Abstract
Silencing within the yeast rDNA repeats inhibits hyperrecombination, represses transcription from foreign promoters, and extends replicative life span. rDNA silencing is mediated by a Sir2-containing complex called RENT (regulator of nucleolar silencing and telophase exit). We show that the Net1 (also called Cfi1) and Sir2 subunits of RENT localize primarily to two distinct regions within rDNA: in one of the nontranscribed spacers (NTS1) and around the Pol I promoter, extending into the 35S rRNA coding region. Binding to NTS1 overlaps the recombination hotspot and replication fork barrier elements, which have been shown previously to require the Fob1 protein for their activities. In cells lacking Fob1, silencing and the association of RENT subunits are abolished specifically at NTS1, while silencing and association at the Pol I promoter region are unaffected or increased. We find that Net1 and Sir2 are physically associated with Fob1 and subunits of RNA polymerase I. Together with the localization data, these results suggest the existence of two distinct modes for the recruitment of the RENT complex to rDNA and reveal a role for Fob1 in rDNA silencing and in the recruitment of the RENT complex. Furthermore, the Fob1-dependent associations of Net1 and Sir2 with the recombination hotspot region strongly suggest that Sir2 acts directly at this region to carry out its inhibitory effect on rDNA recombination and accelerated aging.
Keywords: Gene silencing, chromatin, Sir2, recombination, Fob1, rDNA
Eukaryotic cells maintain stable genomes despite the presence of repetitive DNA and efficient homologous recombination systems. An example of a highly repetitive locus whose stability and integrity is paramount for survival is ribosomal DNA (rDNA). Arranged in one or more arrays, eukaryotic rDNA is tandemly repeated anywhere from less than 100 times to more than 10,000 times (for review, see Nomura 2001). Although mechanisms that regulate this critical region of the genome are not well understood, significant progress has been made through extensive studies in the budding yeast Saccharomyces cerevisiae. In budding yeast, 100–200 copies of a 9.1-kb rDNA repeat exist as a tandem array (Petes and Botstein 1977). Interestingly, recombination rates are significantly lower than would be expected for such a large and repetitive locus, suggesting that recombination is somehow suppressed (Petes 1980). Furthermore, recombination levels are reduced despite the presence of several sequence elements within rDNA that can stimulate recombination (Keil and Roeder 1984; Voelkel-Meiman et al. 1987; Brewer and Fangman 1988; Kobayashi et al. 1992, 2001; Huang and Keil 1995; Gruber et al. 2000; Ward et al. 2000). These findings suggest that both positive and negative regulatory mechanisms have evolved to properly control recombination levels of rDNA.
Positive regulation of rDNA recombination occurs through a mechanism(s) that requires FOB1, a “fork blocking less” gene that promotes a polar replication fork barrier (RFB) that is located within the nontranscribed spacer (NTS) of an rDNA repeat (see Fig. 1A; Kobayashi and Horiuchi 1996). The RFB has been proposed to stimulate recombination directly (Kobayashi and Horiuchi 1996; Johzuka and Horiuchi 2002; Benguria et al. 2003). The FOB1 gene is required for most recombination within rDNA including recombination that results in the expansion or contraction of the array (Kobayashi and Horiuchi 1996; Kobayashi et al. 1998; Johzuka and Horiuchi 2002). Furthermore, FOB1 is required for hotspot (HOT1) activity, a phenomenon in which specific rDNA sequences can stimulate homologous recombination when placed outside of the array (Lin and Keil 1991). Significantly, the cis-element sequences required for establishing the RFB and stimulating recombination are overlapping and are found within a region originally identified as an enhancer of RNA polymerase I (Pol I) transcription outside the rDNA array (Elion and Warner 1984; Wai et al. 2001). How Fob1 functions to generate an RFB or stimulate recombination is not known.
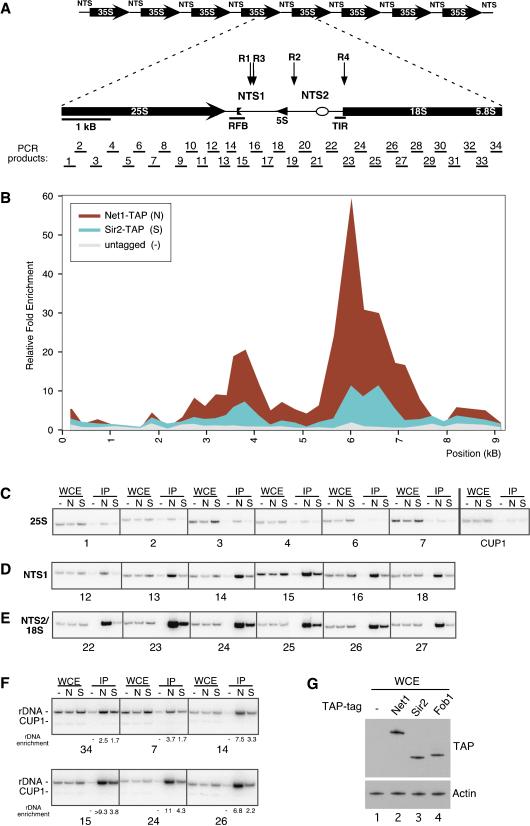
Figure 1.
Net1 and Sir2 associate primarily with the NTS1 and NTS2/18S regions of
rDNA. (A) The physical structure of the tandemly repeating rDNA of
S. cerevisiae is shown above, and a single 9.1-kb rDNA unit
is shown expanded below. Each repeat yields a Pol I-transcribed 35S
precursor rRNA (shown as a divided thick arrow) and a Pol III-transcribed 5S
rRNA (arrowhead). The 35S coding regions are separated by a nontranscribed
spacer (NTS), which is divided by the 5S gene into NTS1 and NTS2. Solid bars
indicate the replication fork block region (RFB) and the Pol I transcription
initiator region (TIR; Elion and Warner
1984,
1986;
Brewer and Fangman 1988;
Kobayashi et al. 1992). The
locations of the replication fork barrier
( ) and autonomously
replicating sequences (
) and autonomously
replicating sequences ( ) are indicated. Vertical arrows indicate
insertion sites of silencing reporters. (R1)
NTS1::Ty1–mURA3; (R2)
NTS2::Ty1–mURA3; (R3) NTS1::mURA3;
(R4) NTS2::mURA3. PCR products analyzed in ChIP assays are
indicated below the rDNA unit. (B) Representative graph
showing the associations of Net1–TAP and Sir2–TAP within the rDNA
repeat. Relative fold enrichment refers to the relative ratio of PCR products
amplified from immunoprecipitated DNA to products from whole-cell extract DNA
(see Materials and Methods). Positions along rDNA correspond to the physical
map shown in A. (C) Examples of the ChIP data used to
calculate enrichment at the 25S region of rDNA. The numbers below the
panels refer to the PCR products shown in A. CUP1 primers were used
as a control. WCE and IP refer to products amplified from whole-cell extracts
and immunoprecipitated DNA, respectively. Untagged control (–),
Net1–TAP (N), and Sir2–TAP (S) cells are indicated above
lanes. (D) Same as in C but showing amplified DNA from the
NTS1 region. (E) Same as in C but showing amplified DNA from
the NTS2/18S region. (F) Multiplex PCR with rDNA and CUP1
primers showing rDNA enrichment. (G) Western blot showing the
relative abundance of TAP-tagged proteins in whole-cell extracts (WCE)
prepared from untagged (lane 1) or Net1–TAP, Sir2–TAP, or
Fob1–TAP strains (lanes 2–4).
) are indicated. Vertical arrows indicate
insertion sites of silencing reporters. (R1)
NTS1::Ty1–mURA3; (R2)
NTS2::Ty1–mURA3; (R3) NTS1::mURA3;
(R4) NTS2::mURA3. PCR products analyzed in ChIP assays are
indicated below the rDNA unit. (B) Representative graph
showing the associations of Net1–TAP and Sir2–TAP within the rDNA
repeat. Relative fold enrichment refers to the relative ratio of PCR products
amplified from immunoprecipitated DNA to products from whole-cell extract DNA
(see Materials and Methods). Positions along rDNA correspond to the physical
map shown in A. (C) Examples of the ChIP data used to
calculate enrichment at the 25S region of rDNA. The numbers below the
panels refer to the PCR products shown in A. CUP1 primers were used
as a control. WCE and IP refer to products amplified from whole-cell extracts
and immunoprecipitated DNA, respectively. Untagged control (–),
Net1–TAP (N), and Sir2–TAP (S) cells are indicated above
lanes. (D) Same as in C but showing amplified DNA from the
NTS1 region. (E) Same as in C but showing amplified DNA from
the NTS2/18S region. (F) Multiplex PCR with rDNA and CUP1
primers showing rDNA enrichment. (G) Western blot showing the
relative abundance of TAP-tagged proteins in whole-cell extracts (WCE)
prepared from untagged (lane 1) or Net1–TAP, Sir2–TAP, or
Fob1–TAP strains (lanes 2–4).
Several lines of evidence suggest that recombination in the yeast rDNA repeats is also negatively regulated through a mechanism that resembles heterochromatic gene silencing and is referred to as rDNA silencing. First, recombination levels in S. cerevisiae are down-regulated by Sir2 (Gottlieb and Esposito 1989), an NAD-dependent deacetylase originally identified for its role in chromatin silencing at the mating-type loci and telomeres (for review, see Moazed 2001; Rusche et al. 2003). Second, Sir2 is required for silencing of Ty1 transposition and transcription of Pol II-dependent reporter genes that are inserted within rDNA (Bryk et al. 1997; Fritze et al. 1997; Smith and Boeke 1997). Finally, the altered sensitivity of rDNA to micrococcal nuclease and dam methyltransferase in sir2Δ cells, as well as loss of rDNA silencing in strains with histone mutations, supports the idea that a Sir2-based silencing mechanism inhibits rDNA recombination by altering chromatin structure (Fritze et al. 1997; Bryk et al. 2002; Hoppe et al. 2002; Park et al. 2002).
Regulation of recombination at rDNA by Fob1 and Sir2 is also a major determinant of budding yeast life span. The accumulation of extrachromosomal rDNA circles (ERCs) excised from the rDNA array can lead to premature cellular senescence in S. cerevisiae (Sinclair and Guarente 1997). Accordingly, loss of silencing in sir2Δ cells results in an increased rate of Fob1-dependent recombination and ERC accumulation and reduces average life span, whereas increasing the dosage of SIR2 suppresses recombination and prolongs average life span (Kaeberlein et al. 1999). In contrast, FOB1 deletion cells display the opposite aging phenotype. The absence of FOB1 reduces recombination and the formation of ERCs, extends average life span, and as expected, suppresses premature aging in cells lacking SIR2 (Defossez et al. 1999).
In yeast, silencing is best understood at the silent mating-type loci and telomeric regions (Rine and Herskowitz 1987; Gottschling et al. 1990). Initiation of silencing at these regions involves the association of DNA-binding proteins with cis-acting silencer elements. The silencer-binding factors then recruit a second class of proteins to DNA to form the SIR complex, consisting of Sir2, Sir3, and Sir4 proteins (for review, see Rusche et al. 2003). As mentioned above, Sir2 is an NAD-dependent deacetylase (Imai et al. 2000; Landry et al. 2000; Smith et al. 2000), and its activity is necessary for the spreading of silencing complexes along chromatin via interactions with the N termini of histones H3 and H4 (Braunstein et al. 1993; Hecht et al. 1996; Strahl-Bolsinger et al. 1997; Hoppe et al. 2002; Luo et al. 2002; Rusche et al. 2002). Sir2 is also the only SIR protein required for rDNA silencing (Bryk et al. 1997; Fritze et al. 1997; Smith and Boeke 1997), and it is part of an rDNA silencing complex called RENT (regulator of nucleolar silencing and telophase exit; Shou et al. 1999; Straight et al. 1999; Visintin et al. 1999). In addition to Sir2, RENT contains Net1 and Cdc14 (Shou et al. 1999; Visintin et al. 1999). Net1 is required for rDNA silencing and localization of Sir2 to rDNA (Straight et al. 1999). Net1 can also associate with Pol I (Shou et al. 2001), but it is unknown if the entire RENT complex, including Sir2, associates with Pol I. Cdc14 is a phosphatase that regulates exit from mitosis (Shou et al. 1999; Visintin et al. 1999), but whether it plays a role in silencing is not known. Moreover, specific silencer elements or DNA-binding proteins that recruit silencing complexes to rDNA have not been described.
To investigate how silencing is initiated at rDNA, we performed a high-resolution mapping of Net1 and Sir2, two subunits of the RENT silencing complex, along the entire 9.1-kb length of an rDNA repeat by chromatin immunoprecipitation (ChIP). Each rDNA unit yields a 35S precursor rRNA and a 5S rRNA, separated by two nontranscribed spacers, NTS1 and NTS2 (see Fig. 1A). Our data show that both silencing proteins are associated primarily with two regions: one region within NTS1 and a second that overlaps the Pol I promoter and part of the 35S rRNA gene. The NTS1 region associated with silencing proteins includes sequences necessary for FOB1-dependent replication fork block and recombination activities. Surprisingly, we find that FOB1 is required for rDNA silencing at NTS1, and we show that Fob1 is primarily associated with this region of rDNA. In contrast, lower levels of Fob1 are localized to the Pol I promoter/35S region, and deletion of FOB1 has no effect on silencing at this location. Consistent with these observations, in fob1Δ cells, we detect dramatically reduced associations of Net1 and Sir2 with NTS1 but unaffected or increased associations with the Pol I promoter/35S region. Both Fob1 and Pol I physically interact with the RENT complex, suggesting two distinct pathways recruit the RENT complex to rDNA.
Results
Net1 and Sir2 are preferentially associated with two regions within rDNA
The structure of a 9.1-kb rDNA repeat unit and important functional elements are shown in Figure 1A. The association of Net1 or Sir2 with rDNA by ChIP has been studied previously using a limited number of sites (ranging from 1 to 7; Gotta et al. 1997; Straight et al. 1999; Armstrong et al. 2002; Bryk et al. 2002; Buck et al. 2002; Hoppe et al. 2002). Because Sir2 associates preferentially with a few sites in the nontranscribed spacer (NTS) as compared with a few sites within the 35S coding region (Gotta et al. 1997; Hoppe et al. 2002), it might be expected that silencing complexes would be found mainly at the NTS. To obtain a more comprehensive picture of where Net1 and Sir2 are associated with rDNA, we designed a panel of 68 primers to amplify fragments of ∼0.25 kb in length that spanned an rDNA repeat (Fig. 1A).
We constructed yeast strains in which the endogenous copy of the NET1 or SIR2 gene was modified to encode a protein with the TAP tag at its C terminus (Net1–TAP or Sir2–TAP). The TAP tag is a dual epitope tag consisting of a calmodulin-binding peptide separated from two Protein A repeats by a TEV-protease cleavage site. Both NET1–TAP and SIR2–TAP strains exhibited the same levels of rDNA silencing as the parental untagged strain, suggesting that the modified proteins were fully functional (Supplementary Fig. 1). Cells were cross-linked with formaldehyde, and Net1–TAP or Sir2–TAP was immunoprecipitated from extracts containing sheared chromatin using an IgG resin. Whole-cell extract chromatin (WCE) or immunoprecipitated chromatin (IP) from untagged or TAP-tagged strains was used as template DNA for quantitative PCR, using the panel of primers shown in Figure 1A.
We generated a graphical representation of Net1 and Sir2 association across an rDNA repeat (Fig. 1B) by using the ratio of IP material to WCE (input) material to calculate relative fold enrichment values for each DNA fragment. Background binding was defined by immunoprecipitation of rDNA fragments from an untagged strain and immunoprecipitation of DNA fragments from CUP1, a repetitive, nonsilenced locus. Unexpectedly, we found that Net1 and Sir2 were not found in a single peak spanning the NTS region. Instead, the data showed two major peaks that overlapped most of NTS1 but only part of NTS2 (Fig. 1B). Notably, the NTS1 peak coincided with the replication fork block region (RFB; Fig. 1B). This region contains a polar replication fork block and a number of cis-elements required for FOB1-dependent rDNA recombination (Lin and Keil 1991; Kobayashi and Horiuchi 1996; Kobayashi et al. 1998; Johzuka and Horiuchi 2002). The NTS2 peak overlapped the Pol I transcription initiation region (TIR; Fig. 1B), but the majority of the peak was in fact spread toward the 18S region of the transcribed 35S rRNA gene. We also observed a much smaller peak at the 5S rRNA gene. The relative associations of Net1 and Sir2 with rDNA fragments closely mirrored each other, but more material was consistently immunoprecipitated by Net1–TAP than Sir2–TAP. Because levels of Net1–TAP were higher than those of Sir2–TAP (Fig. 1G, lanes 2,3), and Sir2 is also required for silencing at other loci, this observation may reflect higher levels of Net1 association with rDNA. Additionally, whereas Net1 is associated with rDNA throughout the cell cycle, Sir2 is partially released toward the end of mitosis (Straight et al. 1999). Finally, differences in cross-linking efficiency can also explain the difference in association of rDNA fragments with Net1 and Sir2 in our experiments.
Examples of the ChIP data used to obtain the graph in Figure 1B are shown in Figure 1C–E. The enrichment of DNA regions corresponding to most of the 25S rRNA by Net1 or Sir2 was near the background level of binding (Fig. 1C). Similarly, DNAfrom CUP1 was not significantly enriched in Net1–TAP and Sir2–TAP immunoprecipitations (Fig. 1C). In contrast, high levels of DNA fragments from NTS1 and NTS2/18S regions were immunoprecipitated by Net1–TAP and Sir2–TAP (Fig. 1D,E). As another way of quantifying relative binding, we performed multiplex PCR with primers for both rDNA and CUP1, using CUP1 as an internal control for calculating relative fold enrichment. Examples of the data and the calculated relative fold enrichment are shown in Figure 1F and are consistent with the two peaks of Net1/Sir2 association in Figure 1B.
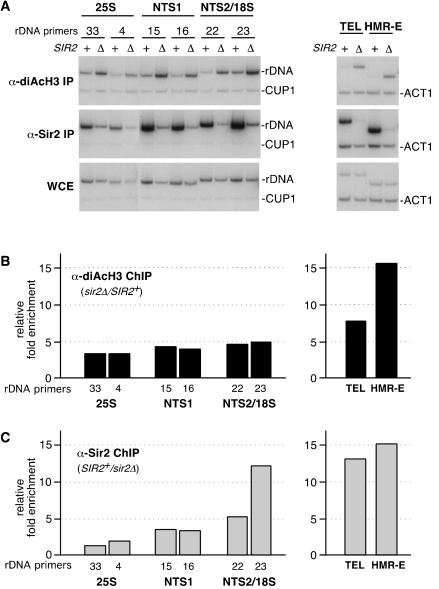
Previous ChIP experiments have shown that in sir2Δ cells, the acetylation levels of histones H3 and H4 associated with the mating-type and telomeric silent chromatin as well as with a few sites in rDNA are increased (Braunstein et al. 1993; Suka et al. 2001; Armstrong et al. 2002; Bryk et al. 2002; Hoppe et al. 2002). We used an antibody that recognizes histone H3 diacetylated at Lys 9 and 14 to examine H3 acetylation levels across the rDNA repeat in SIR2+ versus sir2Δ cells. Figure 2 shows the effect of deleting SIR2 on H3 acetylation levels for rDNA regions corresponding to the 25S rRNA, NTS1, and NTS2/18S. We observed an increase in H3 acetylation at each of the above rDNA regions in the range of threefold to fivefold in sir2Δ compared with SIR2+ cells but no effect on H3 acetylation at the control CUP1 locus (Fig. 2A,B, left panels). Furthermore, in four independent experiments, deletion of SIR2 resulted in an increase in H3 acetylation in the range of twofold to sixfold for all rDNA fragments tested (data not shown). As controls, we tested changes in acetylation of H3 at silent chromatin regions at the HMR silent mating-type locus and a telomeric DNA region. As expected (Suka et al. 2001; Bryk et al. 2002; Hoppe et al. 2002), deletion of SIR2 resulted in a large increase in H3 acetylation at both loci but had no effect on H3 acetylation at the control ACT1 locus (Fig. 2A,B, right panels). As an additional control, we performed ChIP with an anti-Sir2 antibody and with the same cross-linked chromatin used above for immunoprecipitation of acetylated H3 to directly compare H3 acetylation with Sir2 occupancy. As expected (Strahl-Bolsinger et al. 1997), the anti-Sir2 antibody efficiently immunoprecipitated HMR and telomeric DNA fragments but not the unsilenced ACT1 locus (Fig. 2A,C, right panels). Consistent with the results presented in Figure 1, we observed the highest levels of Sir2 binding at the NTS1 and NTS2/18S regions and less binding at regions corresponding to the 25S rRNAcoding sequences (Fig. 2A,C; data not shown). Therefore, although the highest levels of Sir2 binding are observed at the NTS1 and NTS2/18S regions, deletion of SIR2 causes an increase in H3 acetylation throughout rDNA.
Figure 2.
H3 acetylation levels throughout rDNA are increased in sir2Δ cells. (A) Examples of the ChIP data used to determine the associations of diacetylated (K9/K14) histone H3 or Sir2 with rDNA (left panels) or telomeric (TEL) and mating-type loci (HMR-E) regions (right panels). (+) SIR2+ cells; (Δ) sir2Δ cells; numbers above the left panels refer to rDNA primers as indicated in Figure 1A. CUP1 and ACT1 primers were used as internal controls. (B, left) Quantification of ChIP experiments shows that the relative fold enrichment of diacetylated H3 increases throughout the rDNA in sir2Δ cells as compared with SIR2+ cells. rDNA primers are indicated below the graph and correspond to Figure 1A. (Right) H3 acetylation increases at telomeres and the silent mating-type loci in sir2Δ cells. (C, left) Association of Sir2 with rDNA in SIR2+ and sir2Δ cells showing that the highest levels of Sir2 are present at NTS1 and NTS2/18S. (Right) Association of Sir2 with telomeres and the silent mating-type loci is shown for comparison.
Silencing at NTS1 requires Fob1
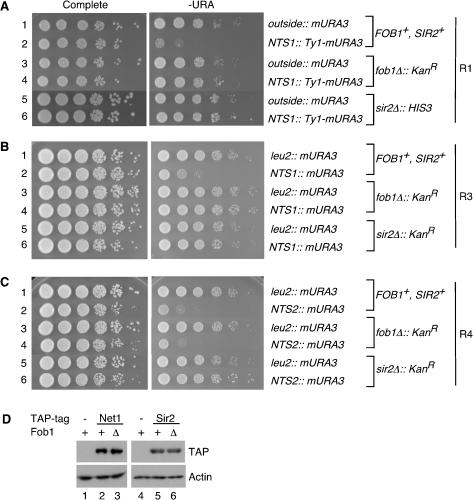
The association of silencing proteins with the RFB region of NTS1 was especially intriguing; this section of rDNA is required for FOB1-dependent recombination and contains a replication fork block. Furthermore, the FOB1 and SIR2 genes oppositely affect the formation of extrachromosomal rDNA circles and the rates of cellular senescence (Kaeberlein et al. 1999). These observations suggested that Fob1 might block or negatively regulate silencing, but instead we found that silencing at NTS1 was abolished in the absence of FOB1 (Fig. 3A,B). To assess silencing, we deleted FOB1 in strains that contained a Ty1 transposon element bearing a Pol II-transcribed reporter gene (Ty1–mURA3) inserted either outside the rDNA array or within NTS1 (Fig. 1A, R1 reporter; Smith and Boeke 1997). Cells were 10-fold serially diluted and spotted on complete medium as a plating control and on medium without uracil to monitor silencing of mURA3. Consistent with previous observations, the reporter gene was not silenced outside the array but was efficiently silenced within NTS1, as indicated by poor growth on URA– medium (Fig. 3A, cf. rows 1 and 2; Smith and Boeke 1997). However, in fob1Δ cells, the reporter gene at NTS1 was completely derepressed (Fig. 3A, cf. rows 2 and 4). The extent of derepression in fob1Δ cells was comparable to what has been observed in sir2Δ cells (Fig. 3A, row 6). Western analysis of whole-cell extracts indicated that the levels of Net1–TAP and Sir2–TAP proteins were not altered in fob1Δ cells (Fig. 3D), ruling out the possibility that deletion of FOB1 disrupts silencing at NTS1 by reducing the cellular levels of these silencing proteins. The loss of silencing was specific to FOB1, as addition of a single-copy plasmid containing the FOB1 gene under the control of its own promoter restored silencing to fob1Δ cells (data not shown).
Figure 3.
Fob1 is required for rDNA silencing at NTS1 but not at NTS2/18S. Silencing was assessed by monitoring the growth of 10-fold serial dilutions of cells on –URA medium. Complete medium was used as a plating control. (A) Both FOB1 and SIR2 are required for Ty1–mURA3 silencing at NTS1. Silencing was assayed using strains containing a Ty1–mURA3 insertion either outside rDNA or at NTS1 (Smith and Boeke 1997). The approximate location of this reporter (R1) within rDNA is shown in Figure 1A. (B,C) In another reporter gene system, FOB1 and SIR2 are both required for silencing at NTS1 (B, R3 reporter), but only SIR2 is required for silencing at NTS2, near the 35S coding region (C, R4 reporter). See Figure 1A for the locations of R3 and R4 reporter genes. (D) The levels of Net1–TAP and Sir2–TAP proteins do not change in the absence of Fob1 as shown by Western blotting of whole-cell extracts. Actin is shown as a loading control.
We next tested if FOB1 influences silencing in regions located farther from the NTS1 enhancer. We deleted the FOB1 gene in a strain in which the Ty1–mURA3 reporter gene is inserted in NTS2, adjacent to the 5S gene (Fig. 1A, R2 reporter). The R2 reporter is typically poorly silenced (Smith and Boeke 1997), and no loss of silencing was detected in fob1Δ cells (data not shown). To further test the role of FOB1 in silencing at the NTS2/18S region, we integrated a plasmid containing the mURA3 reporter gene without Ty1 elements into three sites: outside the rDNA at the LEU2 gene and inside rDNA at two locations corresponding to peaks of Net1 and Sir2 association (Fig. 1A, R3 and R4 reporters). As expected, the mURA3 gene inserted at either NTS1 or NTS2/18S was strongly silenced (Fig. 3B,C, cf. rows 1 and 2), and this silencing was SIR2-dependent (Fig. 3B,C, rows 5 and 6). The greater concentration of silencing proteins at NTS2/18S compared with NTS1 (Fig. 1B) was consistent with the approximately twofold to fivefold stronger silencing for the R4 reporter in NTS2 compared with the R3 reporter in NTS1 (Fig. 3B,C, cf. row 2). However, although silencing at NTS1 was abolished in fob1Δ cells, deletion of FOB1 had no effect on silencing of an identical reporter gene at NTS2/18S (Fig. 3B,C, cf. row 4). Thus, we have identified a novel role for the fork-blocking protein Fob1 and determined that the requirements for silencing of Pol II promoters across an rDNA repeat are not uniform.
Fob1 colocalizes with Net1 and Sir2 to the RFB region of NTS1
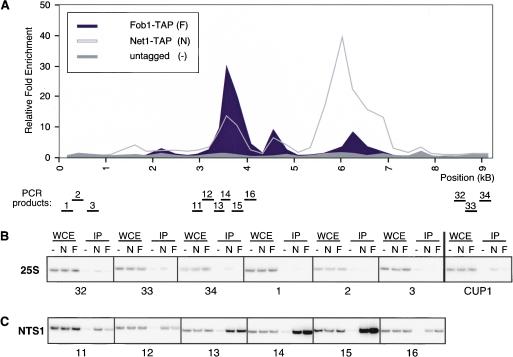
Fob1–GFP has been shown to localize to the nucleolus (Defossez et al. 1999), but it is not known if Fob1 actually associates with rDNA. To determine if Fob1 is associated with the NTS1 region, we mapped the association of Fob1 with rDNA by ChIP, using a strain in which the endogenous FOB1 gene was modified to produce a protein with a C-terminal TAP tag (Fob1–TAP; Fig. 1G, lane 4). Cells expressing Fob1–TAP displayed wild-type levels of silencing, suggesting that the modified protein was functional (Supplementary Fig. 1). A graph representing the enrichment of rDNA regions in Fob1–TAP immunoprecipitations is shown in Figure 4A. We observed a major peak of Fob1 association, which precisely overlapped the NTS1 RFB region and the peak of Net1–TAP association with this rDNA region (Fig. 4A,C). Two smaller peaks of binding occurred at the 5S gene and near the NTS2/18S region (Fig. 4A). In contrast, Fob1 did not associate with the 25S region or the CUP1 locus (Fig. 4B). In summary, Fob1, Net1, and Sir2 displayed similar association profiles with respect to the NTS1 region.
Figure 4.
Fob1 is associated primarily with the NTS1 region. (A) Representative graph showing the association of Fob1–TAP across an rDNA repeat. Most of the protein is concentrated within NTS1. Two smaller peaks are observed at the 5S and near the start of the 35S rRNA genes. (B) Examples of ChIP data showing the association of Fob1–TAP with the 25S of rDNA. CUP1 is not significantly enriched in Fob1–TAP immunoprecipitations. (C) ChIP experiments showing the association of Fob1–TAP with the RFB region of NTS1. Labels are as in Figure 1. (F) Fob1–TAP cells. Primer reference numbers below the panels correspond to PCR products in A.
Fob1 is required for localization of Net1 and Sir2 to NTS1
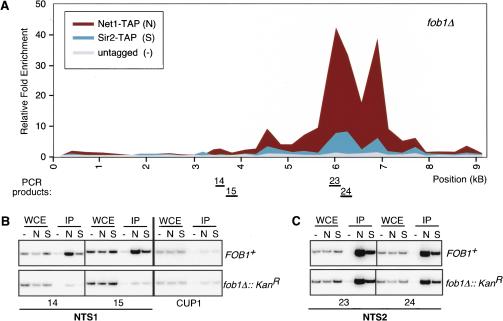
To further investigate the nature of the silencing defect in cells lacking Fob1, we mapped the associations of Net1–TAP and Sir2–TAP along an rDNA repeat in fob1Δ cells by ChIP. The enrichment of NTS1 DNA by Net1–TAP or Sir2–TAP was greatly reduced in fob1Δ cells (Fig. 5A,B). However, similar levels of Net1–TAP and Sir2–TAP were associated with the NTS2/18S region in FOB1+ and fob1Δ cells (Fig. 5C). In addition, there was a reproducible increase in relative association toward the interior of 35S (cf. Figs. 5A and 1B). These findings are consistent with the observed loss of silencing specifically at NTS1 but not NTS2/18S in fob1Δ cells (Fig. 3), and show that Fob1 is required for the proper localization of Net1 and Sir2 to rDNA.
Figure 5.
Fob1 is required for the association of Net1 and Sir2 with NTS1. (A) Graph showing the association of Net1–TAP and Sir2–TAP with rDNA in fob1Δ cells. Net1 and Sir2 associate with the Pol I transcription initiation region and part of the 35S coding region. (B) Examples of ChIP experiments in fob1Δ cells showing reduced binding of both Net1–TAP and Sir2–TAP to the RFB region of NTS1. Panels show examples from both FOB1+ (upper set) and fob1Δ cells (lower set). CUP1 primers were used as a negative control. (C) Net1–TAP and Sir2–TAP associate with the NTS2/18S in both FOB1+ (upper set) and fob1Δ cells (lower set). Labels are as described in Figure 1, and locations of PCR products are shown below the graph in A.
Fob1 specifically associates with the RENT complex
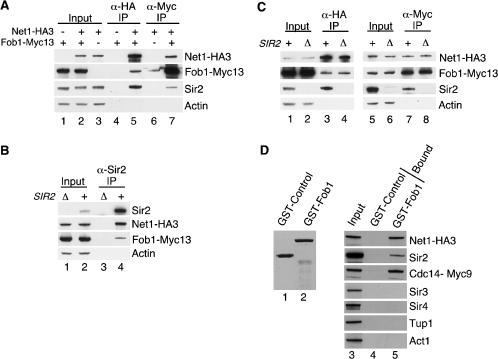
The precise colocalization of Fob1 with Net1 and Sir2 at NTS1, as well as its requirement for Net1 and Sir2 to associate with this region, suggested that Fob1 might physically associate with the RENT complex. To test this possibility, we performed coimmunoprecipitation experiments from extracts prepared from yeast strains in which the endogenous copies of Net1 and Fob1 were modified to produce Net1–HA3 and Fob1–Myc13. Cells expressing Fob1–Myc13 displayed wild-type levels of silencing, suggesting that the modified protein was functional (Supplementary Fig. 1). Immunoprecipitation of Net1–HA3 or Sir2 resulted in coprecipitation of Fob1–Myc13 (Fig. 6A, lane 5, 6B, lane 4), and immunoprecipitation of Fob1–Myc13 resulted in coprecipitation of both Net1–HA3 and Sir2 (Fig. 6A, lane 7). Deletion of SIR2 had no effect on the amount of Net1–HA3 and Fob1–Myc13 that coprecipitated together, indicating that Net1 and Fob1 can associate independently of Sir2 (Fig. 6C, cf. lanes 3 and 4, 7 and 8). However, we consistently coprecipitated more Sir2 than Fob1 with Net1 (Fig. 6A–C), suggesting that the Fob1–Net1 interaction was weaker than the Sir2–Net1 interaction. We also tested the interaction of Fob1 with RENT using GST pull-down assays. We purified bacterially expressed GST–Fob1 and a GST–control protein (Fig. 6D, lanes 1,2), incubated them with whole-cell yeast extracts, and analyzed the bound fractions by Western blotting. We found that GST–Fob1 associated with all three subunits of RENT (Net1–3HA, Sir2, and Cdc14–Myc9), whereas the GST–control protein did not (Fig. 6D, cf. lanes 4 and 5). Furthermore, GST–Fob1 did not bind Sir3 or Sir4 (silencing proteins not required for rDNA silencing), Tup1 (a general transcriptional corepressor), or Act1 (an abundant cytoskeletal protein; Fig. 6D, cf. lanes 4 and 5), suggesting that the association between Fob1 and the RENT complex was specific.
Figure 6.
The RENT complex physically associates with Fob1. (A) Western blots showing that Net1–HA3 coprecipitates with Sir2 and Fob1–Myc13 (lane 5) from whole-cell extracts. Fob1–Myc13 also coprecipitates Net1–HA3 and Sir2 (lane 7). Actin serves as a loading control. (–) Untagged; (+) tagged. One percent of whole-cell extract (input) and 25% of bound material is shown for all panels. (B) Immunoprecipitation of Sir2 coprecipitates Net1–HA3 and Fob1–Myc13 (lane 4). (Δ) sir2Δ cells; (+) SIR2+ cells. (C) Western blots showing that Fob1 and Net1 can physically associate in vivo in the absence of Sir2 (lanes 3,4,7,8). (D, left) A Coomassie-stained gel of purified GST–Control (lane 1) and GST–Fob1 (lane 2) proteins. The control protein (UAP56) is a human protein involved in mRNA splicing. (Right) Western blot indicating that subunits of RENT from whole-cell yeast extracts associate specifically with GST–Fob1 (lane 5) but not with the GST–Control protein (lane 4). Neither GST fusion protein interacts with actin (Act1) or transcriptional repressors that do not participate in rDNA silencing (Sir3, Sir4, and Tup1).
RNA polymerase I interacts with Net1 and Sir2
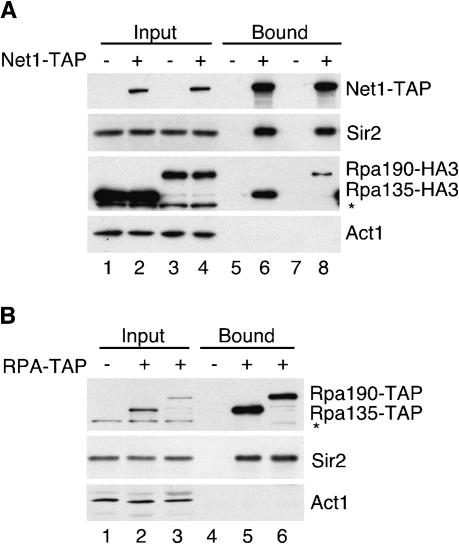
Our ChIP data indicated that Net1 and Sir2 associated with the NTS2/18S region of rDNA in a Fob1-independent manner. Noticeably, this peak of association with rDNAoverlaps the Pol I transcription initiation region. Previous work had demonstrated that the Net1 subunit of RENT is associated with Pol I and is required for optimal Pol I transcription in vitro (Shou et al. 2001). Our data raise the possibility that this interaction may also be important for rDNA silencing and the association of RENT with rDNA. To provide further support for this idea and to specifically address whether other subunits of the RENT complex, in particular Sir2, were also associated with Pol I, we carried out a series of immunoprecipitations. As expected from previous studies (Straight et al. 1999; Shou et al. 2001), immunoprecipitation of Net1–TAP from whole-cell extracts showed that Net1–TAP associated with Sir2 (Fig. 7A, lanes 6,8) and the two largest subunits of Pol I, Rpa135 and Rpa190 (Fig. 7A, lanes 6,8). Furthermore, we found that these interactions were not affected by the presence of ethidium bromide, indicating that they were not DNA-dependent (data not shown). To determine whether Sir2 is also associated with Pol I, the coding regions of RPA135 and RPA190 were modified to express TAP-tagged proteins, and these subunits were immunoprecipitated from whole-cell yeast extracts. Both Pol I subunits associated with Sir2 in the presence of ethidium bromide (Fig. 7B, lanes 5,6). These results show that both the Net1 and Sir2 subunits of RENT are associated with Pol I and suggest that the localization of Net1 and Sir2 to the NTS2/18S region may result from their physical association with Pol I.
Figure 7.
The RENT complex physically associates with RNA polymerase I. (A) Western blots showing that Net1–TAP coimmunoprecipitates with Sir2 (lanes 6,8) and the largest subunits of RNA polymerase I (Rpa135 and Rpa190, lanes 6,8) from whole-cell extracts. Act1 serves as a loading control. (–) Untagged; (+) tagged; (*) a cross-reacting band. One percent of whole-cell extract (input) and 25% of bound material are shown in A and B. (B) Western blots showing that Rpa135–TAP (lane 5) and Rpa190–TAP (lane 6) coimmunoprecipitate with Sir2.
Discussion
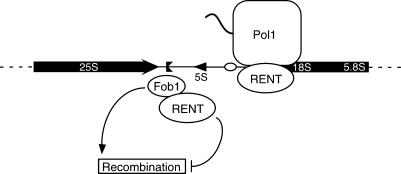
We have shown that the Net1 and Sir2 subunits of the rDNA-specific silencing complex RENT are associated primarily with two regions within an rDNA repeat that overlap the NTS1/RFB and transcription initiation regions. The presence of the RENT complex at these functional elements is consistent with a role for silencing in regulation of specific rDNA activities. We show that Fob1 also associates with the NTS1 replication fork block region and is required for silencing of Pol II-transcribed genes at this location. In the absence of Fob1, Net1 and Sir2 are no longer associated with the NTS1 RFB region but are still present at the NTS2/18S region, where silencing occurs independently of Fob1. Both Fob1 and Pol I physically associate with the RENT complex, suggesting two mechanisms for recruitment of silencing proteins to rDNA (Fig. 8) and providing additional evidence that the integrity and function of rDNA is regulated by silencing proteins. Because the overall structural organization of rRNA genes is conserved in organisms ranging from yeast to humans (for review, see Reeder 1999), the intersection of silencing, recombination, and transcriptional pathways described here may also be conserved in other eukaryotes.
Figure 8.
Two pathways for the recruitment of the RENT complex to rDNA. Fob1 recruits the RENT complex to the replication fork block region of NTS1. Silent chromatin generated by RENT then inhibits Fob1-dependent recombination. A Fob1-independent pathway, involving Pol I, recruits RENT to the NTS2/18S region. See text for details. Labels are as in Figure 1.
Distribution of silencing complexes in rDNA
Binding of the RENT complex to the NTS and throughout the 35S rRNA region is not uniform. Net1 and Sir2 are not exclusively or continuously associated with the nontranscribed region, displaying instead two distinct peaks of association, separated by 1 kb of relatively low occupancy (Fig. 1B). The peak within NTS1 is centered on sequences that overlap the RFB region, whereas the other peak overlaps the Pol I transcription initiation region of NTS2 and includes more than 1 kb of the 35S coding region. Notably, these areas overlap with two previously identified SIR2-responsive regions at NTS1 and 18S, which display altered micrococcal nuclease and dam methyltransferase sensitivities in the absence of SIR2 (Fritze et al. 1997). In addition, the Net1 and Sir2 association profiles are consistent with qualitative silencing assays, indicating that silencing of Ty1–mURA3 reporters is stronger at a site within NTS1 and near the start of the 18S region as compared with two sites near the 5S gene (Smith and Boeke 1997). Similarly, the relatively low association of Net1 and Sir2 with certain sections of the 35S RNA coding region is consistent with the relatively weaker silencing of reporter genes inserted at these locations (Smith and Boeke 1997; J. Huang and D. Moazed, unpubl.). The reduced associations of Net1 and Sir2 with the center of the NTS (Fig. 1B) may be the result of the transcriptional activity of Pol III at the 5S gene or the presence of a barrier element. The left boundary of RENT association with the NTS2/18S region coincides with the main binding site of the cohesin complex within rDNA, adjacent to the 5S promoter (Laloraya et al. 2000). Mutations in some cohesin subunits perturb boundary functions that limit the spreading of silencing at the silent mating-type loci (Donze et al. 1999), suggesting that sites of cohensin association may act as barrier elements. Thus, RENT association may be reduced near the 5S because of the presence of cohesins.
The deacetylase activity of the Sir2 subunit of RENT is required for rDNA silencing (Imai et al. 2000). In addition, deletion of SIR2 results in an increase in levels of acetylated histones H3 and H4 at the rDNA 5S region (Armstrong et al. 2002), a threefold increase in the levels of diacetylated H3 (Lys 9 and 14) at one site in the NTS (Bryk et al. 2002), a 1.6- to 2.4-fold increase in acetylated H4 at two sites in NTS2 near the Pol I promoter region (Hoppe et al. 2002), and increases in both H3 (K9/K14) and H4 acetylation at a site within the NTS1 and transcriptional initiation regions (Buck et al. 2002). Our ChIP analysis indicates that although Net1 and Sir2 are localized primarily to two rDNA regions, Sir2 is required for global hypoacetylation of histone H3 associated with rDNA chromatin (Fig. 2; J. Huang, unpubl.).
Recruitment of RENT to the replication fork block region of NTS1 by Fob1
Our data show that Fob1 is required for the association of the RENT complex with the rDNA NTS1 region and for silencing at this location. Our finding that Fob1 is present at the replication fork block region of NTS1 is consistent with Fob1's requirement for the replication fork block and recombination activities of cis-elements within this region. Additionally, we find that Fob1 is physically associated with RENT in vivo, suggesting that Fob1 is a recruitment factor for silencing complexes. Interestingly, Fob1 possesses homology to two highly conserved domains of retroviral integrases, which are known to mediate DNA cleavage and strand-transfer reactions (Dlakic 2002), raising the possibility that Fob1 binds to DNA at or near the RFB directly.
Our finding that Fob1 is required for rDNA silencing was surprising, given its role in promoting recombination. One possible explanation for our results is that silencing complexes are recruited to the RFB region by Fob1 to counteract the recombination potential of replication fork barriers that are also generated by Fob1. In prokaryotic systems, blocked replication forks can lead to double-strand breaks that are substrates for homologous recombination (for review, see Rothstein et al. 2000), and similar processes may occur at the rDNA of yeast. Alternatively, association of RENT with the Fob1 protein may inhibit the replication fork-blocking activity of Fob1 and thereby Fob1-stimulated recombination. Our data are also consistent with a model in which the RENT complex acts directly on Fob1 to inhibit its fork-blocking activity, for example, by deacetylating it. Furthermore, our findings are consistent with the opposing roles of Fob1 and Sir2 in regulation of rDNA recombination and yeast life span (Gottlieb and Esposito 1989; Kobayashi and Horiuchi 1996; Defossez et al. 1999; Kaeberlein et al. 1999; Johzuka and Horiuchi 2002). Fob1, a positive regulator of rDNA recombination, recruits its own inhibitor, Sir2, to rDNA. Therefore, in the absence of the inhibitory function of Sir2, the enhancement of recombination by Fob1 is unopposed, and increased recombination results in increased accumulation of ERCs and reduced life span in sir2Δ cells. Because rDNA recombination requires Fob1, in fob1Δ cells recombination levels are dramatically reduced, and because Sir2 is no longer recruited to the NTS1 region, silencing at this region is also abolished.
It is notable that a positive regulator of recombination is located in a region of the genome that, because of its repetitive organization, is inherently recombinogenic. In wild-type cells, the levels of recombination at rDNA are significantly lower than predicted for a large and repetitive locus, despite the stimulatory presence of Fob1. Moreover, in the absence of Fob1 or both Fob1 and Sir2, rDNA recombination levels remain low (Defossez et al. 1999; Kaeberlein et al. 1999), suggesting that an additional level of regulation is present that suppresses recombination. Some level of recombination is still necessary, which may explain the positive role of Fob1. For example, unequal sister-chromatid exchange during recombination results in either the gain or loss of repeats, allowing the maintenance of a favorable number of repeats as growth conditions dictate. Contraction of repeats also facilitates the removal of dominant-negative mutations, maintaining the integrity of the array. The ability of Fob1 to promote recombination and to recruit the RENT complex may provide a mechanism for fine-tuning rDNA recombination levels. In addition, our findings are consistent with a model that in wild-type yeast cells, rDNA recombination and replicative life span are regulated by the dual silencing and recombination activities of Fob1.
Association of RENT with the NTS2/35S region
Net1 and Sir2 are also associated with an rDNA region of ∼1.5–2 kb spread over part of NTS2 and the 35S rRNA coding region (Fig. 1B). Because association with this region is not Fob1-dependent (Fig. 5A), an as-yet-unidentified protein recruits silencing complexes specifically to this region. We suggest that the most likely candidate is RNA polymerase I and/or its associated transcription factors. Net1 binds purified Pol I complexes and is required for optimal Pol I-dependent transcription in vitro (Shou et al. 2001), and rDNA silencing is impaired in cells that lack a functional Pol I (Buck et al. 2002). Our findings that both the Net1 and Sir2 subunits of RENT associate with Pol I (Fig. 7A,B) support the hypothesis that the polymerase itself recruits the RENT complex to the NTS2/18S region of rDNA (Fig. 8).
It remains unclear why silencing complexes are recruited to an area of heavy transcription initiation. This region, like the NTS1 replication fork block region, may be involved in stimulating recombination. For example, optimal HOT1 activity requires a cis-element located within the Pol I transcription initiator region in addition to the sequences located at NTS1 (Keil and Roeder 1984; Voelkel-Meiman et al. 1987). Thus, the HOT1 element of NTS2 may contribute to hotspot activity within rDNA. Furthermore, there is evidence supporting a link between recombination and Pol I activity, because it has been shown that HOT1 sequences fail to stimulate recombination at an ectopic location in the absence of Pol I (Huang and Keil 1995). This observation suggests that the presence of Pol I or its transcriptional activity may be required for rDNA recombination. Analogous to the situation with Fob1 at NTS1, the RENT complex might associate with Pol I to suppress Pol I-stimulated recombination. Alternatively, Pol I-dependent silencing in the NTS2 region may influence recombination rates by regulating the activity of DNA replication origins within rDNA. Recently, it has been shown that in rDNA, functional replication origins are clustered and separated by large regions where initiation firing is suppressed in a SIR2-dependent manner (Pasero et al. 2002). In principle, clustering of origin-firing would reduce the number of active replication fork blocks within NTS1 regions, thereby minimizing the likelihood of recombination (Pasero et al. 2002).
The association of Net1 and Sir2 with the Pol I transcription initiation region is strikingly polar, with the highest area of occupancy located toward the 35S coding region and decreasing association toward the middle of the 35S gene (Figs. 1B, 5A). This association pattern is consistent with the role of Net1 as a Pol I transcription factor (Shou et al. 2001) and with the spreading of silencing to sequences flanking the rDNA array in the direction of Pol I transcription (Buck et al. 2002). However, because Net1 remains localized to the nucleolus throughout the cell cycle, and Sir2 is required for hypoacetylation of H3 throughout the rDNA, it is unlikely that low levels of RENT association with rDNA regions corresponding to the middle and 3′-end of 35 rRNA reflect dissociation from Pol I during transcription elongation (Fig. 2; Shou et al. 1999; Straight et al. 1999; Visintin et al. 1999). We favor the possibility that this polar pattern of localization mirrors the mode of association of Pol I or one of its associated factors with rDNA (Figs. 1, 8). Finally, we note that the recruitment of RENT to rDNA by either Pol I or Fob1 may provide redundant and/or novel opportunities for regulation of the cell cycle functions of this complex.
Materials and methods
Yeast strains and plasmids
A strain table is included in the Supplemental Material. NET1, SIR2, FOB1, RPA135, and RPA190 genes were modified with the C-terminal TAP tag as described (Rigaut et al. 1999), and the NET1 gene was modified with a C-terminal HA3 tag by integrating the plasmid pDM239 as described (Straight et al. 1999). The Myc9-tagged Cdc14 strain was a kind gift from R. Deshaies (California Institute of Technology, Pasadena, CA). The RPA190 and RPA135 genes were modified with C-terminal HA3 tags, the FOB1 gene was modified with the C-terminal Myc13 tag, and the FOB1 and SIR2 deletion strains were generated by replacing their open reading frames with the KANR marker as described (Longtine et al. 1998). The SIR2 gene was disrupted with the HIS3 marker using the EcoRV–SphI fragment from plasmid pJR531 (the kind gift of J. Rine, University of California at Berkeley, Berkeley, CA). Ty1–mURA3 silencing reporter strains have been described previously (Smith and Boeke 1997). The mURA3 gene contains the TRP1 promoter instead of the URA3 promoter (Smith and Boeke 1997). mURA3 silencing strains were generated by transformation with pDM316 cut with BstEII to integrate at LEU2, pDM312 cut with SmaI to integrate at NTS2, and pDM704 cut with HindIII to integrate at NTS1. All transformations were performed with the lithium acetate method (Guthrie and Fink 1991), and proper integration was confirmed by PCR.
pDM316 (LEU2::mURA3) was constructed by ligation of a 1.6-kb BamHI–EagI PCR product containing the mURA3 gene into pRS305. pDM312 (RDN1–NTS2::mURA3–LEU2) was generated by ligation of a 1-kb XhoI–EcoRI PCR product of NTS2 and 1.6-kb EcoRI–EagI PCR product containing the mURA3 gene into pRS305 digested with XhoI and EagI. pDM704 (RDN1–NTS1::mURA3–LEU2) was created similarly to pDM312 except using the 0.5-kb XhoI–EcoRI PCR product of NTS1. The mURA3 gene was amplified from pJSS60-2 (Smith and Boeke 1997), and NTS regions were amplified from genomic DNA.
ChIP assays
ChIP assays were carried out as described (Suka et al. 2001), with modifications. Yeast cultures (50 mL) were grown to an OD600 of 1.5–1.8 and cross-linked with 1% formaldehyde at room temperature for 15 min. The reaction was quenched with glycine at a final concentration of 125 mM for 5 min. Cells were washed twice with cold TBS (20 mM Tris-HCl at pH 7.6 and 150 mM NaCl) and frozen at –80°C. Cell pellets were resuspended in 400 μL of lysis buffer (50 mM HEPES-KOH at pH 7.5, 500 mM NaCl, 1 mM EDTA at pH 8.0, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, and protease inhibitors) and bead-beat with glass beads (beads and Mini Beadbeater, Biospec Products) twice for 30 sec. Lysates were sonicated three times for 20 sec at 40% amplitude (Branson Digital Sonifer), and centrifuged at 13,000 rpm for 5 and 15 min. To obtain input DNA, 50 μL of clarified lysate was used, and for each immunoprecipitation reaction.
Thirty microliters of a 50% slurry of prewashed IgG-agarose beads (Sigma) was incubated with each lysate at 4°C for 2 h. For Sir2 or diAcH3 ChIP experiments, 150 μL of lysate was incubated at 4°C overnight with 1.5 or 2.0 μg of antibody (polyclonal anti-Sir2, Hoppe et al. 2002; anti-AcK9/AcK14 H3, Upstate Biotechnology), and further incubated with Protein A Sepharose beads at 4°C for 2 h. Beads were washed three times in lysis buffer, once with 10 mM Tris-HCl (pH 8.0), 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, and 1 mM EDTA, and once with TE (10 mM Tris-HCl at pH 8.0 and 1 mM EDTA at pH 8.0) at room temperature. Beads were eluted by incubating with 100 μL of 50 mM Tris-HCl (pH 8.0), 10 mM EDTA(pH 8.0), and 1% SDS at 65°C for 15 min. Eluate was transferred to a fresh tube and pooled with a final bead wash of 150 μL of TE with 0.67% SDS. For input DNA, 200 μL of TE with 1% SDS was added to 50 μL of lysate. All samples were incubated at 65°C overnight, combined with 250 μL of TE, 15 μg of glycogen, and 100 μg of Proteinase K, and incubated at 37°C for 2 h. After addition of 55 μL of 4 M LiCl, samples were extracted once with phenol:chloroform:isoamyl alcohol and once with chloroform. Precipitated and washed DNA was resuspended in 50 μL of TE with 10 μg of RNase A and incubated at 37°C for 1 h. PCR reactions (12.5 μL) contained 2 μL of template DNA(1:8 dilution of IP and 1:20,000 dilution of WCE for TAP-tag strains; 1:8 dilution of Sir2 and 1:16 dilution of di-AcH3 IPs, and 1:25,000 dilution for WCE), and 1.25 μCi of [α-32P]dCTP. PCR parameters were 1 cycle of 95°C for 2 min, 55°C for 30 sec, and 72°C for 1 min, followed by 21 (multicopy genes) or 28 (single-copy genes) cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min, and a final step of 72°C for 4 min. Primers of 20-nt oligonucleotides were designed with Primer3 software (http://www.basic.nwu.edu/biotools/Primer3.html) to amplify products 250 bp for rDNA or 150 bp for ACT1 or CUP1. Sequences are available upon request. Primers for HMR-E and telomeric sequences (0.6 kb from the end of Chromosome VI-R) have been previously described (Hoppe et al. 2002).
Samples were run on 6% polyacrylamide gels at 100 V for 45 min. PCR products were quantitated using QuantityOne software. Relative fold enrichment was determined by calculating the ratio of rDNA(IP) to rDNA(WCE) and normalizing the data such that the untagged background ratio in Figures 1B, 4A, and 5A is ∼1. Each set of experiments was performed at least three times and produced similar binding profiles. For multiplex PCR, fold enrichment values for each strain were calculated as follows: [rDNA(IP)/CUP1(IP)]/[rDNA(WCE)/CUP1(WCE)]. The untagged strain value was normalized to 1, yielding the rDNA fold enrichment for the tagged strains. In Figure 2B, the relative fold enrichment is defined as the ratio of SIR2+ to sir2Δ for values from the following calculation: [rDNA(IP)/CUP1(IP)]/[rDNA(WCE)/CUP1(WCE)]. In Figure 2C, the relative fold enrichment is defined as the ratio of sir2Δ to SIR2+ for values from the same calculation. For enrichment of TEL or HMR-E sequences, ratios were determined identically using ACT1 instead of CUP1 as an internal control.
Silencing assays
Cells were grown in YEPD to an OD600 of 1.6–1.8 and concentrated to two-thirds the original volume. Three microliters each of 10-fold serial dilutions were spotted on appropriate media. Plates were incubated at 30°C for 2–3 d.
Immunoprecipitation reactions
Reactions were performed essentially as described previously (Straight et al. 1999). One percent of input whole-cell extract and 25% of bound fractions were run on SDS-PAGE gels, blotted, and probed with the indicated antibodies.
Purification of GST fusion proteins
GST–Fob1 protein was expressed from pDM708, which was constructed by ligation of a 1.7-kb BclI–XhoI PCR product containing the FOB1 gene (amplified from genomic DNA) with pGEX-6P-1 (Amersham Pharmacia). The GST–control protein was expressed from the plasmid pGEX-UAP56 (pDM549), a kind gift from R. Reed (Harvard Medical School, Boston, MA). DH5α cells containing expression vectors were grown in 1.5 L of 1.5× LB media to an OD600 of 0.6 and induced with 0.1 mM IPTG at 30°C for 2 h. Cells were washed with cold PBS and stored at –80°C. All subsequent steps were performed at 4°C. The cell pellet was resuspended in lysis buffer (PBS with 1 mM EDTA, 1 mM PMSF, 200 μg/mL lysozyme) and stirred for 10 min. NP-40 and NaCl were added to final concentrations of 0.5% and 300 mM, respectively. The lysate was sonicated to reduce viscosity, and DTT was added to 15 mM. The lysate was centrifuged for 10 min at 10,000 rpm in an SA-600 rotor and for 60 min at 35,000 rpm in a Ti-70 rotor. The supernatant was incubated with glutathione agarose (Sigma) for 1.5 h. The resin was loaded on a column and washed with wash buffer (PBS with 250 mM NaCl, 0.5 mM DTT, and 0.1% NP-40) and with wash buffer without NP-40. Column was eluted with 50 mM Tris-HCl (pH 8.0), 250 mM NaCl, and 10 mM glutathione. Peak fractions were pooled and dialyzed against 50 mM HEPES-NaOH (pH 7.6), 300 mM NaCl, and 30% glycerol and stored at –80°C.
GST pull-downs
Purified GST fusion proteins (5 μg) were bound to glutathione agarose (50 μL of packed resin) at 4°C for 1 h in 200 μL of yeast lysis buffer (50 mM HEPES-NaOH at pH 7.5, 100 mM NaCl, 10% glycerol, 1 mM EDTA, 2 mM DTT, 1 mM PMSF, 0.1% NP-40, and protease inhibitors). Beads were washed three times with lysis buffer, and 20% of bound protein was visualized by Coomassie staining of an SDS-PAGE gel. For this, 10 μL of beads (1 μg of protein) was incubated with 90 μL of whole-cell yeast extract at 4°C for 2 h. Beads were washed three times with lysis buffer and resuspended in SDS sample buffer. One percent of input and 25% of bound proteins were run on 4%–12% gradient gels, transferred to PVDF membranes, and blotted with mouse anti-HA, anti-MYC, or anti-Actin (Sigma), and rabbit anti-Sir2, anti-Sir3, anti-Sir4, or anti-Tup1 (Redd et al. 1997) at 1:5000 or 1:10,000 dilutions. Whole-cell yeast extracts from strains DMY631 and DMY761 were prepared as described previously (Straight et al. 1999).
Acknowledgments
We thank J. Boeke, R. Deshaies, R. Reed, J. Rine, and J. Smith for plasmids and strains; members of the Moazed Lab for helpful discussions; and B. Hall, M. Motamedi, A. Rudner, J.C. Tanny, A. Verdel, and F. Winston for comments on the manuscript. This work was supported by grants from the NIH and the Ellison Medical Foundation (D.M.) and an NIH training grant to Harvard Medical School (J.H.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1108403.
References
- Armstrong C.M., Kaeberlein, M., Imai, S.I., and Guarente, L. 2002. Mutations in Saccharomyces cerevisiae gene SIR2 can have differential effects on in vivo silencing phenotypes and in vitro histone deacetylation activity. Mol. Biol. Cell 13: 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benguria A., Hernandez, P., Krimer, D.B., and Schvartzman, J.B. 2003. Sir2p suppresses recombination of replication forks stalled at the replication fork barrier of ribosomal DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 31: 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M., Rose, A.B., Holmes, S.G., Allis, C.D., and Broach, J.R. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes & Dev. 7: 592–604. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman, W.L. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643. [DOI] [PubMed] [Google Scholar]
- Bryk M., Banerjee, M., Murphy, M., Knudsen, K.E., Garfinkel, D.J., and Curio, M.J. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes & Dev. 11: 255–269. [DOI] [PubMed] [Google Scholar]
- Bryk M., Briggs, S.D., Strahl, B.D., Curcio, M.J., Allis, C.D., and Winston, F. 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12: 165–170. [DOI] [PubMed] [Google Scholar]
- Buck S.W., Sandmeier, J.J., and Smith, J.S. 2002. RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell 111: 1003–1014. [DOI] [PubMed] [Google Scholar]
- Defossez P.A., Prusty, R., Kaeberlein, M., Lin, S.-J., Ferrigno, P., Silver, P.A., Keil, R.L., and Guarente, L. 1999. Elimination of replication fork block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3: 447–455. [DOI] [PubMed] [Google Scholar]
- Dlakic M. 2002. A model of the replication fork blocking protein Fob1p based on the catalytic core domain of retroviral integrases. Protein Sci. 11: 1274–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D., Adams, C.R., Rine, J., and Kamakaka, R.T. 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes & Dev. 13: 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E.A. and Warner, J.R. 1984. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell 39: 663–673. [DOI] [PubMed] [Google Scholar]
- ____. 1986. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol. Cell. Biol. 6: 2089–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritze C.E., Verschueren, K., Strich, R., and Esposito, R.E. 1997. Direct evidence for Sir2 modulation of chromatin structure in yeast rDNA. EMBO J. 16: 6495–6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Strahl-Bolsinger, S., Renauld, H., Laroche, T., Kennedy, B.K., Grunstein, M., and Gasser, S.M. 1997. Localization of Sir2p: The nucleolus as a compartment for silent information regulators. EMBO J. 16: 3243–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S. and Esposito, R.E. 1989. Anew role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56: 771–776. [DOI] [PubMed] [Google Scholar]
- Gottschling D.E., Aparicio, O.M., Billington, B.L., and Zakian, V.A. 1990. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- Gruber M., Wellinger, R.E., and Sogo, J.M. 2000. Architecture of the replication fork stalled at the 3′ end of yeast ribosomal genes. Mol. Cell. Biol. 20: 5777–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. and Fink, G.R. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego.
- Hecht A., Strahl-Bolsinger, S., and Grunstein, M. 1996. Spreading of transcriptional repressor Sir3 from telomeric heterochromatin. Nature 383: 92–96. [DOI] [PubMed] [Google Scholar]
- Hoppe G.J., Tanny, J.C., Rudner, A.D., Gerber, S.A., Danaie, S., Gygi, S.P., and Moazed, D. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22: 4167–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.S. and Keil, R.L. 1995. Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple cis-acting sequences. Genetics 141: 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S.-I., Armstrong, C.M., Kaeberlein, M., and Guarente, L. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800. [DOI] [PubMed] [Google Scholar]
- Johzuka K. and Horiuchi, T. 2002. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes to Cells 7: 99–113. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., McVey, M., and Guarente, L. 1999. The Sir2/3/4 complex and Sir2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Dev. 13: 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil R.L. and Roeder, G.S. 1984. Cis-acting, recombination stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell 39: 377–386. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. and Horiuchi, T. 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes to Cells 1: 465–474. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Hidaka, M., Nishizawa, M., and Horiuchi, T. 1992. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol. Gen. Genet. 233: 355–362. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Heck, D.J., Nomura, M., and Horiuchi, T. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: Requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes & Dev. 12: 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Nomura, M., and Horiuchi, T. 2001. Identification of DNA cis elements essential for expansion of ribosomal DNA repeats in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloraya S., Guacci, V., and Koshland, D. 2000. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 151: 1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J., Sutton, A., Tafrov, S.T., Heller, R.C., Stebbins, J., Pillus, L., and Sternglanz, R. 2000. The silencing protein Sir2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. 97: 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-H. and Keil, R.L. 1991. Mutations affecting RNA polymerase I-stimulated exchange and rDNA recombination in yeast. Genetics 127: 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Luo K., Vega-Palas, M.A., and Grunstein, M. 2002. Rap1–Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes & Dev. 16: 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8: 489–498. [DOI] [PubMed] [Google Scholar]
- Nomura M. 2001. Ribosomal RNA genes, RNA polymerases, nucleolar structures, and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harbor Symp. Quant. Biol. 66: 555–565. [DOI] [PubMed] [Google Scholar]
- Park J.-H., Cosgrove, M.S., Youngman, E., Wolberger, C., and Boeke, J.D. 2002. A core nucleosome surface crucial for transcriptional silencing. Nat. Gen. 32: 273–279. [DOI] [PubMed] [Google Scholar]
- Pasero P., Bensimon, A., and Schwob, E. 2002. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes & Dev. 16: 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T.D. 1980. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell 19: 765–774. [DOI] [PubMed] [Google Scholar]
- Petes T.D. and Botstein, D. 1977. Simple Mendelian inheritance of the reiterated ribosomal DNA of yeast. Proc. Natl. Acad. Sci. 74: 5091–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd M.J., Arnaud, M.B., and Johnson, A.D. 1997. A complex composed of Tup1 and Tsn6 represses transcription in vitro. J. Biol. Chem. 272: 11193–11197. [DOI] [PubMed] [Google Scholar]
- Reeder R.H. 1999. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog. Nucleic Acid Res. Mol. Biol. 62: 293–327. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotech. 17: 1030–1032. [DOI] [PubMed] [Google Scholar]
- Rine J. and Herskowitz, I. 1987. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R., Michel, B., and Gangloff, S. 2000. Replication fork pausing and recombination or “gimme a break.” Genes & Dev. 14: 1–10. [PubMed] [Google Scholar]
- Rusche L.N., Kirchmaier, A.L., and Rine, J. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 481–516. [DOI] [PubMed] [Google Scholar]
- Shou W., Seol, J.H., Shevchenko, A., Baskerville, C., Moazed, D., Chen, Z.W.S., Jang, J., Shevchenkno, A., Charbonneau, H., and Deshaies, R.J. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97: 233–244. [DOI] [PubMed] [Google Scholar]
- Shou W., Sakamoto, K.M., Keener, J., Morimoto, K.W., Traverso, E.E., Azzam, R., Hoppe, G.J., Feldman, R.M.R, DeModena, J., Moazed, D., et al. 2001. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell 8: 45–55. [DOI] [PubMed] [Google Scholar]
- Sinclair D.A. and Guarente, L. 1997. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell 91: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Smith J.S. and Boeke, J.D. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes & Dev. 11: 241–254. [DOI] [PubMed] [Google Scholar]
- Smith J.S., Brachmann, C.B., Celic, I., Kenna, M.A., Muhammad, S., Starai, V.J., Avalos, J.L., Escalante-Semerena, J.C., Grubmeyer, C., Wolberger, C., et al. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. 97: 6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht, A., Luo, K., and Grunstein, M. 1997. Sir2 and Sir4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 11: 83–93. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Shou, W., Dowd, G.J., Turck, C.W., Deshaies, R.J., Johnson, A.D., and Moazed, D. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97: 245–256. [DOI] [PubMed] [Google Scholar]
- Suka N., Suka, Y., Carmen, A.A., Wu, J., and Grunstein, M. 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8: 473–479. [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang, E.S., and Amon, A. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398: 818–823. [DOI] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Keil, R.L., and Roeder, G.S. 1987. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell 48: 1071–1079. [DOI] [PubMed] [Google Scholar]
- Wai H., Johzuka, K., Vu, L., Eliason, K., Kobayashi, T., Horiuchi, T., and Nomura, M. 2001. Yeast RNA polymerase I enhancer is dispensable for transcription of the chromosomal rRNA gene and cell growth, and its apparent transcription enhancement from ectopic promoters requires Fob1 protein. Mol. Cell. Biol. 21: 5541–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T.R., Hoang, M.L., Prusty, R., Lau, C.K., Keil, R.L., Fangman, W.L., and Brewer, B.J. 2000. Ribosomal DNA replication fork barrier and HOT1 recombination hot spot: Shared sequences but independent activities. Mol. Cell. Biol. 20: 4948–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]