Abstract
The X-inactive-specific transcript (Xist) locus is a cis-acting switch that regulates X chromosome inactivation in mammals. Over recent years an important goal has been to understand how Xist is regulated at the initiation of X inactivation. Here we report the analysis of a series of targeted mutations at the 5′ end of the Xist locus. A number of these mutations were found to cause preferential inactivation, to varying degrees, of the X chromosome bearing the targeted allele in XX heterozygotes. This phenotype is similar to that seen with mutations that ablate Tsix, an antisense RNA initiated 3′ of Xist. Interestingly, each of the 5′ mutations causing nonrandom X inactivation was found to exhibit ectopic sense transcription in embryonic stem (ES) cells. The level of ectopic transcription was seen to correlate with the degree of X inactivation skewing. Conversely, targeted mutations which did not affect randomness of X inactivation also did not exhibit ectopic sense transcription. These results indicate that X chromosome choice is determined by the balance of Xist sense and antisense transcription prior to the onset of random X inactivation.
Keywords: Xist, Tsix, X chromosome inactivation, nonrandom X inactivation
X chromosome inactivation in mammals provides dosage compensation for X-linked genes between XX females and XY males (Lyon 1961). One of the two X chromosomes is transcriptionally silenced early in female development, leading to mosaic expression of either the maternal (Xm) or paternal (Xp) X chromosome in different cell populations. Although initiation of X inactivation is generally random, there are cases of either complete or partial skewing. In marsupials (Cooper et al. 1971, 1990) and in certain extraembryonic lineages of rodents (Takagi and Sasaki 1975; Wake et al. 1976; Zakian et al. 1991), there is imprinted inactivation of the paternally derived X chromosome. Partial nonrandom X inactivation occurs in mice heterozygous at the Xce locus (Cattanach et al. 1969), in interspecific vole hybrids (Zakian et al. 1991), and also in humans (Naumova et al. 1996; Plenge et al. 1997).
X inactivation is regulated by a cis-acting master switch locus, classically referred to as the X inactivation center (Xic). The Xic functions in the in cis propagation of silencing over the entire chromosome, and is required at the initiation of X inactivation to determine how many (counting), and which (choice) X chromosomes are inactivated (for review, see Rastan and Brown 1990).
The Xic has been defined at the molecular level and shown to comprise the X-inactive-specific transcript (Xist) gene and associated regulatory elements (for review, see Avner and Heard 2001). The Xist gene produces a large noncoding RNA that localizes to the inactive X chromosome in cis and induces chromosome-wide silencing (Brockdorff et al. 1992; Brown et al. 1992; Clemson et al. 1996; Wutz et al. 2002). The counting and choice functions of the Xic are thought to be attributable to cis-acting sequences that regulate Xist expression.
X chromosome counting has been proposed to involve a blocking factor that binds to a single Xic, marking it to be the active X chromosome, in both XY and XX cells (Rastan 1983). Thus Xist expression and resulting X inactivation would occur as a default pathway on all except one X chromosome. Consistent with this view, deletion of 65 kb of sequence immediately downstream of Xist results in an allele from which X inactivation is initiated by default, in both XX cells and X0 cells (Clerc and Avner 1998). This indicates that the cis-acting sequences required for counting lie within the 65-kb deletion.
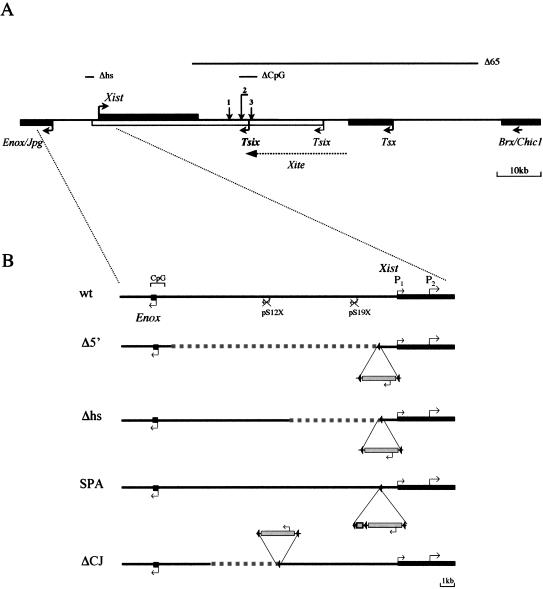
Recent progress towards understanding the molecular basis of choice in X chromosome inactivation has come through the discovery of a noncoding gene antisense to Xist, termed Tsix (Fig. 1A; Debrand et al. 1999; Lee et al. 1999; Mise et al. 1999). Deletion of the major Tsix promoter on one allele in XX cells was shown to result in complete nonrandom X inactivation of the targeted allele (Lee and Lu 1999). X chromosome counting was unaffected, however. Based on these findings, Lee and Lu (1999) suggested that Tsix mediates X chromosome choice by antagonizing Xist expression in cis. A number of studies have tested this idea further (see Fig. 1A). Introduction of a transcriptional stop signal, causing a truncation of Tsix RNA, recapitulated the extreme non-random X inactivation phenotype, suggesting that it is the Tsix transcript, rather than any associated DNA elements, that influences Xist expression (Luikenhuis et al. 2001; Sado et al. 2001). Conversely, nonrandom inactivation of the wild-type versus targeted allele was reported when either constitutive or inducible promoters were used to replace the endogenous Tsix allele (Luikenhuis et al. 2001; Stavropoulos et al. 2001). Thus Tsix must be down-regulated in order for accumulation of Xist RNA to occur in cis. A more recent study has demonstrated that deletion of elements mapping upstream of the major Tsix promoter results in partially skewed X inactivation (Ogawa and Lee 2003). It was suggested that these elements, termed Xite (X inactivation intergenic transcription elements), are involved in regulating the persistence of Tsix RNA in cis.
Figure 1.
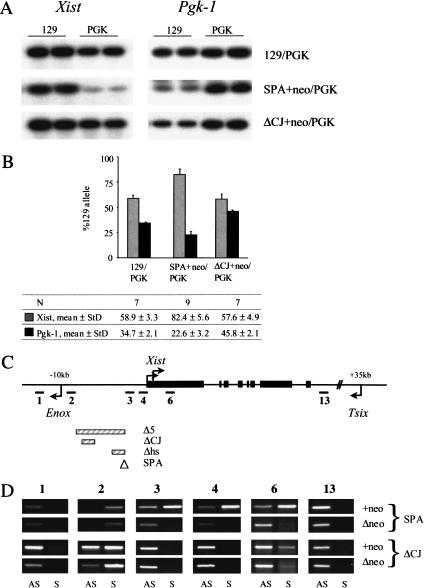
Targeted mutagenesis of the Xist region. (A) Schematic representation of the Xic between Enox/Jpg and Brx/Chic1 genes. Relative positions of Xist, Tsix, and Tsx, as well as Xite elements are shown. Transcriptional start sites and the direction of transcription are indicated by arrows. The Tsix major promoter is shown in bold. Lines above the schematic indicate the position of the targeted deletions: Δhs (Newall et al. 2001), Δ65 (Clerc and Avner 1998), and ΔCpG (Lee and Lu 1999). Vertical arrows show the integration positions of a transcriptional stop signal or inducible promoter (1; Luikenhuis et al. 2001), an IRESβgeo cassette, which blocks transcription from both Tsix promoters (2; Sado et al. 2001), and insertion of a constitutive promoter (3; Stavropoulos et al. 2001). (B) The wild-type (wt) locus and four different mutations in the Xist upstream region (Δ5′, Δhs, SPA, and ΔCJ) are presented. (wt) Schematic representation of the region from –10 kb to +2 kb relative to the Xist P1 start site. The relative position of two pseudogenes, pS12X and pS19X, as well as the position of the Enox CpG island are also shown. Deleted regions in Δ mutants are indicated by a broken line; the site of floxed PGKneo cassette integration is shown as insertion below or above the schematic with the direction of neo transcription indicated by arrows. SPA insertion is shown as a box.
Although it is clear that Tsix is a key regulator of X chromosome choice, the molecular mechanism of Tsix action is not well understood. Prior to X inactivation, Tsix expression is accompanied by low levels of unstable Xist RNA expression from all X chromosomes (Panning et al. 1997; Sheardown et al. 1997). Thus an interaction between sense and antisense RNAs, for example involving a double-stranded RNA (dsRNA) intermediate, has been proposed to be important (Maxfield Boumil and Lee 2001). Involvement of Xist sense transcription in choice is supported by the observation that insertion of a PGKneo cassette at a specific site upstream of Xist gives rise to preferential X inactivation of the targeted allele in XX heterozygotes (Newall et al. 2001). In the present study, we extended this observation through analysis of a series of targeted insertions and deletions between Xist and the closest 5′ flanking gene, Enox/Jpx (Chureau et al. 2002; Johnston et al. 2002). A number of these mutations were found to cause preferential X inactivation of the targeted allele in XX heterozygotes. In all of these cases, the targeted embryonic stem (ES) cell lines exhibited ectopic sense transcription initiating immediately upstream of Xist. Moreover, the level of sense transcription was seen to broadly correlate with the degree of skewing of X inactivation choice. Our results support the view that the likelihood that a given X chromosome will be selected as the active or inactive X is governed by the balance of sense and antisense transcription prior to the onset of X inactivation.
Results
Targeted mutagenesis of the Xist 5′ region affects X chromosome choice
To investigate the role of the Xist 5′ region in the regulation of X chromosome inactivation, we constructed a targeting vector designed to delete a sequence between 1 and 9 kb upstream of the Xist P1 start site (XistΔ5′, hereinafter referred to as Δ5′; Fig. 1B). This mutation was introduced into the 129/1 XY ES cell line, and correctly targeted clones were selected and verified by Southern blot hybridization and PCR analyses (data not shown). We also generated cell lines in which the PGKneo cassette was deleted by Cre-mediated recombination.
To analyze the effect of these mutations on X inactivation in vivo, targeted cell lines were injected into blastocysts, and chimeric animals were obtained for neo+ and Δneo mutants. Mutations at the Xist locus, transmitted via XY ES cells, can have a range of effects, depending on whether they affect counting, choice, imprinting, in cis inactivation, or combinations thereof. Ablation of the counting function is predicted to cause inactivation of the single X chromosome, and hence cell lethality, in the ES-cell-derived component of chimeric embryos (Penny et al. 1996; Clerc and Avner 1998). Such mutations should therefore not produce adult chimeras. Chimeric males transmitting mutations that ablate in cis inactivation have a daughterless phenotype due to failure of paternally imprinted X inactivation in the trophectoderm (Marahrens et al. 1997). Lee (2000) demonstrated that a mutation that affects the maternal imprint on Xist is transmitted to daughters, but that these animals are generally unable to transmit the mutant allele, either to male or female progeny. Finally, a mutation that only affects choice was transmitted stably and propagated through both male and female germlines (Newall et al. 2001). In the case of the Δ5′ and Δ5′+neo mutations, chimeras were obtained and were able to transmit the mutated X chromosome through the germline to produce females heterozygous for the mutation. The female heterozygotes were able to transmit the mutant X chromosome through either male or female germlines in subsequent generations, and the resultant progeny were normal and fertile. These observations suggest that counting, in cis inactivation, and Xist imprinting are not significantly affected by the mutations. However, they leave open the possibility that X inactivation choice is affected.
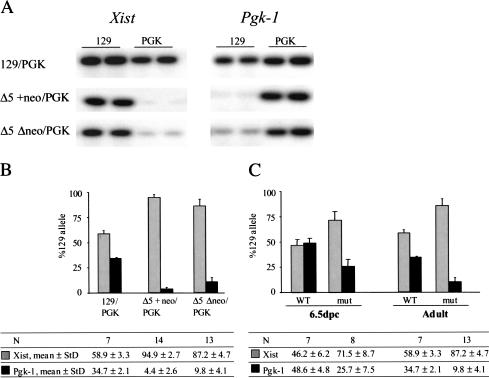
To determine whether the mutations skew X chromosome choice, we analyzed females heterozygous for the Δ5′ mutations using quantitative single nucleotide primer extension (SNuPE) assays. Xist and Pgk-1 alleles were distinguished by characterized polymorphisms between the 129 and PGK mouse strains (Singer-Sam et al. 1992; Penny et al. 1996). Representative examples are shown in Figure 2A. A quantification of the data for [Δ5′+neo × PGK]F1 mice indicates that the mutation causes highly skewed Xist expression, with the targeted allele being expressed in the majority of cells (Fig. 2B). Pgk-1 expression exhibited a reciprocal bias, confirming extreme nonrandom X inactivation. Importantly, the Δ5′Δneo heterozygotes also exhibited nonrandom X inactivation, albeit at a moderately reduced level (Fig. 2B).
Figure 2.
In vivo analysis of X inactivation randomness. (A) Representative examples of SNuPE data for the expression of polymorphic Xist and Pgk-1 alleles in wild-type control [129 × PGK]F1 female and heterozygous mutant animals. (B) Quantitation of Xist and Pgk-1 SNuPE data for control and mutant mice. The precise percentages as well as the number of animals analyzed are given in the table. (C) Quantitation of Xist and Pgk-1 SNuPE data for control [129 × PGK]F1 female and mutant [Δ5′Δneo × PGK]F1 female 6.5-dpc embryos. Data for adult mice of the same genotype are given for comparison.
To confirm that nonrandom X inactivation is attributable to skewed choice, and not to a cell selection event, we analyzed embryos immediately after the onset of random X inactivation, at 6.5 days postcoitum (dpc; Rastan 1982). We dissected embryos from matings between [129 × Δ5′Δneo]F1 females and PGK strain males. Extraembryonic regions were separated and used for genotyping. The embryo proper was analyzed for allele-specific Xist and Pgk-1 expression. We found that the level of Xist expression from the 129 (mutant) allele in [Δ5′Δneo × PGK]F1 embryos was significantly higher compared to [129 × PGK]F1 littermates (Fig. 2C). The level of Pgk-1 transcript from the targeted X chromosome was reciprocal to Xist. These results suggest that skewed X inactivation occurs at the level of choice rather than secondary selection events. The extent of skewing was slightly lower in embryos compared to adults. This is attributable to the contribution of visceral endoderm cells which undergo imprinted inactivation of the paternal (PGK strain) X chromosome. A similar result was obtained previously for the XistΔhs+neo (hereinafter referred to as Δhs+neo) mutation, which also gives rise to primary nonrandomness (Newall et al. 2001).
Mutagenesis of Xist 5′ region activates sense transcription upstream of Xist
To examine how the Δ5′ deletion affects general transcription around the Xist locus in undifferentiated ES cells, we performed strand-specific reverse transcriptase PCR (RT–PCR) analysis on Xist and adjacent upstream and downstream sequences (Fig. 3A). Control samples showed the expected patterns of transcription, that is, Xist sense transcription (amplicon 6) in ES cells and XX somatic cells, and antisense Tsix transcription (amplicons 3, 4, 6, and 13) in ES cells only (Fig. 3B).
Figure 3.
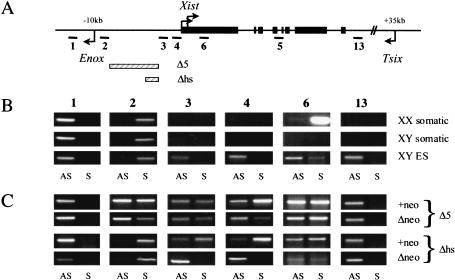
In vitro analysis of transcriptional activity at Xist and adjacent regions. (A) Schematic representation of the Xist locus; the regions analyzed by RT–PCR (1–4, 6, 13), and the positions of Tsix (+35 kb) and Enox (–10 kb) promoters relative to the Xist P1 start site are shown. (B,C) Strand-specific RT–PCR across Xist and its adjacent regions in control samples and targeted cell lines. The numbers above correspond to the amplicons shown in the schematic in A. cDNA made from undifferentiated XY ES cells and XX and XY somatic cells was used as a template for amplification. (AS) Antisense transcript; (S) sense transcript. Gray bars show relative size and position of the targeted deletions.
Analysis of Δ5′+neo undifferentiated ES cells revealed that Tsix expression was not affected by the deletion. Interestingly, ectopic sense (Xist) transcription was detected upstream of Xist (amplicons 3 and 4), and transcription within the Xist locus was seen to be elevated (amplicon 6; Fig. 3C). Importantly, ectopic sense transcription, albeit at an apparently lower level (see also below), was observed for the Δneo mutants.
Upstream sense transcription in the Δ5′+neo mutation is most likely attributable to insertion of the PGK promoter, as the same effect was observed with the Δhs+neo mutation, and the PGKneo cassette was inserted in an identical position (Newall et al. 2001). Previously we suggested that this was due to activation of an endogenous cryptic promoter at position –550 bp. However with the Δ5′+neo allele, we detected ectopic sense transcription immediately upstream of this point and adjacent to the PGK promoter (amplicon 3, Fig. 3C). We confirmed that sense transcription at amplicon 3 also occurs in the Δhs+neo mutation (Fig. 3C). These results suggest that upstream sense transcription is in fact resulting from bidirectional activity of the PGK promoter. Importantly, in the Δ5′Δneo mutant the PGKneo cassette has been deleted, and therefore sense transcription upstream of Xist must have a different origin.
Bidirectional activity of the Enox promoter accounts for the Δ5′Δneo phenotype
To find the basis for ectopic sense transcription in Δ5′Δneo XY ES cells, we further analyzed the Xist 5′ region. A prominent CpG island associated with the Enox gene, 10 kb upstream of Xist (Johnston et al. 2002), is brought into close proximity to Xist by the Δ5′ deletion, and we suspected that this could be the site of origin of sense transcription. Since the Enox gene is transcribed in the opposite direction to Xist (Johnston et al. 2002), ectopic sense transcripts should result from bidirectional activity of the Enox CpG island. To test this we carried out strand-specific analysis using amplicons 1 and 2, located on either side of the CpG island (Fig. 3). As expected, Enox transcription (amplicon 1) was detectable in all control cells (Fig. 3B). Importantly though, transcripts were also detected antisense to Enox (and therefore sense with Xist) using amplicon 2 (Fig. 3B).
Bearing in mind this pattern of expression in control samples, we proceeded with the analysis of the Δ5′ mutant cell lines. The pattern of Enox expression was not affected in either mutant (amplicon 1); however, transcription from both strands was detected for amplicon 2 (Fig. 3C). Thus, as predicted, transcription in the Xist sense orientation, arising from the bidirectional activity of the Enox CpG island, accounts for ectopic sense transcription across Xist. The transcription antisense to Xist, which is detectable in Δ5′+neo and Δ5′Δneo, presumably arises from run-through transcription from the PGKneo cassette and/or Tsix. We also analyzed amplicons 1 and 2 in the previously characterized Δhs+neo and ΔhsΔneo mutants. Here the antisense band for amplicon 2 was seen to be very weak (Fig. 3C). Thus sequences deleted in Δ5′ normally function to terminate transcription between Xist and Enox in both directions.
In summary, these results indicate that the Enox CpG island has bidirectional promoter activity, and that the Δ5′ deletion has removed sequence that normally insulates Xist from transcriptional interference.
Levels of ectopic sense transcripts correlate with the degree ofX inactivation skewing
Our initial PCR analyses indicated that the level of ectopic transcripts may be lower in Δ5′Δneo compared to Δ5′+neo ES cells. As the Δ5′Δneo mutation also results in a reduced level of X inactivation skewing in heterozygous female mice compared to Δ5′+neo (Fig. 2B), we went on to investigate this further. To determine the levels of Xist and Tsix RNA, we carried out quantitative real-time RT–PCR in both wild-type and mutant XY ES cell lines. For Xist we used primers that span exons IV and V of the gene in order to exclude any contribution from antisense Tsix RNA. To quantitate Tsix RNA, we used primer pair 13 located downstream of the Xist transcription unit (Fig. 3A). A β-actin amplimer provided a control.
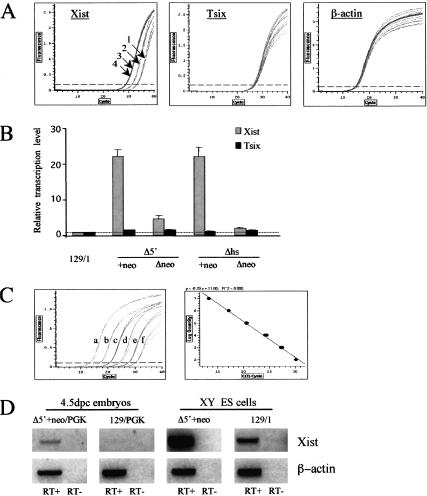
Results from a typical experiment are illustrated in Figure 4A. Average values obtained from independent experiments are summarized in Figure 4B. Xist RNA levels assayed at the exon IV/V boundary were clearly elevated in mutant ES cell lines in which ectopic upstream transcripts were detected (Δhs+neo, Δ5′Δneo, and Δ5′+neo). Moreover, the level of Xist RNA correlates with the extent of X inactivation skewing seen in female animals heterozygous for the corresponding mutation. Thus Δ5′+neo and Δhs+neo mutations (extreme skewing; Fig. 2B; Newall et al. 2001) have the highest levels of Xist RNA (20–25 times above wild type), the Δ5′Δneo mutation (moderate skewing, Fig. 2B) has an intermediate level of RNA (4–5 times above wild type), and ΔhsΔneo (no skewing; Newall et al. 2001) is indistinguishable from wild-type cells. Levels of Tsix RNA, in contrast, were similar in all cell lines.
Figure 4.
Quantitative analysis of ectopic Xist transcription. (A,B) Strand-specific real-time RT–PCR on mutant (Δ5′+neo, Δ5′Δneo, Δhs+neo, ΔhsΔneo) and control (129/1) XY ES cell lines. (A) Representative examples of Xist (amplicon 5 in Fig. 3A), Tsix (amplicon 13 in Fig. 3A), and β-actin amplification plots are shown. Note that Tsix and β-actin amplicons have similar CT values for all cell lines analyzed, whereas the CT values for Xist amplicons vary significantly between 129/1 (1), Δ5′Δneo (2), Δ5′+neo (3), and Δhs+neo (4) ES cell lines. (B) Relative transcription levels in control and mutant ES cell lines are shown for Xist and Tsix amplicons. (C,D) RT–PCR analysis of ectopic Xist transcription in 4.5-dpc heterozygous embryos. β-actin was used as an internal standard to normalize the cDNA concentration between different embryonic samples by real-time PCR. (C) An example of amplification plots (left) and a standard curve of CT value vs. molecule number (right) for a serial dilution of β-actin plasmid DNA are shown. Amplification curves a–f correspond to the 10× serial dilution of plasmid DNA from 107 to 102 molecules/reaction, respectively. (D) Strand-specific RT–PCR on Xist upstream region (amplicon 4 in Fig. 3A) in [Δ5′ + neo × PGK]F1 and [129 × PGK]F1 4.5-dpc embryos. XY ES cell samples are shown for comparison. The SYBR green-stained gel image is shown as a negative for better contrast.
Our results indicate that elevated levels of sense transcription across Xist at the onset of X inactivation could increase the probability of that allele being chosen as the inactive X chromosome in heterozygous females. This assumes that the data obtained from analysis of XY ES cells can be extrapolated to a physiological context. To test this we assayed ectopic sense transcription upstream of Xist in Δ5′+neo and wild-type embryos. Random X inactivation occurs in epiblast cells between 5.5 and 6 dpc (Rastan 1982), and we therefore analyzed embryos immediately prior to this, at 4.5 dpc. The results are shown in Figure 4C and D. Upstream sense transcripts were clearly detectable in mutant embryos (Fig. 4D, left panel). A weak band was also detected for wild-type embryos (Fig. 4D, second panel from left). This finding has been reported previously (Debrand et al. 1999), and is attributable to the increased number of PCR cycles used. Consistent with this, under these conditions we could detect upstream sense transcripts in 129/1 XY ES cells, albeit at a lower level than in the mutant XY ES cells (Fig. 4D, left two panels). In summary, these results show that elevated sense transcription occurs in mutant embryos in vivo, and therefore support the idea that this could underlie skewed X inactivation in heterozygous females.
RNA FISH analysis of the Δ5′ mutations in undifferentiated ES cells
We established by strand-specific RT–PCR analysis that the Δ5′ mutations lead to elevated levels of Xist expression in undifferentiated XY ES cells. To determine how this is manifested on the single-cell level, we carried out an RNA fluorescence in situ hybridization (FISH) analysis on control and mutant cells. In undifferentiated ES cells, nascent Xist and Tsix RNA can be detected as a pinpoint signal. These transcripts are produced from the single Xist allele in XY ES cells (Fig. 5A), and from both alleles in XX ES cells (data not shown). This contrasts with XX somatic cells, where accumulation of stable Xist RNA over the inactive X chromosome is seen as a single large compact domain (Fig. 5B). Accumulation of Xist RNA is observed only very rarely in wild-type XY ES cells (Fig. 5G).
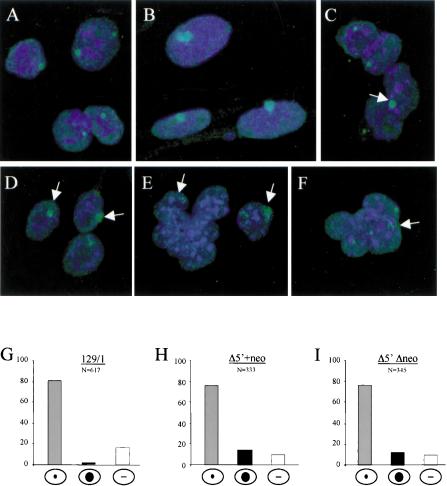
Figure 5.
FISH analysis of Δ5′ undifferentiated ES cells. RNA FISH in normal undifferentiated XY ES cells (A), normal female fibroblasts (B), and Δ5′+neo XY ES cells (C–F) using a double-stranded probe for Xist exon 1. Xist signal is either compact and similar to female somatic signal (arrow; C,D) or more dispersed and scattered around the site of transcription (arrow; E,F) in a portion of cells. Quantitation of FISH data (G–I) shows the percentage of cells with punctate signal, accumulated Xist signal, or no signal.
In the majority of Δ5′ mutant XY ES cells, both neo+ and Δneo, we observed a single pinpoint signal, as seen in control XY ES cells. However, in 10%–15% of the cells, we observed accumulation of Xist RNA (Fig. 5C–I). In a small number of the cells, the accumulated signal was compact and appeared similar to that seen in XX somatic cells (Fig. 5C,D). More typically the signal was weaker and relatively diffuse (Fig. 5E,F). No noticeable difference was observed between Δ5′Δneo and Δ5′+neo, either in the frequency of cells showing this pattern, or qualitatively in the appearance of the Xist signal. In light of the quantitative analysis showing a marked difference between these mutants (Fig. 4B), atypical Xist localization patterns do not appear to simply reflect increased Xist RNA levels. Indeed, similar findings were reported previously for the ΔhsΔneo mutation (Newall et al. 2001), and also for XY ES cells in which antisense Tsix transcription was ablated by insertion of a terminator sequence immediately downstream of the promoters (Sado et al. 2001; see Discussion).
Analysis of ectopic sense transcription and nonrandom X inactivation in XistSPA and XistΔCJ mutants
To substantiate the data obtained for Δ5′ and Δhs deletions, we analyzed two other targeted mutations; the first is an insertion of a transcriptional terminator site, SPA-MAZ4 (Levitt et al. 1989; Ashfield et al. 1994), in the region –1.1 kb upstream of the Xist P1 start site, with the PGKneo cassette in the same position as in Δ5′+neo and Δhs+neo mutants. No endogenous sequence was deleted in this mutation, but it was designed to disrupt any transcription between the Xist promoter and upstream regions (XistSPA, hereinafter referred to as SPA; Fig. 1). The second targeted mutation was a deletion of a 2.5-kb region between 5.5 and 8 kb upstream, spanning a site which previously was thought to include an upstream promoter for the Xist gene (Johnston et al. 1998). This deletion also removes the pS12X pseudogene (XistΔCJ, hereinafter referred to as ΔCJ; Fig. 1B).
SNuPE analysis of Xist and Pgk-1 allele-specific expression demonstrates nonrandom inactivation of the targeted allele for [SPA+neo × PGK]F1 heterozygous females (Fig. 6A,B). However, in [ΔCJ+neo × PGK]F1 females, random X inactivation was observed. A slight skewing from 50:50 found in this experiment is caused by Xce allelic differences between 129 and PGK strains, and is similar to the skewing observed for the control [129 × PGK]F1 littermates (Fig. 6B). These data demonstrate that the deletion of the 5.5–8-kb upstream region does not affect X inactivation choice. Moreover, the results from these two mutations show that the effect of PGKneo cassette on the randomness of X inactivation is position-dependent and does not interfere with Xist regulation when inserted 5.5 kb upstream.
Figure 6.
In vivo and in vitro analyses of SPA and ΔCJ mutants. (A) Representative examples of SNuPE data for the expression of polymorphic Xist and Pgk-1 alleles in [129 × PGK]F1 female controls and SPA and ΔCJ heterozygous mutants. (B) Quantitation of Xist and Pgk-1 SNuPE data for control and mutant mice. The precise percentages as well as the number of animals analyzed are given in the table. (C,D) Schematic representation of the Xist locus (C) and strand-specific RT–PCR across Xist and its adjacent regions in targeted cell lines (D). Gray bars show relative size and position of the targeted deletions. Triangle in C indicates the site of SPA-MAZ4 insertion (see Fig. 3 for more details).
A detailed strand-specific RT–PCR analysis was undertaken next for SPA and ΔCJ mutant XY ES cell lines, both +neo and Δneo. No significant differences from the control XY ES cells were detected for amplicons 1, 3, 4, 6, and 13 for ΔCJ+neo, ΔCJΔneo, and SPAΔneo cell lines (Fig. 6C). However, the SPA+neo has ectopic sense transcription upstream of Xist, similar to the Δ5′+neo, Δhs+neo, and Δ5′Δneo ES cell lines. Thus, again, only alleles with ectopic sense transcription are seen to undergo preferential X inactivation in heterozygous females. No antisense transcription was detected for amplicon 2 in wild-type cells (Fig. 3B), but strong bands were detected for this amplicon in ΔCJ mutants. This latter result indicates that the 5.5–8-kb upstream region is important for terminating transcription antisense to Xist.
Discussion
In this study we investigated the function of Xist upstream sequences in the regulation of X chromosome inactivation. Analysis of a series of targeted mutations, both in the presence and absence of associated PGKneo selection cassettes, demonstrated that certain mutations result in preferential X inactivation of the targeted X chromosome in heterozygous female mice. Analysis of transcription of the mutant alleles in undifferentiated XY ES cells demonstrated elevated sense transcription across Xist, arising from inserted or juxtaposed upstream promoters, exclusively in mutations associated with nonrandom X inactivation. In the Δ5′Δneo mutation, elevated sense transcription was attributable to deletion of sequences that normally function to terminate sense transcripts arising from bidirectional activity of the promoter of the Enox gene, located 10 kb upstream of Xist. Quantitative analysis indicates that the level of ectopic sense transcription determines the degree of X inactivation skewing. Thus as proposed previously (Maxfield Boumil and Lee 2001), our data indicate that sense and antisense transcripts interact to set the probability of a given allele being chosen as the active or inactive X chromosome.
Effects of targeted mutagenesis upstream of Xist on X chromosome choice
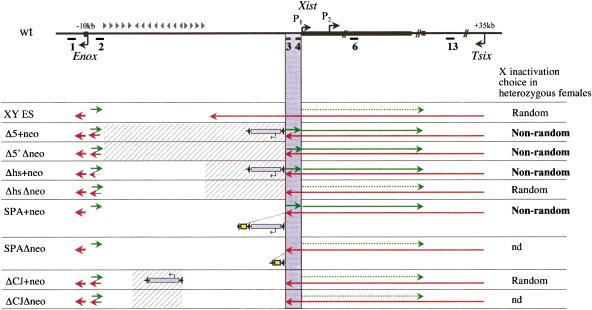
Figure 7 summarizes our observations of transcriptional activity of the Xist locus in undifferentiated XY ES cell lines carrying different targeted mutations. Four of the mutant alleles (SPA+neo, Δ5′+neo, Δ5′Δneo, and Δhs+neo) give rise to preferential X inactivation in heterozygous XX females. In XY ES cells, only these alleles have elevated sense transcription across the locus as a consequence of upstream promoter activity. This strong correlation suggests that nonrandom X inactivation may be directly attributable to the elevated sense transcription.
Figure 7.
Sense transcription across the Xist promoter modulates X inactivation choice. Summary of strand-specific transcriptional activity across the Xist locus in control and mutant cell lines. A schematic of the region analyzed and the relative positions of Xist, Tsix, and Enox promoters is shown above. Cross-hatched areas correspond to the regions deleted in the different mutations. The position of floxed PGKneo cassette integration and the direction of neo transcription in each mutant are shown (shaded rectangle). Yellow rectangles represent SPA termination sites. Green arrows show sense transcription and red arrows antisense. The thickness of the arrows reflects the transcriptional activity. Shaded area highlights the region where ectopic Xist transcription was detected. Gray triangles above the schematic show the position and directionality of the putative insulator/terminator elements upstream of Xist. The choice status of X inactivation in heterozygous females is shown alongside each genotype.
In support of a direct role for elevated sense transcription, the greatest degree of skewing observed occurs with the Δ5′+neo mutation (Fig. 2B). Here, the combined proximity of the Enox CpG island and the PGKneo promoter may be expected to act in combination to enhance levels of sense transcription. Indeed, sense transcription is clearly higher in Δ5′+neo (and also in Δhs+ neo, which in a previous study was shown to cause a high degree of skewing; Newall et al. 2001), compared to Δ5′ Δneo XY ES cells (Fig. 3C). This correlation is not absolute, as the SPA mutation results in an intermediate skewing level (Fig. 6B), but has elevated Xist RNA levels similar to Δ5′+neo (data not shown). This could be attributable to an added effect resulting from insertion of the SPA-MAZ4 terminator sequence. Binding of the MAZ factor has been shown to cause DNA bending (Ashfield et al. 1994), and such an effect could be antagonistic to the increased sense transcription.
An alternative explanation for the observed effects on X chromosome choice is that the mutations have interfered with transcription and/or processing of Tsix transcripts. A number of observations argue against this possibility. Antisense transcription close to the Tsix promoter and across the Xist locus is apparently unaffected in mutant XY ES cells (Figs. 3, Fig. 6D, amplicons 3, 4, 6, and 13), and this is supported by quantitative analysis (Fig. 4). The fact that the Δhs mutation does not cause skewing when PGKneo is removed demonstrates that deletion of specific regions at the 3′ end of Tsix does not correlate with skewed inactivation. Finally, incorrect termination of Tsix transcription does not appear to be a factor, as the ΔCJ mutations allow Tsix transcripts to run through at least as far as the Enox promoter (see below), and this again does not result in skewed X inactivation.
Dissection of regulatory elements upstream of Xist
The recently discovered Enox gene lies in close proximity 10 kb upstream of Xist. Enox is subject to X inactivation, albeit with low-level partial escape (Johnston et al. 2002), raising the question of whether these oppositely regulated loci are separated by specific insulatory sequences. Clerc and Avner (1998) demonstrated that deletion of intervening sequences between the 3′ end of Xist and the downstream gene, Brx, does not affect X inactivation of Brx, indicating that specific boundaries between the Xist 3′ end and X inactivated loci are not required. In contrast to this, our results demonstrate that deletion of upstream sequences results in transcription from the bidirectional Enox promoter interfering with Xist regulation. We have also found that Xist/Tsix transcription interferes with normal Enox regulation in the Δ5′ mutations, with an increase in escape from inactivation being apparent specifically on the Δ5′Δneo allele (data not shown). Thus, the sequences deleted in Δ5′ function as a bidirectional insulator between the Xist and Enox loci.
Comparison of expression from the different mutant alleles in XY ES cells allows us to define these insulatory sequences more precisely. Previously we reported that the Δhs deletion in itself does not affect transcription in the upstream region (Newall et al. 2001). The results presented here further demonstrate, in light of the discovery of bidirectional Enox promoter activity, that sequences deleted in Δhs are not required to terminate upstream sense transcripts. However we did observe low levels of antisense transcription at amplicon 2 in Δhs cell lines (Fig. 3C), suggesting that the region does contribute in part to terminating Tsix RNA. The effect however is far less apparent than that seen with the ΔCJ mutations. Here strong antisense transcription is seen with amplicon 2, suggesting that Tsix transcripts have not been correctly terminated. This was also apparent with the Δ5′ mutations which cover the same region. Thus the region 5.5–8 kb upstream of Xist appears to function in terminating Tsix transcription.
As with Δhs, the ΔCJ deletion does not affect termination of sense transcripts arising from bidirectional Enox promoter activity. The sequences responsible for this activity must therefore lie either between the 5′ ends of the ΔCJ and Δ5′ deletions (8–9 kb upstream of Xist), or alternatively between the 5′ end of the Δhs deletion and the 3′ end of the ΔCJ deletion (3.6–5.5 kb upstream of Xist).
Taken together our results highlight the importance of sequences that buffer juxtaposed genes from transcriptional interference. Assuming that bidirectional activity from CpG island promoters is relatively widespread, then sequences with similar functions will probably have been selected for at many sites throughout the genome. In this context it is interesting to note that the region upstream of Xist has a high density of inter-spersed repetitive elements and pseudogenes. This is also true in other species, although the specific mobile elements involved are often different (Nesterova et al. 2001; Chureau et al. 2002). Thus insertion of mobile elements may be selected for at specific sites to facilitate the buffering of transcriptional units from one another.
Implications for understanding X chromosome choice
Extreme nonrandom X inactivation has been reported to occur as a consequence of deleting the major promoter for the Tsix gene (Lee and Lu 1999), and also as a result of premature termination of Tsix transcripts (Luikenhuis et al. 2001; Sado et al. 2001). Similar to the results reported here, the chromosome bearing the targeted allele is preferentially inactivated in heterozygous females (or XX ES cells). Furthermore, premature termination of Tsix was reported to result in partial Xist RNA accumulation in a proportion of XY ES cells (Sado et al. 2001), again mirroring the phenotype observed in the present study. These similarities raise the intriguing possibility that X chromosome choice is modulated by the balance between sense and antisense Xist transcription at the onset of random X inactivation. Thus we can envisage that there is a competition between agonistic (sense) and antagonistic (antisense) transcription at each allele. Any advantage offered to one allele over the other would skew the probability of that X chromosome up-regulating Xist and being inactivated. Other observations on Tsix function are consistent with this view. An X chromosome on which the Tsix promoter has been replaced with a heterologous strong constitutive promoter is always chosen as the active X (Luikenhuis et al. 2001). Ogawa and Lee (2003) described intermediate skewing of X inactivation occurring as a result of a deletion upstream of the major Tsix promoter. Although this effect was attributed to decreased persistence of Tsix RNA following the onset of random X inactivation, it is interesting to note that a small reduction in relative levels of antisense transcription from the mutant allele was seen in heterozygous ES cells prior to X inactivation.
Although loss of Tsix transcription and increased sense Xist transcription have very similar effects on random X inactivation, Tsix mutations also affect imprinted X inactivation when inherited on Xm (Lee 2000). Additionally, Tsix homozygotes (obtained using rare animals that survive inheritance of the mutation on Xm) show effects in random X inactivation resulting in sex ratio distortion (Lee 2002). None of our mutations, including the Δ5′+neo which causes near complete nonrandom X inactivation, showed a parent of origin effect, and females homozygous for all of the mutations are normal, fertile, and show no sex ratio distortion in their progeny (N. Brockdorff, T.B. Nesterova, A.E.T. Newall, C.M. Johnston, and R. Appanah, unpubl.). Thus we can conclude that in imprinted X inactivation, increasing sense transcription is not equivalent to deleting Tsix.
What are the possible mechanisms whereby the balance of Xist and Tsix transcription could influence X chromosome choice? One idea is that Xist and Tsix form a dsRNA that triggers an RNA interference (RNAi) response that represses Xist transcription in cis. Transcriptional gene silencing has been linked to RNAi in plants (Hamilton and Baulcombe 1999; Mette et al. 2000), and recent studies have demonstrated that RNAi can induce silent chromatin in cis at centromeres (Reinhart and Bartel 2002; Volpe et al. 2002). A second possibility is that Tsix transcripts directly interfere with PolII recruitment at the Xist promoters. Increasing sense transcription would be expected to antagonize this effect. Finally, sense transcription could serve to potentiate the locus, for example by recruiting chromatin-modifying enzymes such as HATs, similar to the proposed function of intergenic transcripts in the β-globin locus (Gribnau et al. 2000). Tsix RNA would counteract this by interfering with PolII loading at the Xist promoter. These possibilities are not mutually exclusive. Experiments that differentiate between these models will be a priority for future studies.
In summary, the results of this study indicate that the balance of sense and antisense RNAs at the Xist locus act as a molecular rheostat regulating X chromosome choice. Fine tuning of the probability of a given allele being activated can occur as a result of relatively subtle changes in the level of sense, and possibly antisense RNAs. This mechanism may have parallels in other circumstances involving developmentally regulated mono-allelic gene expression. Most notably, sense and antisense RNAs have been described at a number of loci subject to genomic imprinting.
Materials and methods
Construction of targeting vectors
129Ola genomic sequences for vector construction were isolated from a λDASHII genomic library. For the Δ5′+neo vector, the 3′ homology region (3′HR, see Table 1) was inserted into EcoRV/HindIII-digested pBluescript SK+, carrying a PGKneo cassette flanked by loxP sites, giving the plasmid pL2NB3′. 3′HR was obtained by cloning of a 1.1-kb SacII-digested PCR fragment (primers Sx48 and MX32b, Table 2) with a 2.4-kb SacII/EcoR1 fragment from Xist exon 1. An XhoI/SalI fragment of the diptheria-toxin A (DT-A) gene, used for negative selection of non-homologous recombinants (Yagi et al. 1990), was inserted into SalI-digested pL2NB3′ at the 3′ end of 3′HR. A plasmid (pDT14) was selected, with the direction of DT-A transcription colinear with Xist. NotI/HpaI 5′HR (see Table 1) was isolated from cloned phage DNA and ligated into NotI/SmaI-digested pDT14 vector, yielding the final construct Δ5′+neo.
Table 1.
Summary of the targeting experiments
| Δ5′ | SPA | ΔCJ | |
|---|---|---|---|
| 5′HR | 8.4 kb (-17.6 kb - 9.22 kb) | 4.62 kb (-5.72 kb - 1.1 kb) | 7 kb (-15.31 kb - 8.232 kb) |
| 3′HR | 3.48 kb (-1.1 kb + 2.38 kb) | 3.48 kb (-1.1 kb + 2.38 kb) | 5.683 kb (-5.717 kb - 0.034 kb) |
| Mutation | Deletion of 8.12 kb | Insertion of SPAtetraMAZ sequence | Deletion of 2.5 kb |
| Targeted clones | 5 | 5 | 6 |
| % homologous recombinants | 0.5% | 1% | 6.7% |
| Clones with neo Credeletion | 8 in 380 | 6 in 550 | 1 in 80 |
Table 2.
Primers and PCR conditions for genotyping and strand-specific RT–PCR
| Mutation/region | Primer, sense | Primer, antisense | PCR conditions |
|---|---|---|---|
| Δ5′ | CJx37, GCTGCCAGCATTGCTGTAGC | TNTR1, AATTGGTTTGCTTATGGACGAT | 94°C, 30 sec; 54°C, 30 sec; 72°C, 30 sec for 35 cycles |
| neo | NeoTN2, TCCCCTCAGAAGAACTCGTCAA | NeoTN5, TCCTGCCGAGAAAGTATCCATCAT | 94°C, 30 sec; 60°C, 30 sec; 72°C, 30 sec for 33 cycles |
| wt | Sx53, TCTCTTTGTACAGCTCATTTAG | TNTR1, AATTGGTTTGCTTATGGACGAT | 94°C, 30 sec; 54°C, 30 sec; 72°C, 30 sec for 33 cycles |
| Amp 2 | EST58, ATAGAAGACCAGCTTGTGTTTTG | EST59, GTGTAGTTCTCTGCCTTTATTGA | 94°C, 30 sec; 58°C, 30 sec; 72°C, 30 sec for 35 cycles |
| Amp 3 | Sx48, GTAACCTTTACTTCTCAATTCAG | TNTR1, AATTGGTTTGCTTATGGACGAT | 94°C, 30 sec; 59°C, 30 sec; 72°C, 30 sec for 35 cycles |
| Amp 5 | Qmex4, GCAAGGAAGACAAAGGCTCAAAGAAT | Qmex52 GGAGAGAGAACCAAATAGAGCAGAAT | See Materials and Methods |
| Amp 13 | T13s, TATACAATACGCATCACACAAAAAG | T13as, TACCACTGAGCCACATCCTTAC | 94°C, 30 sec; 60°C, 30 sec; 72°C, 30 sec for 35 cycles |
| Part of 3′ HR, Δ5′ | Sx48, GTAACCTTTACTTCTCAATTCAG | Mx32b, TAAGCCATAAGGCTTGGTGG | 94°C, 30 sec; 58°C, 30 sec; 72°C, 30 sec for 30 cycles |
| Part of 3′HR, SPA | SX43, GTGAGTACACTGTCGGTCACTGTT | TT2, TTCCTGCAGGTCTAACTTTAGTTGACAGAGC | 94°C, 30 sec; 54°C, 30 sec; 72°C, 1 min for 30 cycles |
| β-actin | BA11, GATATCGCTGCGCTGGTCGT | BA2, AGATCTTCTCCATGTCGTCC | See Materials and Methods |
To create the SPA+neo targeting vector, the SPA-MAZ4 transcriptional terminator [a gift from Dr. N. Proudfoot (Sir William Dunn School of Pathology, University of Oxford, Oxford, UK); Levitt et al. 1989; Ashfield et al. 1994] was directionally cloned into EcoRV/XhoI sites of a plasmid, immediately downstream of a single loxP site, yielding pLoxP-SPA. 5′HR (see Table 1) was obtained by cloning an Xist λDASHII-derived NotI/EcoRV fragment into a NotI/EcoRV-digested vector, carrying an EcoRV/SbfI PCR fragment (primers Sx43 and TT2, engineered SbfI site at the 5′end), giving plasmid pTNT5′. The NotI/SbfI fragment was excised from pTNT5′ and inserted into NotI/PstI-digested pLoxP-SPA, giving pTNT5′SPA. The final construct, SPA+neo, was obtained by cloning the XhoI(blunt)/NotI fragment from pTNT5′SPA into NotI/SmaI-digested DT14.
The ΔCJ+neo targeting vector was constructed by cloning 3′HR and 5′HR from genomic phage λDASHII clones into pL2NB3′. The λ phage clone containing 5′HR was cut with ApaI, blunt-ended, and then digested with BamHI. The released 7-kb fragment (see Table 1) was cloned into SmaI/BamHI-cut pL2NB3′, giving CJT3. Another genomic clone containing 3′HR was digested with NotI, blunt-ended, and cut with ClaI. Clone CJT4 was obtained by cloning of a 5.7-kb fragment into EcoRV/ClaI-cut CJT3. Finally, the XhoI/SalI fragment of the DT-A gene was inserted into SalI-digested CJT4, yielding the targeting vector ΔCJ+neo.
Generation of targeted ES cells and mutant mice
For generation of targeted ES cells and mutant mice, 129/1 XY ES cells (Kay et al. 1993) were grown as described (Penny et al. 1996). Homologous recombination was essentially as described by Newall et al. (2001). The number of colonies screened and the efficiency of targeting for each construct are shown in Table 1. The PGKneo cassette was removed by Cre-induced recombination as described (Newall et al. 2001).
All targeted ES clones, selected by primary Southern analysis, were grown and characterized extensively by Southern blot analysis using DNA probes for both 3′ and 5′ ends. Genomic PCR analysis with primers across the deletion/mutation was performed to verify that the clones are correct (data not shown). Finally, each clone chosen for blastocyst injection was characterized cytogenetically to ensure karyotypic integrity.
Male chimeras were mated to C57BL/6 females, and F1 daughters which received the mutated allele were used for further in vivo analysis and for establishing colonies. Mice were genotyped by PCR analysis of tail-tip genomic DNA. Specific primers and PCR conditions used for each mutant type are summarized in Table 2. For embryo analysis, timed matings were set up between Δ5′Δneo heterozygous females and PGK males. Day of plug detection was designated as 0.5 dpc. Embryos were dissected at 6.5 dpc. Extraembryonic and embryonic ectoderm regions were separated. The former were used for DNA genotyping, and the latter for RNA extraction and SNuPE. The sex of the embryos was determined by PCR using primers that give distinct bands for the Ube1X and Ube1Y genes (Chuma and Nakatsuji 2001).
SNuPE analysis
Females heterozygous for the targeted mutation were mated with PGK males, and the genotyped progeny were used for quantitative analysis. DNA and RNA were isolated from kidneys of adult females as described (Penny et al. 1996). Allele-specific expression of Xist and Pgk-1 genes was analyzed, using polymorphisms between laboratory-strain mice and PGK strain. PCR and SNuPE conditions were as described (Singer-Sam et al. 1992; Newall et al. 2001).
RT–PCR and real-time PCR analyses
RNA was isolated from ES cells as described (Penny et al. 1996). cDNA synthesis was primed from random hexamers (Pharmacia) with Superscript II reverse transcriptase (GIBCO-BRL) according to the manufacturer's instructions. Strand-specific RT–PCR was performed according to the method described (Newall et al. 2001). Xist/Tsix primer pairs 4 and 6 were as described (Lee et al. 1999). Primers T13s and T13as were from the same region as amplicon 13 (Lee et al. 1999), but give cleaner product in our hands. Enox primers for amplicon 1 were EP31 and EP2 (Johnston et al. 2002). Primer pairs and PCR conditions for amplicons 2 and 3 are given in Table 2.
Real-time PCR analysis of gene expression was performed on strand-specific cDNA templates. Concentration of total RNA for cDNA synthesis was accurately quantified using RiboGreen RNA Quantitation Kit (Molecular Probes), according to the manufacturer's instructions. One microgram of RNA was used as a template for cDNA synthesis with a mixture of strand-specific primers for Xist (amplicon 5, antisense primer VMex5, GGCATGAGTAGGGTAGCAGT), Tsix (amplicon 13, sense primer T13s, see Table 2), and β-actin (BAss, ACACGCAGCT CATTGTAG; Shibata and Lee 2003) as described (Newall et al. 2001). Real-time PCR was performed with the SYBR Green PCR Master Mix (Applied Biosystems) on a DNA Engine Opticon System (MJ Research) using the following conditions: 95°C, 10 min; (95°C, 15 sec; 61°C, 15 sec; 72°C, 1 min) × 40 cycles. PCR primers are listed in Table 2. A melting curve test was performed at the end of each experiment to ensure the specificity of amplification, and the products were also resolved by gel electrophoresis. The comparative CT method was used for relative quantitation of gene expression (User Bulletin #2, Applied Biosystems, 2001). Each amplicon was analyzed at least three times in triplicate with independent cDNA preparations.
For the expression analysis of 4.5-dpc embryos, β-actin was used as the internal normalization control. Plasmid DNA, containing exons 1–4 of mouse β-actin cDNA, was serially diluted 10-fold times from 107 to 102 molecules/reaction, and used to create a standard curve. The concentration of β-actin cDNA in each sample was estimated by comparison with the standard curve, and normalized accordingly.
Analysis of ectopic sense transcripts in vivo
Late blastocysts at 4.5 dpc were obtained by flushing uteri from crosses between homozygous females carrying either mutant (Δ5′+neo) or 129/1 alleles with PGK males. Pools of 50–60 blastocysts were collected, washed in 2 drops of PBS, transferred into thin-wall 0.2-mL PCR tubes in a small volume of PBS, and collected at the bottom of the tubes by centrifugation at 1300g for 1 min. Embryos were then snap-frozen in 5μL of PBS and kept at –20°C until use.
A cell-to-cDNA kit (Ambion) was used to prepare RNA for cDNA synthesis. All procedures were in accordance with the manufacturer's instructions, except for the volume of the lysis solution, which was reduced twice to avoid sample dilution. The lysates were treated with 2 U of RNase-free DNase for 30 min at 37°C. DNase was subsequently removed by incubation with 5 μL of DNase removal reagent (Ambion) for 3 min at room temperature. Half of the volume of extracted RNA was used for strand-specific cDNA synthesis according to the conditions described above. The cDNA concentration in different embryo preparations was normalized to the β-actin internal control (see above), and strand-specific RT–PCR was performed for Xist upstream region. Each PCR reaction used cDNA from the equivalent of two to three embryos. The PCR conditions were as follows: 94°C, 5 min; (94°C, 30 sec; 52°C, 30 sec; 72°C, 45 sec) × 40 cycles; 72°C, 7 min. PCR products were resolved on an agarose gel and stained with SYBR green (Molecular Probes) to increase the sensitivity of detection.
FISH analysis
RNA FISH was performed as described (Lawrence et al. 1989; Clerc and Avner 1998). GPT16, a 6-kb DNA fragment spanning most of Xist exon 1, was labeled with digoxygenin-16-dUTP (Boehringer) and detected with antidigoxygenin fluorescein isothiocyanate (AD-FITC) raised in sheep (Boehringer), followed by anti-sheep FITC (Vector Laboratories). Images were acquired on a Leica DMRB fluorescence microscope using a Photometrics CCD camera and Quips Software (Applied Imaging).
Acknowledgments
We thank the X inactivation group and Suren Zakian for helpful comments and critical reading of the manuscript. This work was supported by the Medical Research Council, UK. T.B.N. was supported by an International Development Award from the Wellcome Trust.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.271203.
References
- Ashfield R., Patel, A.J., Bossone, S.A., Brown, H., Campbell, R.D., Marcu, K.B., and Proudfoot, N.J. 1994. MAZ-dependent termination between closely spaced human complement genes. EMBO J. 13: 5656–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avner P. and Heard, E. 2001. X-chromosome inactivation: Counting, choice and initiation. Nat. Rev. Genet. 2: 59–67. [DOI] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth, A., Kay, G.F., McCabe, V.M., Norris, D.P., Cooper, P.J., Swift, S., and Rastan, S. 1992. The product of the mouse Xist gene is a 15-kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526. [DOI] [PubMed] [Google Scholar]
- Brown C.J., Hendrich, B.D., Rupert, J.L., Lafreniere, R.G., Xing, Y., Lawrence, J., and Willard, H.F. 1992. The human XIST gene: Analysis of a 17-kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542. [DOI] [PubMed] [Google Scholar]
- Cattanach B.M., Pollard, C.E., and Perez, J.N. 1969. Controlling elements in the mouse X-chromosome. I. Interaction with the X-linked genes. Genet. Res. 14: 223–235. [DOI] [PubMed] [Google Scholar]
- Chuma S. and Nakatsuji, N. 2001. Autonomous transition into meiosis of mouse fetal germ cells in vitro and its inhibition by gp130-mediated signaling. Dev. Biol. 229: 468–479. [DOI] [PubMed] [Google Scholar]
- Chureau C., Prissette, M., Bourdet, A., Barbe, V., Cattolico, L., Jones, L., Eggen, A., Avner, P., and Duret, L. 2002. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 12: 894–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C.M., McNeil, J.A., Willard, H.F., and Lawrence, J.B. 1996. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132: 259–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc P. and Avner, P. 1998. Role of the region 3′ to Xist exon 6 in the counting process of X-chromosome inactivation. Nat. Genet. 19: 249–253. [DOI] [PubMed] [Google Scholar]
- Cooper D.W., VandeBerg, J.L., Sharman, G.B., and Poole, W.E. 1971. Phosphoglycerate kinase polymorphism in kangaroos provides further evidence for paternal X inactivation. Nat. New Biol. 230: 155–157. [DOI] [PubMed] [Google Scholar]
- Cooper D.W., Johnston, P.G., VandeBerg, J.L., and Robinson, E.S. 1990. X-chromosome inactivation in marsupials. In Mammals from pouches and eggs: Genetics, breeding and evolution ofmarsupials and monotremes (eds. J.A.M. Graves, R.M. Hope, and D.W. Cooper). CSIRO, Australia.
- Debrand E., Chureau, C., Arnaud, D., Avner, P., and Heard, E. 1999. Functional analysis of the DXPas34 locus, a 3′ regulator of Xist expression. Mol. Cell Biol. 19: 8513–8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribnau J., Diderich, K., Pruzina, S., Calzolari, R., and Fraser, P. 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell 5: 377–386. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe, D.C. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952. [DOI] [PubMed] [Google Scholar]
- Johnston C.M., Nesterova, T.B., Formstone, E.J., Newall, A.E., Duthie, S.M., Sheardown, S.A., and Brockdorff, N. 1998. Developmentally regulated Xist promoter switch mediates initiation of X inactivation. Cell 94: 809–817. [DOI] [PubMed] [Google Scholar]
- Johnston C., Newall, A., Brockdorff, N., and Nesterova, T. 2002. Enox, a novel gene that maps 10 kb upstream of Xist and partially escapes X inactivation. Genomics 80: 236–244. [DOI] [PubMed] [Google Scholar]
- Kay G.F., Penny, G.D., Patel, D., Ashworth, A., Brockdorff, N., and Rastan, S. 1993. Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell 72: 171–182. [DOI] [PubMed] [Google Scholar]
- Lawrence J.B., Singer, R.H., and Marselle, L.M. 1989. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell 57: 493–502. [DOI] [PubMed] [Google Scholar]
- Lee J.T. 2000. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell 103: 17–27. [DOI] [PubMed] [Google Scholar]
- ____. 2002. Homozygous Tsix mutant mice reveal a sex-ratio distortion and revert to random X-inactivation. Nat. Genet. 32: 195–200. [DOI] [PubMed] [Google Scholar]
- Lee J.T. and Lu, N. 1999. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99: 47–57. [DOI] [PubMed] [Google Scholar]
- Lee J.T., Davidow, L.S., and Warshawsky, D. 1999. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 21: 400–404. [DOI] [PubMed] [Google Scholar]
- Levitt N., Briggs, D., Gil, A., and Proudfoot, N.J. 1989. Definition of an efficient synthetic poly(A) site. Genes & Dev. 3: 1019–1025. [DOI] [PubMed] [Google Scholar]
- Luikenhuis S., Wutz, A., and Jaenisch, R. 2001. Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol. Cell. Biol. 21: 8512–8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M.F. 1961. Gene action in the X chromosome of the mouse (Mus musculus L.). Nature 190: 372–373. [DOI] [PubMed] [Google Scholar]
- Marahrens Y., Panning, B., Dausman, J., Strauss, W., and Jaenisch, R. 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes & Dev. 11: 156–166. [DOI] [PubMed] [Google Scholar]
- Maxfield Boumil R. and Lee, J.T. 2001. Forty years of decoding the silence in X-chromosome inactivation. Hum. Mol. Genet. 10: 2225–2232. [DOI] [PubMed] [Google Scholar]
- Mette M.F., Aufsatz, W., van der Winden, J., Matzke, M.A., and Matzke, A.J. 2000. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19: 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mise N., Goto, Y., Nakajima, N., and Takagi, N. 1999. Molecular cloning of antisense transcripts of the mouse Xist gene. Biochem. Biophys. Res. Commun. 258: 537–541. [DOI] [PubMed] [Google Scholar]
- Naumova A.K., Plenge, R.M., Bird, L.M., Leppert, M., Morgan, K., Willard, H.F., and Sapienza, C. 1996. Heritability of X chromosome-inactivation phenotype in a large family. Am. J. Hum. Genet. 58: 1111–1119. [PMC free article] [PubMed] [Google Scholar]
- Nesterova T.B., Slobodyanyuk, S.Y., Elisaphenko, E.A., Shevchenko, A.I., Johnston, C., Pavlova, M.E., Rogozin, I.B., Kolesnikov, N.N., Brockdorff, N., and Zakian, S.M. 2001. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res. 11: 833–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newall A.E., Duthie, S., Formstone, E., Nesterova, T., Alexiou, M., Johnston, C., Caparros, M.L., and Brockdorff, N. 2001. Primary non-random X inactivation associated with disruption of Xist promoter regulation. Hum. Mol. Genet. 10: 581–589. [DOI] [PubMed] [Google Scholar]
- Ogawa Y., and Lee, J.T. 2003. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol. Cell. 11: 731–743. [DOI] [PubMed] [Google Scholar]
- Panning B., Dausman, J., and Jaenisch, R. 1997. X chromosome inactivation is mediated by Xist RNA stabilization. Cell 90: 907–916. [DOI] [PubMed] [Google Scholar]
- Penny G.D., Kay, G.F., Sheardown, S.A., Rastan, S., and Brockdorff, N. 1996. Requirement for Xist in X chromosome inactivation. Nature 379: 131–137. [DOI] [PubMed] [Google Scholar]
- Plenge R.M., Hendrich, B.D., Schwartz, C., Arena, J.F., Naumova, A., Sapienza, C., Winter, R.M., and Willard, H.F. 1997. A promoter mutation in the XIST gene in two unrelated families with skewed X-chromosome inactivation. Nat. Genet. 17: 353–356. [DOI] [PubMed] [Google Scholar]
- Rastan S. 1982. Timing of X-chromosome inactivation in postimplantation mouse embryos. J. Embryol. Exp. Morphol. 71: 11–24. [PubMed] [Google Scholar]
- ____. 1983. Non-random X-chromosome inactivation in mouse X-autosome translocation embryos—Location of the inactivation centre. J. Embryol. Exp. Morphol. 78: 1–22. [PubMed] [Google Scholar]
- Rastan S. and Brown, S.D. 1990. The search for the mouse X-chromosome inactivation centre. Genet. Res. 56: 99–106. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J. and Bartel, D.P. 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831. [DOI] [PubMed] [Google Scholar]
- Sado T., Wang, Z., Sasaki, H., and Li, E. 2001. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128: 1275–1286. [DOI] [PubMed] [Google Scholar]
- Sheardown S.A., Duthie, S.M., Johnston, C.M., Newall, A.E., Formstone, E.J., Arkell, R.M., Nesterova, T.B., Alghisi, G.C., Rastan, S., and Brockdorff, N. 1997. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell 91: 99–107. [DOI] [PubMed] [Google Scholar]
- Shibata S., and Lee, J.T. 2003. Characterization and quantitation of differential Tsix transcripts: Implications for Tsix function. Hum. Mol. Genet. 12: 125–136. [DOI] [PubMed] [Google Scholar]
- Singer-Sam J., LeBon, J.M., Dai, A., and Riggs, A.D. 1992. A sensitive, quantitative assay for measurement of allele-specific transcripts differing by a single nucleotide. PCR Methods Appl. 1: 160–163. [DOI] [PubMed] [Google Scholar]
- Stavropoulos N., Lu, N., and Lee, J.T. 2001. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc. Natl. Acad. Sci. 98: 10232–10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi N. and Sasaki, M. 1975. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256: 640–642. [DOI] [PubMed] [Google Scholar]
- Volpe T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Wake N., Takagi, N., and Sasaki, M. 1976. Non-random inactivation of X chromosome in the rat yolk sac. Nature 262: 580–581. [DOI] [PubMed] [Google Scholar]
- Wutz A., Rasmussen, T.P., and Jaenisch, R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30: 167–174. [DOI] [PubMed] [Google Scholar]
- Yagi T., Ikawa, Y., Yoshida, K., Shigetani, Y., Takeda, N., Mabuchi, I., Yamamoto, T., and Aizawa, S. 1990. Homologous recombination at c-fyn locus of mouse embryonic stem cells with use of diphtheria toxin A-fragment gene in negative selection. Proc. Natl. Acad. Sci. 87: 9918–9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian S.M., Nesterova, T.B., Cheryaukene, O.V., and Bochkarev, M.N. 1991. Heterochromatin as a factor affecting X-inactivation in interspecific female vole hybrids (Microtidae, Rodentia). Genet. Res. 58: 105–110. [Google Scholar]