correct export and localization of p25. We suggest that once p25 is correctly localized to the outer membrane, proteolytic cleavage of p25 results in synthesis of p17. This, in turn, suggests that the serine protease, which cleaves p25, is either secreted to the medium or associated with the outer membrane.
The regulated accumulation of the C-signal during development is critical for the correct temporal and spatial coordination of aggregation and sporulation. Transcription of the csgA gene is regulated by the stringent response (Crawford and Shimkets 2000), the proteins in the act operon (Gronewold and Kaiser 2001), and by the C-signal itself (Kim and Kaiser 1991). We suggest that proteolytic cleavage of p25 to the active p17 C-signal protein by a serine protease, which displays increased activity during development, provides an additional control mechanism, which ensures that the C-signal is synthesized appropriately during development.
Activation of key regulators by regulated proteolysis is an important regulatory mechanism in prokaryotes and eukaryotes. For example, during sporulation in Bacillus subtilis, σE and σK are processed to their active forms by proteolytic processing of inactive precursors (Stragier and Losick 1996). Similarly, small secreted peptides produced by proteolytic processing of larger precursors have important functions in controlling development of genetic competence (Lazazzera and Grossman 1998) and sporulation (Perego and Brannigan 2001) in B. subtilis, and heterocyst formation in Anabaena (Yoon and Golden 1998). Compelling evidence for the importance of proteolytic processing in the generation of key regulators in eukaryotic systems is provided by the processing of the Notch ligand Delta (Qi et al. 1999) and proteolytic processing of the transcription factor Cubitus interruptus in Drosophila (Aza-Blanc et al. 1997).
Typically, extracellular signaling molecules in bacteria are small diffusible compounds such as homoserine lactones (Fuqua and Greenberg 2002) and peptides (Lazazzera and Grossman 1998). Compared to these signaling molecules, p17 is unusual with respect to size and because it is not freely diffusible. In this regard C-signal is functionally analogous to eukaryotic signaling proteins such as Delta (Mumm and Kopan 2000) and Boss (Kramer et al. 1991), both of which are membrane-bound. Moreover, the homology of p25 to an enzyme family, synthesis by proteolysis, the cell-surface association, and morphogenetic properties of p17 are analogous to the Hedgehog family of extracellular signal proteins in eukaryotes (Ingham and McMahon 2001). Thus, prokaryotes and eukaryotes employ functionally analogous mechanisms to synthesize extracellular signaling proteins.
Materials and methods
All M. xanthus strains used were derived from DK1622 (wild type; Kaiser 1979). DK5208 (csgA::Tn5-132 ΩLS205) and DK5253 (csgA::Tn5-132 ΩLS205, Tn5 lac Ω4435; Kroos and Kaiser 1987); DK1622/pTK98-10 (attB::pTK98-10; Kruse et al. 2001); DK10407 (pilA::TcR; Wu and Kaiser 1996), DK3470 (dsp1693, Tn5 Ω1407; Shimkets 1986) and HK1321 (wzm::ΩKanR; Bowden and Kaplan 1998). E. coli Top10 [F–, λ–, mcrA, Δ(mrr-hsdRMS-mcrBC), Φ80ΔlacZM15, ΔlacX74, deoR, recA1, araD139, Δ(ara, leu)7697, galU, galK, rpsL, endA1, nupG; Invitrogen] was used in cloning experiments; BL21 [DE3; F–, ompT gal (dcm) (lon) hsdS; rB– mB–; an E. coli B strain; Novagen] was used to express recombinant MalE proteins and for the preparation of cell extracts.
To express p25 and the 170 C-terminal residues of p25 in M. xanthus, the corresponding csgA alleles were cloned downstream of the pilA promoter. This strategy was chosen because the native csgA mRNA is leaderless (Lee et al. 1995). First, the start codon of the pilA gene and 1 kb upstream were amplified from wild-type chromosomal DNA using the primers oPpilA-1 (5′-CGGAATTCGCGGCGTTGAACGAGGGG-3′) and oPpilA-2 (5′-GCTCTAGACATGGGGGTCCTCAGAGAAGG-3′). The PCR-product was cut with EcoRI and XbaI and cloned in pSWU19, giving rise to pSL626. pSWU19 is a pBGS18 (Spratt et al. 1986) derivative containing the Mx8 attP region (S. Wu, pers. comm.). The reading frames corresponding to p25 or the 170 C-terminal residues were amplified from pTK100 (Kruse et al. 2001) using the 5′-end primer op25-Xba (5′-GCTCTAGACGCTACGTCATCACCGG-3′) or op17-Xba (5′-GCTCTAGAGACGACGACAGCGTGCGC-3′) and the 3′-end primer ocsgA-8A (5′-CCCAAGCTTCTACTACCAGGGCACTTC-3′). The PCR products and pSL626 were cut with XbaI and HindIII, and ligated to give pSL627 and pSL628. Plasmids were integrated at the Mx8 attB site after electroporation (Kashefi and Hartzell 1995) in DK5208. Strains were verified by PCR using four primers specific for the 5′ and 3′ ends of the attP and attB regions.
To construct plasmids encoding MalE–p25, the entire csgA gene in pTK100 was amplified using the primers ocsgA-7A (5′-TCCCCCGGGATGCGCTACGTCATCACC-3′) and ocsgA-8A. The PCR product was cut with SmaI and HindIII, cloned in pBluescript II SK(–) (Stratagene), and sequenced. This fragment was cloned in pMAL-c2x (NEB) cut with XmnI and HindIII, resulting in pSL580. The plasmid was sequenced with the malE-primer (5′-GGTCGTCAGACTGTCGATGAAGCC-3′) and transferred into BL21 (DE3). The same procedure was used to construct pSL581 (MalE–p18.1) and pSL584 (MalE–p17.8) except that the 5′-primers were op17-C1 (5′-TCCCCCGGGGACGACGACAGCGTGCGC-3′) and op17-C4 (5′-TCCCCCGGGAGCGTGCGCGCGTTCGCC-3′).
Growth and development
E. coli cells were grown in LB in the presence of relevant antibiotics (Sambrook et al. 1989). M. xanthus was grown in CTT medium in liquid cultures at 32°C or on CTT agar plates with relevant antibiotics (Hodgkin and Kaiser 1977). M. xanthus was developed on TPM-agar plates or in submerged MC7-culture as described (Søgaard-Andersen et al. 1996). Briefly, cells were grown in CTT medium to a density of 5 × 108 cells/mL, harvested, and resuspended in either TPM-buffer (10 mM Tris-HCl at pH 7.6, 1 mM KPO4 at pH 7.6, 8 mM MgCl2) or in MC7 buffer (10 mM MOPS at pH 7.0, 1 mM CaCl2) to a cell density of 5 × 109 cells/mL. For starvation on TPM-agar, 50 μL concentrated cells were spotted and incubated at 32°C. In submerged culture, 50 μL concentrated cells were mixed with 350 μL MC7, transferred to a microtiter well with a diameter of 15 mm, and incubated at 32°C. When cells were developed in shaking MC7-suspension, concentrated cells were incubated at 32°C at 230 rpm. Development was followed visually as described (Kruse et al. 2001).
Immunoblot assays and SDS-PAGE
Materials to be analyzed in immunoblots were resuspended in Tris/Tricine-lysis buffer (TTL buffer; Schägger and von Jagow 1987), boiled for 5 min, and loaded on a 10% Tris/Tricine–SDS–polyacrylamide gel (Schägger and von Jagow 1987; prepared from a 40:2 acrylamide:bis-acrylamide stock solution). Immunoblots were prepared by standard procedures (Sambrook et al. 1989) using anti-FruA antibodies (S. Lobedanz, unpubl., anti-p25 antibodies (Kruse et al. 2001), anti-FrzCD antibodies (McCleary et al. 1990), or AglU antibodies (Thomasson et al. 2002). The secondary antibody was peroxidase-conjugated goat-anti-rabbit immunoglobulin G (DAKO). Immunoblots were developed with Renaissance Plus chemiluminescence reagent (NEN Life Science Products). Antibodies against the N and C termini of p25 were generated by immunizing rabbits with PepN (GVR SPEGARRLEPLKQKAC) and PepC (CNPEHSGRFFDYQGTEV PW; Fig. 3), respectively (Genosys Biotechnologies). The Cys residues were included to couple the peptides to the keyhole limpet hemacyanin carrier protein.
Biochemical fractionation
Biochemical fractionation of cells was done essentially as described (Thomasson et al. 2002). Briefly, 5 × 109 cells were developed for 24 h on TPM-agar plates, harvested, resuspended in 1 mL of 50 mM Tris-HCl at pH 7.6 and lysed by sonication. Cell debris was removed by centrifugation at 5000g for 10 min at 4°C. The cell-free supernatant was centrifuged at 45,000g for 30 min at 4°C. The resulting supernatant is enriched in soluble proteins. The pellet containing the crude envelope fraction was resuspended in 0.8 mL of 50 mM Tris-HCl at pH 7.6, 2% Triton X-100, 10 mM MgCl2, and subjected to ultracentrifugation as described. The resulting supernatant is enriched for inner membrane proteins, whereas the pellet is enriched for outer membrane proteins (Sprott et al. 1994). The pellet was resuspended in 0.2 mL of 50 mM Tris-HCl at pH 7.6, 2% Triton X-100, 10 mM MgCl2. Samples were acetone-precipitated and resuspended in equal volumes of TTL buffer. The same volume of each sample was loaded on an SDS–polyacrylamide gel.
Triton X-114 phase separation was done essentially as described (Brusca and Radolf 1994). DK1622/pTK98-10 cells were starved for 24 h on TPM-agar, harvested, resuspended in PBS (154 mM NaCl, 10 mM NaPO4 at pH 7.2, and lysed by sonication. Lysed cells were brought to 2% Triton X-114, and incubated in a roller shaker for 1 h at 4°C. The samples were centrifuged at 2400g for 10 min at 4°C to remove insoluble proteins. The supernatant was incubated at 37°C for 10 min, and then the centrifugation step was repeated at room temperature. This step results in phase separation with an aqueous phase and a detergent phase. The aqueous phase was transferred to a new tube; Triton X-114 was added to a final concentration of 2% and then the sample was centrifuged to remove Triton X-114 soluble proteins. The detergent phase from the initial phase separation was mixed with 1 mL ice-cold PBS until the phases merged, then warmed and centrifuged at room temperature as described. This step was repeated five times. Finally, all samples were acetone-precipitated and resuspended in equal volumes of TTL buffer. The same volume of each sample was loaded on the SDS–polyacrylamide gel.
Purification of the C-signal and C-signal bioassay
The purification was done essentially as described (Kim and Kaiser 1990a,b) except that the protease inhibitors PMSF, leupeptin, and pepstatin were included in all buffers (McCleary et al. 1990) and that the strain DK1622/pTK98-10 was developed in shaking MC7-suspension for 12 h. Briefly, harvested cells were resuspended in MC7 buffer, freeze-thawed three times, and then sonicated on ice. The lysate was centrifuged for 1 h at 100,000g. The pellet was resuspended in buffer A (10 mM MOPS at pH 7.2, 1 mM CaCl2, 4 mM MgCl2), brought to 1.2% (w/v) CHAPS, and gently agitated for 18 h, then centrifuged for 1 h at 100000g. The supernatant was concentrated by ultrafiltration through an Amicon YM10 membrane (Millipore) and then loaded on a Mono Q HR 5/5 column (Pharmacia) equilibrated with buffer B (20 mM MOPS at pH 7.0, 2 mM CaCl2, 0.4% CHAPS). Proteins were eluted with a linear NaCl gradient in buffer B. C-signal eluted in a broad peak around 250 mM NaCl. Adjacent fractions from the Mono Q column were pooled. Fractions to be tested in the C-signal bioassay (Kim and Kaiser 1990b) were dialyzed against buffer A supplemented with 50 mM NaCl (buffer A50). Dialyzed samples were serially diluted in A50; each dilution (400 μL) was warmed to 32°C and added to 2.5 × 108 csgA responder cells (DK5253) that had been starved for 6 h at 32°C in submerged culture in A50. Prior to the addition of the samples, the A50 overlying the responder cells was removed. Fruiting bodies and spores were detected as described (Kim and Kaiser 1990a). One unit of C-signal is the amount of C-signal that restores wild-type fruiting body formation and sporulation to 2.5 × 108 responder cells (Kim and Kaiser 1990a). Samples to be tested in immunoblots were acetone precipitated and resuspended in 1/40 of their original volume in TTL buffer. The same volume of each sample was loaded on the SDS–polyacrylamide gel. Protein concentrations were determined using the Bio-Rad Protein Assay (Bio-Rad) with bovine plasma IgG (Bio-Rad) as a standard.
Purification of MalE proteins
MalE–p25, MalE–p18.1, MalE–p17.8, and MalE were purified by affinity chromatography on an amylose resin column as described by the manufacturer (NEB). Proteins were dialyzed against A50 before use.
NAD+ filter binding assay
The assay was performed as described (Lee et al. 1995). Briefly, 2.5 μM MalE–p25, MalE–p18.1, or MalE protein was incubated in binding buffer (buffer A + 0.5 M NaCl) with various concentrations of radiolabeled NAD+ in a total reaction volume of 20 μL for 1 h at room temperature. NAD+ labeled with 32P (1000 Ci/mmole, 10 mM; Amersham) was diluted with NAD+ (Sigma) to a specific activity of 2.0 Ci/mmole. Binding reactions were applied to an Immobilon P membrane (Millipore) under vacuum using the Bio-Dot Microfiltration System (Bio-Rad). The filter was washed twice with 3 mL of binding buffer under vacuum and air-dried. Purified MalE was used to measure the background level of NAD+ binding, which was subtracted from the experimental values.
In vitro protease assay
M. xanthus cell extracts were prepared from DK1622 starved on TPM-agar for 9 h or from vegetative cells. E. coli cell extracts were prepared from exponentially growing BL21 (DE3). Cells were harvested by centrifugation, resuspended to a density of 5 × 1010 cells/mL in A50 and lysed by sonication. Next, 7.5 μg of protein were used directly in a reaction mixture with a total volume of 25 μL. MalE–p25 in A50 was added to a final concentration of 2.5 μM. Reactions were incubated for 2 h on ice. Reactions were stopped by addition of 2× TTL buffer, incubation at 96°C for 3 min, and freezing in liquid nitrogen. The same volume of each sample was loaded on the SDS–polyacrylamide gel. Protease inhibitors were added to the following final concentrations: aprotinin (0.3 μM), benzamidine (0.5 mM), PMSF (1 mM), leupeptin (1 μM), pepstatin (1 μM), and EDTA (1.0 mM; Sigma).
Homology modeling
The homology model of p25 was constructed using the crystal structures of 7-α-hydroxysteroid dehydrogenase complexed with NAD+ (PDB ID 1AHH, chain B; Tanaka et al. 1996) and mannitol dehydrogenase (PDB ID 1H5Q, chain L; Horer et al. 2001) as templates. The structural alignment was constructed with Swiss-PdbViewer, version 3.7 (http://www.expasy.org/spdbv), and then the homology model was calculated by the Swiss Model server (Guex and Peitsch 1997). Stereochemical evaluation of the model was done by the accompanying What-Check report. Binding of NAD+ to p25 was evaluated by superimposing p25 on 1AHH and then transferring the coordinates of NAD+ onto the model structure of p25. Collisions and improper stereochemistry were released by energy minimization in a 6.5 Å water shell using steepest descent to a maximum derivate less than 100 kJ/(mol Å) with the GROMACS software and force field (Lindahl et al. 2001). The topology of NAD+ was calculated by the PRODRG2 server (van Aalten et al. 1996). Contacts between NAD+ and p25 were analyzed in Swiss-PdbViewer according to the structural parameters and definitions of Carugo and Argos (1997a,b).
Acknowledgments
We thank Dale Kaiser and Seung Kim for discussions and Lee Kroos, Heidi Kaplan, and Birgitte H. Kallipolitis for many helpful comments on the manuscript. We also thank Eric Nudleman and Dale Kaiser for the Tgl antibodies, Patricia Hartzell for the AglU antibodies, and Heidi Kaplan for the wzm strain. The Danish Natural Science Research Council and the FREJA research program supported this work.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.274203.
References
- Aza-Blanc P., Ramirezweber, F., Laget, M., Schwartz, C., and Kornberg, T. 1997. Proteolysis that is inhibited by Hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89: 1043–1053. [DOI] [PubMed] [Google Scholar]
- Baker M.E. 1994. Myxococcus xanthus C-factor, a morphogenetic paracrine signal, is similar to Escherichia coli 3-oxoacyl-[acyl-carrier-protein] reductase and human 17 β-hydroxysteroid dehydrogenase. Biochem. J. 301: 311–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden M.G. and Kaplan, H.B. 1998. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol. Microbiol. 30: 275–284. [DOI] [PubMed] [Google Scholar]
- Brusca J.S. and Radolf, J.D. 1994. Isolation of integral membrane proteins by phase partitioning with Triton X-114. Methods Enzymol. 228: 182–193. [DOI] [PubMed] [Google Scholar]
- Carugo O. and Argos, P. 1997a. NADP-dependent enzymes. I: Conserved stereochemistry of cofactor binding. Proteins 28: 10–28. [DOI] [PubMed] [Google Scholar]
- ____. 1997b. NADP-dependent enzymes. II: Evolution of the mono- and dinucleotide binding domains. Proteins 28: 29–40. [DOI] [PubMed] [Google Scholar]
- Crawford E.W. and Shimkets, L.J. 2000. The stringent response in Myxococcus xanthus is regulated by SocE and the CgsA C-signaling protein. Genes & Dev. 14: 483–492. [PMC free article] [PubMed] [Google Scholar]
- Dworkin M. 1996. Recent advances in the social and developmental biology of the Myxobacteria. Microbiol. Rev. 60: 70–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellehauge E., Nørregaard-Madsen, M., and Søgaard-Andersen, M. 1998. The FruA signal transduction protein provides a checkpoint for the temporal coordination of intercellular signals in Myxococcus xanthus development. Mol. Microbiol. 30: 807–817. [DOI] [PubMed] [Google Scholar]
- Fuqua C. and Greenberg, E.P. 2002. Listening in on bacteria: Acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3: 685–695. [DOI] [PubMed] [Google Scholar]
- Gill R.E., Karlok, M., and Benton, D. 1993. Myxococcus xanthus encodes an ATP-dependent protease which is required for developmental gene transcription and intercellular signaling. J. Bacteriol. 175: 4538–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronewold T.M.A. and Kaiser, D. 2001. The act operon controls the level and time of C-signal production for Myxococcus xanthus development. Mol. Microbiol. 40: 744–756. [DOI] [PubMed] [Google Scholar]
- Guex N. and Peitsch, M.C. 1997. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. and Kaiser, D. 1977. Cell–cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. 74: 2938–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horer S., Stoop, J., Mooibroek, H., Baumann, U., and Sassoon, J. 2001. The crystallographic structure of the mannitol 2-dehydrogenase NADP+ binary complex from Agaricus bisporus. J. Biol. Chem. 276: 27555–27561. [DOI] [PubMed] [Google Scholar]
- Ingham P.A. and McMahon, A.P. 2001. Hedgehog signaling in animal development: Paradigms and principles. Genes & Dev. 15: 3059–3087. [DOI] [PubMed] [Google Scholar]
- Inouye M., Inouye, S., and Zusman, D.R. 1979. Gene expression during development of Myxococcus xanthus: Pattern of protein synthesis. Dev. Biol. 68: 579–591. [DOI] [PubMed] [Google Scholar]
- Kaiser D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. 76: 5952–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashefi K. and Hartzell, P.L. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF defect. Mol. Microbiol. 15: 483–494. [DOI] [PubMed] [Google Scholar]
- Kim S.K. and Kaiser, D. 1990a. C-factor: A cell–cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell 61: 19–26. [DOI] [PubMed] [Google Scholar]
- ____. 1990b. Purification and properties of Myxococcus xanthus C-factor, an intercellular signaling protein. Proc. Natl. Acad. Sci. 87: 3635–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1991. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J. Bacteriol. 173: 1722–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14: 51–55. [DOI] [PubMed] [Google Scholar]
- Kramer H., Cagan, R.L., and Zipursky, S.L. 1991. Interaction of bride of sevenless membrane-bound ligand and the sevenless tyrosine-kinase receptor. Nature 352: 207–212. [DOI] [PubMed] [Google Scholar]
- Kroos L. and Kaiser, D. 1987. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes & Dev. 1: 840–854. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kuspa, A., and Kaiser, D. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117: 252–266. [DOI] [PubMed] [Google Scholar]
- ____. 1990. Defects in fruiting body development caused by Tn5 lac insertions in Myxococcus xanthus. J. Bacteriol. 172: 484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T., Lobedanz, L., Berthelsen, N.M.S., and Søgaard-Andersen, L. 2001. C-signal: A cell surface-associated morphogen that induces and coordinates multicellular fruiting body morphogenesis and sporulation in M. xanthus. Mol. Microbiol. 40: 156–168. [DOI] [PubMed] [Google Scholar]
- Lazazzera B.A. and Grossman, A.D. 1998. The ins and outs of peptide signaling. TIM 6: 288–294. [DOI] [PubMed] [Google Scholar]
- Lee B.-U., Lee, K., Mendez, J., and Shimkets, L.J. 1995. A tactile sensory system of Myxococcus xanthus involves an extracellular NAD(P)+-containing protein. Genes & Dev. 9: 2964–2973. [DOI] [PubMed] [Google Scholar]
- Lee K. and Shimkets, L.J. 1994. Cloning and characterization of the socA locus which restores development to Myxococcus xanthus C-signaling mutants. J. Bacteriol. 176: 2200–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1996. Suppression of a signaling defect during Myxococcus xanthus development. J. Bacteriol. 178: 977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Lee, B.-U., and Shimkets, L.J. 1992. csgA expression entrains Myxococcus xanthus development. Genes & Dev. 6: 401–410. [DOI] [PubMed] [Google Scholar]
- Lindahl E., Hess, B., and van der Spoel, D. 2001. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Mod. 7: 306–317. [Google Scholar]
- McBride M.J., Köhler, T., and Zusman, D.R. 1992. Methylation of FrzCD, a methyl-accepting taxis protein of Myxococcus xanthus, is correlated with factors affecting cell behaviour. J. Bacteriol. 174: 4246–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary W.R., McBride, M.J., and Zusman, D.R. 1990. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J. Bacteriol. 172: 4877–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm J.S. and Kopan, R. 2000. Notch signaling: From the outside in. Dev. Biol. 228: 151–165. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Fujitani, S., Mao, X., Inouye, S., and Komano, T. 1996. FruA, a putative transcription factor essential for the development of Myxococus xanthus. Mol. Microbiol. 22: 757–767. [DOI] [PubMed] [Google Scholar]
- Oppermann U., Filling, C., Hult, M., Shafqat, N., Wu, X., Lindh, M., Shafqat, J., Nordling, E., Kallberg, Y., Persson, B., et al. 2003. Short-chain dehydrogenases/reductases (SDR): The 2002 update. Chem. Biol. Interact. 143–144: 247–253. [DOI] [PubMed] [Google Scholar]
- Perego M. and Brannigan, J.A. 2001. Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides 22: 1541–1547. [DOI] [PubMed] [Google Scholar]
- Plamann L., Kuspa, A., and Kaiser, D. 1992. Proteins that rescue A-signal-defective mutants of Myxococcus xanthus. J. Bacteriol. 174: 3311–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A.P. 1993. The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev. 57: 50–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Rand, M.D., Wu, X., Sestan, N., Wang, W., Rakic, P., Xu, T., and Artavanis-Tsakonas, S. 1999. Processing of the Notch ligand Delta by the metalloprotease Kusbanian. Science 283: 91–94. [DOI] [PubMed] [Google Scholar]
- Rodriguez J.P. and Kaiser, D. 1997. Identification and localization of the Tgl protein, which is required for Myxococcus xanthus social motility. J. Bacteriol. 179: 4372–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schägger H. and von Jagow, G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166: 368–379. [DOI] [PubMed] [Google Scholar]
- Shimkets L.J. 1986. Role of cell cohesion in Myxococcus xanthus fruiting body formation. J. Bacteriol. 166: 842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1999. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53: 525–549. [DOI] [PubMed] [Google Scholar]
- Shimkets L.J. and Rafiee, H. 1990. CsgA, an extracellular protein essential for Myxococcus xanthus development. J. Bacteriol. 172: 5299–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L.J., Gill, R.E., and Kaiser, D. 1983. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc. Natl. Acad. Sci. 80: 1406–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søgaard-Andersen L. and Kaiser, D. 1996. C factor, a cell-surface-associated intercellular signaling protein, stimulates the Frz signal transduction system in Myxococcus xanthus. Proc. Natl. Acad. Sci. 93: 2675–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søgaard-Andersen L., Slack, F., Kimsey, H., and Kaiser, D. 1996. Intercellular C-signaling in Myxococcus xanthus involves a branched signal transduction pathway. Genes & Dev. 10: 740–754. [DOI] [PubMed] [Google Scholar]
- Spratt B.G., Hedge, P.J., te Heesen, S., Edelman, A., and Broome-Smith, J.K. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41: 337–342. [DOI] [PubMed] [Google Scholar]
- Sprott G.D., Koval, S.F., and Schmaitman, C.A. 1994. Cell fractionation. In Methods for general and molecular bacteriology (eds. P. Gerhardt, R.G.A. Murray, W.A. Wood, and N.R. Kreig) pp. 72–103. American Society for Microbiology Press, Washington DC.
- Stragier P. and Losick, R. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30: 297–341. [DOI] [PubMed] [Google Scholar]
- Tanaka N., Nonaka, T., Tanabe, T., Yishimoto, T., Tsuru, D., and Mitsui, Y. 1996. Crystal structures of the binary and ternary complexes of 7-α-hydroxysteroid dehydrogenase from Escherichia coli. Biochemistry 35: 7715–7730. [DOI] [PubMed] [Google Scholar]
- Teleman A.A., Strigini, M., and Cohen, S.M. 2001. Shaping morphogen gradients. Cell 105: 559–562. [DOI] [PubMed] [Google Scholar]
- Thomasson B., Link, J., Stassinopoulos, A.G., Burke, N., Plamann, L., and Hartzell, P.L. 2002. MglA, a small GTPase, interacts with a tyrosine kinase to control type IV pili-mediated motility and development of Myxococcus xanthus. Mol. Microbiol. 46: 1399–1413. [DOI] [PubMed] [Google Scholar]
- van Aalten D.M., Bywater, R., Findlay, J.B., Hendlich, M., Hooft, R.W., and Vriend, G. 1996. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J. Comput. Aid. Mol. Des. 10: 255–262. [DOI] [PubMed] [Google Scholar]
- Wu S.S. and Kaiser, D. 1996. Markerless deletions of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacB gene. J. Bacteriol. 178: 5817–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen M.-R., Tseng, Y.-H., Nguyen, E.H., Wu, L.-F., and Saier Jr., M.H. 2002. Sequence and phylogenetic analyses of the twin-arginine targeting (Tat) protein export system. Arch. Microbiol. 177: 441–450. [DOI] [PubMed] [Google Scholar]
- Yoon H.-S. and Golden, J.W. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282: 935–938. [DOI] [PubMed] [Google Scholar]