Abstract
Candida glabrata is an important opportunistic pathogen causing both mucosal and bloodstream infections. C. glabrata is able to adhere avidly to mammalian cells, an interaction that depends on the Epa1p lectin. EPA1 is shown here to be a member of a larger family of highly related genes encoded in subtelomeric clusters. Subtelomeric clustering of large families of surface glycoprotein-encoding genes is a hallmark of several pathogens, including Plasmodium, Trypanosoma, and Pneumocystis. In these other pathogens, a single surface glycoprotein is expressed, whereas other genes in the family are transcriptionally silent. Similarly, whereas EPA1 is expressed in vitro, EPA2-5 are transcriptionally repressed. This repression is shown to be due to regional silencing of the subtelomeric loci. In Saccharomyces cerevisiae, subtelomeric silencing is initiated by Rap1p binding to the telomeric repeats and subsequent recruitment of the Sir complex by protein-protein interaction. We demonstrate here that silencing of the subtelomeric EPA loci also depends on functional Sir3p and Rap1p. This identification and analysis of the EPA gene family provides a compelling example in an ascomycete of chromatin-based silencing of natural subtelomeric genes and provides for the first time in a pathogen, molecular insight into the transcriptional silencing of large subtelomeric gene families.
Keywords: Yeast, subtelomeric, GPI, virulence, fungal pathogen, adhesin
Candida species, normally part of the mucosal flora in immunocompetent individuals, can cause invasive infections in immunocompromised individuals. In the hospital environment, Candida species are responsible for ∼8% of all hospital-acquired bloodstream infections (Schaberg et al. 1991; Gerberding et al. 1999; Trick et al. 2002). The two most frequently encountered species in the United States are Candida glabrata and Candida albicans. C. glabrata, a haploid yeast, accounts for upwards of 15% of isolates from the bloodstream as well as from mucosal sites (Schuman et al. 1998; Pfaller et al. 1999, 2001; Vazquez et al. 1999). Little is known about the virulence factors permitting this organism to colonize the human host and cause disease. As with C. albicans, adherence of C. glabrata to host cells is hypothesized to be important in virulence (Calderone and Fonzi 2001). In vitro adherence of C. glabrata to cultured human epithelial cells is mediated in large part by a single adhesin, encoded by the EPA1 gene, which recognizes host encoded N-acetyl lactosamine-containing glycoconjugates (Cormack et al. 1999). EPA1 is a member of a large class of glycosylphosphatidylinositol-anchored cell wall proteins (GPI-CWPs) found in diverse fungal species including Saccharomyces cerevisiae, C. albicans, and Aspergillus fumigatus (Brul et al. 1997; Kapteyn et al. 2000; Klis et al. 2001). At the sequence level, these proteins are characterized by the presence of a hydrophobic signal sequence, an N-terminal “functional” domain that mediates specific protein-protein or protein-ligand interactions, a hydrophobic C-terminal sequence signaling GPI anchor addition, and (in many cases) a large C-terminal serine/threonine-rich region which functions in localization within different layers of the cell wall (Frieman et al. 2002). GPI-CWPs traffic to the plasma membrane, where they are clipped from the GPI anchor and cross-linked to β1-6 glucan in the cell wall (Lu et al. 1994, 1995; Kapteyn et al. 1996) a form that is often accessible to the extracellular environment (Frieman et al. 2002).
Although EPA1 mediates 95% of in vitro adherence to cultured epithelial cells, deletion of EPA1 results in no significant virulence phenotype in murine models of systemic or vaginal candidiasis (Cormack et al. 1999). By extension from other pathogens, this might be due to the presence of additional adhesins that compensate in vivo for the absence of EPA1. For example, in C. albicans, the ALS1 and ALA1 genes, which encode adhesins for unidentified host ligands, are members of a family of so-called ALS genes (Hoyer 2001). HWP1, which permits the covalent attachment of Candida to host tissue via host transglutaminases, is one of four related genes found in the C. albicans genome sequence (Candida Genome Project, Stanford University). This multiplication of putative adhesins in C. albicans raises the possibility of functional redundancy within the gene families, and may indicate that proteins present at the cell surface are of prime importance in the host-pathogen interaction, providing evolutionary pressure for expansion of the family.
Here we demonstrate that EPA1 is part of a large family of related genes in C. glabrata, and we characterize four additional EPA-related genes. Our data reveal an unexpected complexity in the potential for cell-surface variation in C. glabrata and suggest that a new family of proteins defined by homology to the adhesin EPA1 may be critical for C. glabrata's interaction with mammalian hosts. Strikingly, the EPA genes are present in subtelomeric clusters, similar to the subtelomeric clustering of surface glycoprotein genes in many other pathogens, including Pneumocystis carinii, Trypanosoma brucei, and Plasmodium spp. In addition, we show that most of the EPA genes are transcriptionally silenced by machinery analogous to the silencing machinery of S. cerevisiae, providing a compelling example in an ascomycete of chromatin-based regulation of natural subtelomeric genes.
Results
EPA1 is part of a cluster of C. glabrata-specific cell wall proteins
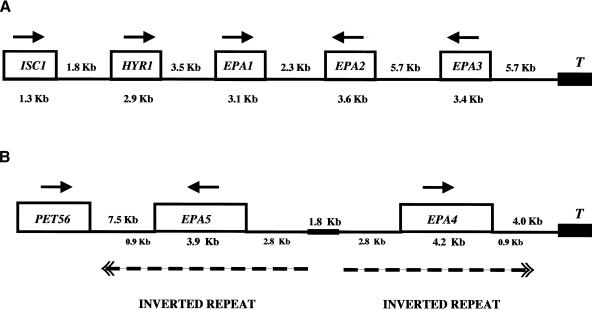
Analysis of C. glabrata genomic DNA revealed that EPA1 (Fig. 1A) is part of a cluster of related genes. We identified and sequenced a fosmid (Castano et al. 2003) containing 40kb of C. glabrata genomic DNA flanking the EPA1 locus (Fig. 1A). Using fosmid sequence, we used chromosome walking to clone and sequence the adjacent genomic region (Materials and Methods). The sequence of the locus has been deposited with GenBank (accession no. AY344226). This analysis showed that the EPA1 locus is adjacent to a telomere in a cluster of four genes encoding predicted GPI-CWPs, each possessing an N-terminal signal sequence, a C-terminal serine/threonine-rich region, and a C-terminal GPI-anchor addition signal. These four genes, which we have named HYR1, EPA1, EPA2, and EPA3 (see below) are clustered in a 24-kb region located 5.7 kb from the telomere. In addition to the clustering of these related genes, the region is remarkable for the large intergenic distances (2.3-5.7 kb) between predicted coding regions. The telomere sequence in our terminal clone is composed of a 16-bp sequence (CTGT-GGGG-TCT-GGG-TG), repeated 30 times in our clone. The sequence of this telomere repeat is in agreement with that previously published (McEachern and Blackburn 1994) and contains a consensus Rap1p binding site (Cohn et al. 1998).
Figure 1.
Structure of the EPA clusters. (A) EPA1 is present in a cluster of four related genes encoding predicted GPI-linked cell wall proteins: HYR1, EPA1, EPA2, and EPA3. This cluster is located 5.7 kb from the telomere (T). (B) EPA4 and EPA5 are essentially identical genes and are present in the genome as an inverted repeat. The repeated sequences include a 2.8-kb upstream noncoding region, the coding region (EPA4, 4.2 kb; EPA5, 3.9 kb), and 0.9 kb of 3′ noncoding region. The inverted repeats (broken line) are separated by a unique region of 1.8 kb. The EPA4/5 cluster is localized 4 kb from the telomere. The numbers in kilobases show the size of the genes (below the gene name) and the intergenic distance (between the genes). Arrows indicate direction of transcription.
Even though S. cerevisiae and C. glabrata are phylo-genetically closely related yeasts, the genes in the cluster are not present in S. cerevisiae. Upstream of the cluster there are orthologs of a number of nontelomeric S. cerevisiae genes, including the S. cerevisiae ISC1 (YER019w), which encodes an inositol phosphosphingolipid phospholipase C (65% identity/similarity), and the S. cerevisiae essential gene YJL010c (83% identity/similarity). Upstream of these two genes, the genome is clearly syntenic to that of S. cerevisiae, with the C. glabrata sequence containing the orthologs of the S. cerevisiae ORFs YKL142w, YKL145w, YKL149w, YKL150w, and YKL151c genes (data not shown).
Structure of EPA2, EPA3, and HYR1
C. glabrata HYR1 is the gene located closest to the centromere in the cluster. From the deduced amino acid sequence, there is an N-terminal signal sequence, and the first 400 amino acids share homology (21% identity/19% similarity) to the C. albicans HYR1 gene (Bailey et al. 1996). Like the C. albicans counterpart, C. glabrata HYR1 is predicted to encode a GPI-anchored cell wall protein (GPI-CWP), as it possesses a signal sequence, a C-terminal GPI addition signal, and a C-terminal serine/threonine-rich region (28% S/T). Its predicted transcription orientation is toward the telomere, and its stop codon is separated from the start codon of EPA1, the second gene in the cluster, by a 3.5-kb intergenic region.
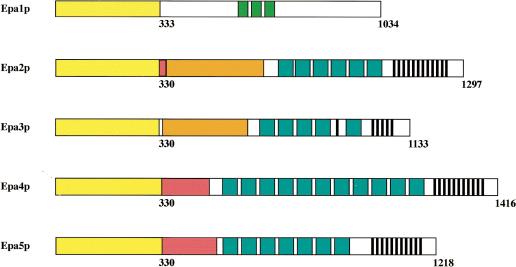
EPA2 and EPA3 are the third and the fourth genes of the cluster separated from EPA1 by a 2.3-kb intergenic region and from each other by a long intergenic region of 5.7 kb (Fig. 1A). They encode predicted cell wall proteins of 1297 amino acids and 1133 amino acids, respectively. The predicted transcription orientation for both EPA2 and EPA3 is toward the centromere, in contrast to that of HYR1 and EPA1. EPA1, EPA2, and EPA3 are clear paralogs. For the three EPA genes, the predicted N-terminal 300-amino acid regions (shown to be the ligand binding domain for Epa1p; Frieman et al. 2002) are highly related (Table 1). The C-terminal regions of EPA2 and EPA3 are homologous to each other but not to the C-terminal region of EPA1, a result of the fact that EPA2 and EPA3 share internal Ser/Thr/Pro-rich repeats with each other but not with EPA1 (Fig. 2).
Table 1.
EPA family homology in the ligand-binding domain
| Epa2p | Epa3p | Epa4p | Epa5p | |
|---|---|---|---|---|
| Epa1p | 46 | 41 | 46 | 45 |
| Epa2p | 40 | 87 | 86 | |
| Epa3p | 39 | 39 | ||
| Epa4p | 99 |
Percent identity of the N-terminal domains (junction of N-terminal and C-terminal domains as defined in Fig. 2) of EPA1-5
Figure 2.
Schematic of domain structure of the EPA1 homologs. The sequences of EPA1-5 have been deposited with GenBank (accession nos. AY344225 and AY344226). The N-terminal domains of EPA1, EPA2, EPA3, EPA4, and EPA5 are indicated in yellow. The exact breakpoint between the N-terminal and C-terminal domains is somewhat arbitrary, but in general is marked by the first sequence that is highly enriched for Ser or Thr residues. The homology between the N-terminal ligand binding domains is detailed in Table 1. The C-terminal domains are characterized by several repeated sequences. For details and full amino acid sequence of the longer repeats, see the GenBank files indicated above. The green boxes in Epa1p indicate a 40-amino acid repeat (VRS.. .SKM) specific for that protein. The red boxes in Epa2p, Epa4p, and Epa5p indicate stretches almost entirely made up of Ser residues. The orange boxes in Epa2p and Epa3p indicate regions composed of three different repeats (HNPSS, VNPSS, and KPADPSPADPS), themselves repeated multiple times. The blue boxes in Epa2p-Epa5p indicate a 56-amino acid motif (DSFSL.. .NNGDS) found in all four proteins; the black stripes in these same genes indicate a 14-amino acid repeat (PSPNGGGSGNNGNT).
EPA4 and EPA5 are encoded as a subtelomeric near-perfect inverted repeat
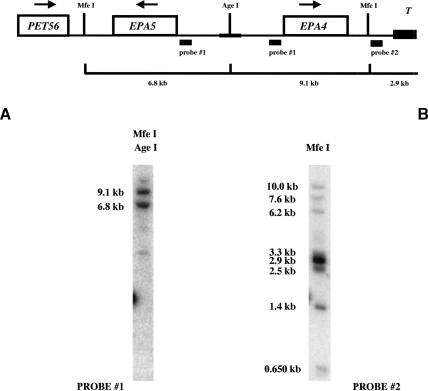
Given the discovery of EPA2 and EPA3, we used low-stringency PCR amplification to determine whether other EPA-related genes are present in the genome. We designed primers (primers #1124/#1125) based on homology among EPA1, EPA2, and EPA3 and amplified a fragment from C. glabrata genomic DNA prepared from strain BG465, which lacks EPA1, EPA2, and EPA3 (see below). The nucleotide sequence was not identical, but showed a high degree of homology, to the EPA2 gene. Three fosmids (F11D8, F23H10, and F29C7) that contained this fragment were identified and used for sequence analysis. The fosmids were found to contain two EPA-related genes, which we named EPA4 and EPA5. Chromosome walking was used to clone and sequence the adjacent genomic region (Materials and Methods). The sequence of the locus has been deposited with Gen-Bank (accession no. AY344225). As for the EPA1-3 locus, the terminal clone identified for this cluster contained multiple 16-bp telomeric repeats, indicating that the EPA4-5 locus, like the EPA1-3 locus, is adjacent to a telomere (Fig. 1B). As diagrammed in Figure 2, EPA4 and EPA5 are essentially identical to each other (99% identity). In the N-terminal ligand binding domain, they are very close relatives of EPA2 (Table 1). EPA4 is slightly longer than EPA5, as it has four additional internal 56-amino acid repeats within the Ser/Thr-rich region. In addition, just after the N-terminal ligand binding domain, each gene has a region of slightly different lengths composed almost entirely of serine residues (153 in EPA4 and 178 in EPA5). Apart from these two differences, they have only a single nucleotide difference (which results in a change at amino acid 95 from glycine for EPA4 to arginine for EPA5). More than 2.8 kb of upstream noncoding region and 0.9 kb of 3′ noncoding region are also identical for the two genes. This is diagrammed in Figure 1B (not drawn to scale). The genes are present in the genome as a large inverted repeat, separated by a unique sequence of 1.8 kb (Fig. 1B). Several additional experiments were carried out to eliminate the possibility that the extensive inverted repeat structure of the locus resulted from a recombination/duplication event of the cloned locus in Escherichia coli. Southern blot analysis using EPA4/EPA5 sequence as probe showed two bands of predicted size after digestion with an enzyme in the unique region between the two inverted repeats (Fig. 3A). PCR amplification and sequencing of the products also showed that EPA4 and EPA5 are both present in the genome. The inverted orientation of these genes with respect to one another was verified using PCR with a single primer which, because of the inverted duplication, could amplify a band of predicted size. Finally, Southern blot analysis using a probe to the unique region telomeric to EPA4 hybridized to a band of predicted molecular weight corresponding to the telomeric fragment (Fig. 3B). This probe also hybridized to multiple other bands, suggesting that the region is repeated multiple times in the genome (as is true for subtelomeric sequences in S. cerevisiae). Thus, EPA4 and EPA5, essentially identical to each other and close homologs of the EPA2 gene, are present as an inverted repeat found within 4 kb of the telomere. As we were unable to distinguish between them in our subsequent transcriptional analyses, we refer to the pair as EPA4/EPA5.
Figure 3.
Structural analysis of the EPA4/5 loci. Southern blot analysis was made to confirm the structure of EPA4 and EPA5. Genomic DNA was isolated from wild-type strain BG14 and cut with AgeI (AgeI recognizes a site in the unique region between the inverted repeats) and MfeI, and the bands were separated in an 0.8% agarose gel (see Materials and Methods). Probe #1 (pAP481) anneals 157 bp upstream from the ATG of both EPA4 and EPA5, giving 6.8-kb and 9.1-kb bands. Probe #2 (pAP479) anneals 1.343 kb from the telomere repeats, showing the expected 2.9-kb band. This Southern blot confirms the presence of EPA4/5 as an inverted repeat and that this subtelomeric region is reiterated in the genome.
Deletion of the EPA1-3 cluster reduces C. glabrata virulence
The EPA1 gene mediates in vitro adherence to epithelial cells through binding to host carbohydrate (Cormack et al. 1999). In a systemic infection model, immunocompetent BalbC mice, an epa1 Δ mutant strain, showed no decrease in virulence. In light of our discovery of additional putative cell wall proteins (HYR1, EPA1, EPA2, and EPA3) in C. glabrata and the extent of homology between EPA1, EPA2, and EPA3, we tested whether these genes play a role in virulence. We deleted the four genes in the EPA1 cluster by a classical two-step deletion procedure (see Materials and Methods), generating two independent strains deleted for all four genes (BG459 and BG461). The deletions were made in a ura3 mutant strain of C. glabrata (BG14) in which the URA3 locus is deleted (see Supplementary Table S1). The chromosomal structure of the deletion mutants was confirmed by PCR and Southern blot. Prior to their analysis in vivo, each independent mutant and the parental ura3 strain were made Ura+ by restoring URA3 at the URA3 locus to generate strains BG462, BG465, and BG475 (see Supplementary Table S1). The three strains grew with identical doubling times in rich media (YPD) or minimal media (YNB) at 30°C or 37°C (data not shown).
To examine virulence of these mutated strains, we infected mice by tail vein injection using 2 × 107 yeast cells with strains BG462 (wild type), BG465 (mutant 1), and BG475 (mutant 2). Mice were sacrificed on day 7 after infection, and the kidneys, spleen, and liver were recovered. The organs were homogenized, and dilutions were plated for colony-forming units (CFUs) on YPD plates supplemented with penicillin and streptomycin. For each strain, 10mice were infected. The average number of yeast recovered from the kidneys was three- to fivefold less for the mutant strains compared to that for the wild-type strain (Table 2). This modest attenuation was not the result of a general inability to persist or grow in vivo, because the recoverable counts for liver and spleen were essentially identical for the two mutant strains and for wild type (Table 2). The kidney colonization was significantly lower for mutant strain BG465 (p = 0.008) and for the independent mutant strain BG475 (p = 0.04) than for the wild-type strain BG462. The experiment was repeated two more times with the consistent finding that the colonization of the kidney was lower for the mutant strains than for the wild-type strain (Table 2). These data suggest that the HYR1/EPA1-3 gene cluster has a role in virulence, albeit a modest one.
Table 2.
Virulence of C. glabrata (hyrl epa1 epa2 epa3)Δ
| Kidney
|
Liver
|
Spleen
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exp. 1
|
Exp 2
|
Exp. 3
|
Exp. 1
|
Exp. 1
|
||||||||||
| Strain | Wild | M465 | M475 | Wild | M475 | Wild | M465 | M475 | Wild | M465 | M475 | Wild | M465 | M475 |
| Sample size | 10 | 10 | 10 | 10 | 10 | 10 | 8 | 9 | 10 | 10 | 10 | 10 | 10 | 10 |
| Min. (×10−2) | 2500 | 100 | 790 | 1900 | 500 | 6400 | 200 | 2700 | 900 | 100 | 520 | 10,600 | 270 | 18,000 |
| Max. (×10−2) | 122,000 | 15,300 | 45,000 | 49,000 | 48,000 | 57,200 | 31,700 | 37,100 | 6500 | 3700 | 5400 | 42,000 | 61,000 | 82,000 |
| Mean (×10−2) | 31,430 | 4520 | 14,219 | 14,100 | 6830 | 22,520 | 9980 | 13,140 | 2570 | 2200 | 2705 | 24,390 | 32,207 | 39,300 |
| P-valuea | — | 0.008 | 0.04 | — | 0.008 | — | 0.05 | 0.12 | — | 0.91 | 1 | — | 0.16 | 0.06 |
Balb/C mice were infected by tail vein injection with 2 × 107 cells per strain and 10 mice were used per experiment per strain. Mice were sacrificed at day 7 after infection and kidney, liver, and spleen were recovered. The organs were homogenized and dilutions were plated on YPD Pen/Str plates, and CFU were scored (see Materials and Methods). Strains BG462 (wt), BG465 (mutant 1), and BG475 (mutant 2) were used in these experiments (see Supplemental Material, Table S1). There is a significant three- to five-fold reduction in kidney colonization, but no difference in colonization of spleen or liver. Representative counts are shown for spleen and liver from the first experiment; in the subsequent experiments as well there was no significant difference in recovered CFUs for the three strains for spleen and liver (data not shown).
Kruskal-Wallis test.
We also deleted the EPA4 and EPA5 genes in the strains already deleted for HYR1 and EPA1, EPA2, and EPA3. This mutant strain also had a moderate decrease (threefold) in colonization specifically of the kidney (data not shown).
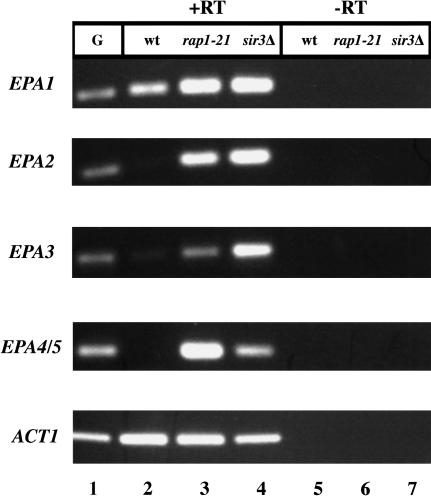
The EPA genes are transcriptionally silent in vitro
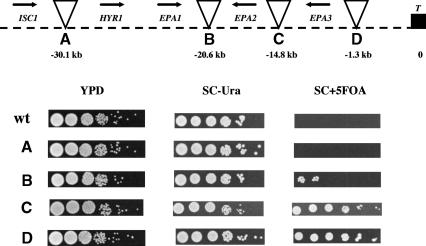
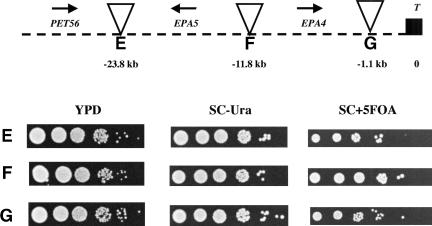
The finding that the EPA1 gene is required to mediate in vitro adherence to epithelial cells suggested that it is expressed in vitro under the conditions of the adherence assay, and indeed we were able to show that EPA1 is expressed in vitro (Fig. 4, lane 2). We used reverse transcriptase PCR (RT-PCR) to determine whether EPA2, EPA3, and EPA4/EPA5 are expressed under the same conditions. We found that there was no detectable transcript for any of the EPA2-5 genes (Fig. 4, lane 2). Because EPA2-5 are so close to the telomere, we considered whether EPA2-5 might be subject to silencing, as are reporter genes placed adjacent to telomere sequences in S. cerevisiae. We used transposon mutagenesis to place the URA3 gene at seven locations in the intergenic regions of HYR1, EPA1, EPA2, EPA3, EPA4, and EPA5. The locations of these seven URA3 markers are shown in Figure 5. These URA3 reporters provided an assay for transcriptional repression across the region independent of measuring transcription of the EPA genes themselves. 5-FOA, an analog of orotate, an intermediate in uracil metabolism, is toxic to cells able to metabolize uracil. In the assay, cells expressing the URA3 gene die in the presence of 5-FOA; in contrast, cells in which the URA3 gene is silent are able to grow in the presence of 5-FOA. Thus, if the URA3 gene at a particular location is subject to transcriptional silencing, the corresponding strain is able to grow on SC + 5-FOA plates. Remarkably, for the EPA1-3 locus, there was a clear silencing effect (as measured by growth on SC + 5-FOA plates) for URA3 placed at telomeric locations, whereas insertions distal (telomeric) to EPA1 or proximal (centromeric) to HYR1 were not appreciably silenced at all. Indeed, there was a gradient of silencing of the EPA1-3 locus, with pronounced silencing of URA3 integrated at the most telomeric site but with decreased silencing as the integration site was moved towards the centromere (Fig. 5, top). At the EPA4/EPA5 locus, URA3 genes placed at all three integration sites were completely silenced (Fig. 5, bottom).
Figure 4.
Expression analysis of EPA1, EPA2, EPA3, and EPA4/5 by RAP1 and SIR3. Wild-type and the mutant strains, rap1-21 and sir3Δ, were assayed for EPA1, EPA2, EPA3, and EPA4/5 expression by RT-PCR (see Materials and Methods). ACT1 was used as the control. (-RT) Controls without RT; (G) genomic control.
Figure 5.
Silencing effect on the URA3 gene located in the intergenic regions at the EPA1 and EPA4/5 clusters. (Top) Strains were constructed carrying Tn7 insertions in the intergenic regions of ISC1 and HYR1 (A), EPA1 and EPA2 (B), EPA2 and EPA3 (C), and EPA3 and telomere (D). (Bottom) Strains were constructed carrying Tn7 insertions in the intergenic regions of PET56 and EPA5 (E), EPA5 and EPA4 (F), and EPA4 and telomere (G). Cells were inoculated at the same O.D.600 nm and grown in YPD for 13 doublings, and 10-fold serial dilutions were spotted on YPD, SC-Ura, and SC + 5FOA (see Materials and Methods). Cells that are Ura+ die on SC + 5FOA plates. Thus only cells in which the URA3 gene is transcriptionally repressed can grow on this medium. Triangles represent Tn7 insertions, arrows indicate direction of transcription (T, telomere), and numbers in kilobases are the distance from telomere to the insertion point.
EPA2-5 are silenced in an SIR3-dependent fashion
To begin to analyze the mechanism by which genes in this subtelomeric family are silenced, we undertook a screen for mutants that de-repress transcription of the silent genes. We had previously constructed a library of 10,000 insertion mutants (Castano et al. 2003) distributed randomly in the C. glabrata genome. EPA1 is expressed at low levels in stationary phase, and the cells are consequently nonadherent. We set out to study EPA family regulation by screening for mutants that were hyper-adherent in stationary phase, potentially due to ectopic expression of EPA1 paralogs. The screen and most of the genes identified in the screen will be described elsewhere. However, one of the mutants found in the screen had an insertion in the promoter region of a gene with homology to SIR3. This insertion resulted in a hyper-adherent phenotype in stationary-phase cells. In S. cerevisiae, SIR3 encodes one of the critical structural components involved in silencing chromatin at the silent mating loci and at telomeres (Rine and Herskowitz 1987; Gottschling et al. 1990; Aparicio et al. 1991; Moretti et al. 1994; Liu and Lustig 1996), suggesting that chromatin-based subtelomeric silencing might have a role in repression of the EPA genes. We cloned the full-length C. glabrata SIR3 gene from our fosmid library and determined its sequence (GenBank accession no. AY344224). Overall sequence homology to the S. cerevisiae SIR3 gene is modest, with 21% identity and 17% similarity over the length of the protein (data not shown). However, within the N-terminal 215 amino acids, there is a high degree of homology (45% identity/22% similarity). In S. cerevisiae, mutations in this conserved region, which contains a bromo-adjacent homology domain (Callebaut et al. 1999; Zhang et al. 2002), suppress the silencing defects of mutations in the tail of histone H4 (Johnson et al. 1990; Liu and Lustig 1996). Furthermore, the C. glabrata SIR3 gene is adjacent to the ECM7 gene in C. glabrata as it is in S. cerevisiae, consistent with the overall synteny between the two genomes.
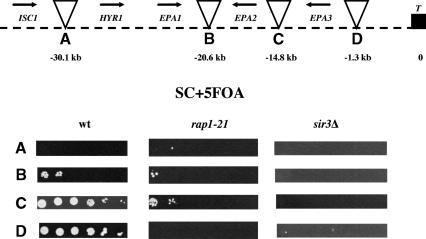
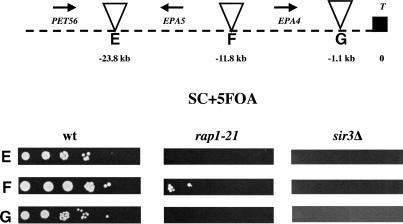
We made a disruption of the entire SIR3 ORF, generating strain BG676. We then analyzed this strain for expression of EPA1-5. EPA1 is expressed in log-phase cells in both the wild type and the sir3Δ strains (Fig. 4, lanes 2,4). However, EPA2, EPA3, and EPA4/5, which were silent in the wild-type strain, were expressed in the sir3Δ strain (Fig. 4, lanes 2,4). This suggested that Sir3p acts normally to repress at least some of the EPA genes. To examine the role of SIR3 in regional repression of the two loci, we assayed expression of the URA3 insertions in this region. We showed above that the URA3 gene was transcriptionally silenced when integrated at various positions within the two EPA clusters. We tested whether that transcriptional repression was dependent on SIR3, by introducing the seven Tn7-URA3 constructs into the identical genomic locations in the sir3Δ background. Silencing of the URA3 gene in all cases was eliminated, showing that the position-dependent silencing of URA3 integrated at both subtelomeric EPA loci was dependent on SIR3 (Fig. 6, top and bottom).
Figure 6.
Silencing of the URA3 gene located in the intergenic regions at the EPA clusters depends on SIR3 and RAP1. The Tn7 insertions from Figure 5 and this figure were introduced in the sir3Δ and rap1-21 strains, and their ability to grow on SC + 5FOA plates was scored. (Top) Insertions at the EPA1 locus. (Bottom) Insertions at the EPA4/5 locus. (See Fig. 5 legend).
RAP1 is required for EPA gene silencing
In S. cerevisiae, Sir3p is not able to bind DNA directly. Rather it is recruited to the DNA by the action of a variety of DNA binding proteins, including Rap1p, Abf1p, and ORC. Silencing at telomeres and at the mating loci in S. cerevisiae specifically requires the DNA binding protein Rap1p, and mutation of RAP1 causes a loss of subtelomeric and mating loci silencing (Buchman et al. 1988; Kurtz and Shore 1991; Sussel and Shore 1991; Kyrion et al. 1993). Because the telomere sequence in C. glabrata contains consensus Rap1p binding sites, it seemed possible that Rap1p would have a role in subtelomeric silencing in C. glabrata as it does in S. cerevisiae. To determine whether RAP1 is required for EPA family silencing in C. glabrata, we set out to disrupt RAP1 function. Because RAP1 is essential in C. glabrata (Haw et al. 2001) as it is in S. cerevisiae, we could not delete it and instead constructed a truncation allele of the chromosomal RAP1 gene, in which the nucleotides corresponding to the C-terminal 28 amino acids were removed. The corresponding allele in S. cerevisiae, rap1-21, is viable but results in a protein unable to interact with Sir3p and which is therefore unable to mediate silencing at subtelomeric loci (Sussel and Shore 1991; Liu et al. 1994). In C. glabrata, the equivalent rap1-21 mutation de-repressed transcription of EPA2, EPA3, and EPA4/EPA5 (Fig. 4, lanes 2,3). This suggested that functional Rap1p is required for silencing of the EPA2, EPA3 and EPA4/5 loci. Consistent with this, the rap1-21 mutation also relieved silencing of the URA3 gene integrated at all subtelomeric locations tested (Fig. 6, top and bottom). Silencing of URA3 integrated between EPA2 and EPA3 was only partially relieved in the rap1-21 background (Fig. 6, top).
Discussion
EPA genes are present in silent subtelomeric clusters
The EPA1 gene of C. glabrata encodes a lectin mediating adherence to mammalian epithelial cells (Cormack et al. 1999). We describe here the cloning and initial characterization of four homologs of EPA1. The five EPA genes described here are encoded in two subtelomeric clusters. Under a variety of conditions we examined in vitro, EPA1 is expressed and EPA2, EPA3, EPA4, and EPA5 are transcriptionally repressed. We present clear evidence that this repression reflects chromatin-based regional silencing, in as much as transcriptional silencing of the native EPA2, EPA3, EPA4, and EPA5 loci or of heterologous URA3 reporter genes integrated at a variety of positions within both loci depends on functional SIR3. EPA1 is normally expressed at low levels, and these levels increase in the sir3Δ background (Fig. 4), suggesting that the EPA1 locus, although not efficiently silenced, is perhaps subject to some repressive chromatin effects. Mechanistic details of this regional silencing are apparently conserved between S. cerevisiae and C. glabrata, in as much as the rap1-21 mutant, which fails to recruit the Sir complex to telomeres in S. cerevisiae and is therefore nonfunctional for silencing, also displays a loss of silencing in C. glabrata. Although loss of URA3 silencing in the rap1-21 background was complete for most of the insertion sites, there was still some residual URA3 silencing for insertion C (between EPA2 and EPA3) and insertion F (between EPA4 and EPA5). This suggests that, for these two regions at least, there might be mechanisms partially redundant with Rap1p for recruiting the Sir complex to chromatin. To further examine this possibility, we are currently exploring whether the EPA4/EPA5 and EPA2/EPA3 intergenic regions contain silencer elements.
The conditions under which the silent EPA genes are normally expressed remain to be defined. One attractive hypothesis is that EPA gene expression varies during infection with different EPA genes expressed at different times. In this regard, a recent report showing that subtelomeric silencing in S. cerevisiae might be regulated in response to a variety of cell stresses (Ai et al. 2002) seems pertinent—cell stress encountered during the course of an infection might signal derepression of some of the silenced EPA genes.
The degree of silencing differs at two different telomeres
The extent of silencing at the two telomeric EPA loci studied here apparently differs. Silencing of the EPA1 cluster apparently spreads from the telomere itself or a site just adjacent to the telomere and results in a gradient of silencing, with URA3 positioned at the telomere being completely silenced and the degree of silencing falling off as its position is moved away from the telomere. In contrast, URA3 placed upstream, downstream, or in the middle of the EPA4/EPA5 inverted repeat are all equally silenced. At that locus, far from there being a gradient of silencing emanating from the telomere, the entire EPA4/EPA5 locus seems to be silenced as a block. This difference in silencing at the two telomeres is reminiscent of the situation in S. cerevisiae. Most studies of subtelomeric silencing in S. cerevisiae have focused on marker genes placed immediately adjacent to truncated telomeres. However, when URA3 was placed at different native telomeric sites, the strength of silencing was found to vary dramatically depending on the structure of the individual telomere, due in part to differences in the X and Y′ element structure of the individual telomeres (Pryde and Louis 1999). Based on our results with the EPA4/EPA5 and EPA1-3 loci, such telomere to telomere variation likely applies in C. glabrata as well.
The organization of EPA4 and EPA5 may play a role in the robust silencing of this locus. These genes are present as an essentially perfect inverted repeat that varies at only three positions (a single nucleotide change, and two differences in the number of internal direct repeats) across 7.8 kb, including 3.5 kb of identical 5′ and 3′ untranslated regions. This striking organization is similar to the genomic organization of the PAI gene family in Arabidopsis thaliana. In that organism, there are four copies of the PAI gene, two of which are present in an inverted repeat. The inverted repeat is required for methylation and silencing of the two PAI genes making up the repeat as well as the unlinked PAI genes (Luff et al. 1999). By extension, perhaps the inverted repeat organization of EPA4 and EPA5 has a direct role in transcriptional silencing of EPA4/EPA5 or even of other EPA genes in trans.
Subtelomeric surface glycoproteins in other pathogens
The EPA1-3 cluster is located 5.7 kb, and the EPA4-5 cluster 4.0kb from their respective telomeres, This organization resembles several telomere-localized multigene families in S. cerevisiae such as the SUC, MAL, and MEL genes (carbon utilization), the PAU genes (induced by hypoxia), and the COS genes (for review, see Pryde and Louis 1997). A variety of human pathogens also encode large families of surface glycoproteins, with functions in cytoadhesion and antigenic variation, encoded in subtelomeric clusters. In this category are the MSG genes of the ascomycetous fungal pathogen P. carinii, the causative agent of pneumocystis pneumonia (for review, see Stringer and Keely 2001), the var, rifn, and stevar genes of Plasmodium falciparum, the vir genes of Plasmodium vivax (del Portillo et al. 2001), and the VSG genes of Trypanosoma brucei, (Borst and Rudenko 1994), all localized in subtelomeric regions. The ALS genes of C. albicans encode a family of cell wall proteins, but their genomic localization has not been reported.
In a comparison of P. vivax and P. falciparum, genes present in subtelomeric regions were species-specific, in marked contrast to the overall chromosome structure, which is highly syntenic between different plasmodial species (del Portillo et al. 2001). This may be similar to the situation in C. glabrata and its close relative S. cerevisiae. The region upstream of the EPA1-3 cluster is essentially syntenic with S. cerevisiae (see Results). Upstream of the EPA4/5 cluster, the genome is also syntenic to that of S. cerevisiae, with the C. glabrata sequence containing the orthologs of the S. cerevisiae PET56 (YOR201C), BFR1 (Y0R198C), and SLK19 (YOR195W). Other groups have commented on the significant overall synteny between C. glabrata and S. cerevisiae (Walsh et al. 2002; Wong et al. 2002, 2003). From our analysis here and our ongoing studies of other C. glabrata telomeres, this synteny clearly does not extend to the subtelomeric regions. Perhaps virulence genes in C. glabrata, which have evolved in response to the host, are enriched in subtelomeric locations.
In T. brucei and P. carinii, as in C. glabrata, variant surface glycoproteins are tightly transcriptionally regulated: for both species, a given cell transcribes only one member of the gene family from a telomeric expression site. In T. brucei, which has on the order of 1000 VSG genes and some 20telomeric expression sites, only one gene is expressed (Borst et al. 1996). In P. carinii, which has between 60and 144 MSG genes, all but one seem to be silent in a given cell (for review, see Stringer and Keely 2001). In both species, recombination between silent loci and a telomeric expression locus results in expression of a new family member. We have not tested whether the transcriptional status of the silent EPA genes can be altered by the kind of DNA re-arrangements that occurs in P. carinii or T. brucei. In this regard, it may be relevant that different clinical isolates do show some differences in the expression pattern of the EPA genes as measured by RT-PCR (data not shown). These expression differences might be due to epigenetic differences between strains; alternatively, genetic changes, analogous to the recombination events in P. carinii or T. brucei, might underlie the EPA expression differences we have noticed between strains.
EPA family evolution
In addition to the five EPA genes described here, the EPA family includes many other members. In a Southern analysis of BG2, a clinical C. glabrata isolate, numerous bands hybridized at low stringency to probes from EPA1, EPA2, and EPA3 (data not shown). Furthermore, in an 8× genome sequence of C. glabrata carried out by Bernard Dujon (Pasteur Institute), there are conservatively an additional 11 sequences which are closely related to the EPA genes in the N-terminal ligand binding domain (B. Dujon, pers. comm.). Based on preliminary physical mapping, at least six of the additional EPA genes are also subtelomeric (data not shown).
Why should such diverse pathogens as C. glabrata, P. falciparum, P. vivax, T. brucei, and P. carinii locate families of genes encoding surface proteins at subtelomeric locations? Apart from well defined roles of telomeres in chromosome replication and stability (Lee et al. 1998; Hackett et al. 2001) and transcriptional silencing (Aparicio et al. 1991), subtelomeric regions display increased levels of homologous and ectopic nonallelic recombination (Freitas-Junior et al. 2000). One advantage of locating large gene families in subtelomeric loci may therefore lie in the efficient expansion of, and generation of diversity within, surface glycoprotein-encoding gene families. It has been shown in both S. cerevisiae and P. falciparum that telomeres form clusters localized at the nuclear periphery, generating distinct nuclear compartments (Freitas-Junior et al. 2000). In P. falciparum isolates with spontaneous deletions of telomere-associated sequences, chromosome ends fail to form clusters (Scherf et al. 2001). The extent of the chromosome end alignment is unclear, but there is evidence that it spreads up to 30-40 kb, enough to include the regions encoding the variant surface antigen families (Scherf et al. 2001). DNA in this region has been shown to undergo ectopic nonallelic recombination at a much elevated rate. In P. falciparum, chimeric var genes have been found that were generated by gene conversion involving two var genes on two different chromosomes (Freitas-Junior et al. 2000). The physical alignment of heterologous chromosome ends presumably brings together regions of sequence homology and allows efficient DNA exchange.
Thus, in addition to any role in transcriptional silencing, the subtelomeric localization of EPA family members may simply reflect the evolutionary history and expansion of the family by efficient nonallelic recombination. Indeed, the C-terminal Ser/Thr-rich domains of EPA2-5 show evidence of domain shuffling. Thus, EPA2 and EPA4/5 are extremely closely related (Fig. 2; Table 1); however, EPA2 and EPA3 share a domain absent from EPA4/5 (diagrammed as an orange box in Fig. 2). A reasonable hypothesis would be that the ancestral parent of EPA2 and EPA4/5 underwent a duplication, and one of the two resulting genes replaced a portion of the Ser-rich region (red box in Fig. 2) with the EPA2/3-specific domain (orange box), generating EPA2. This modular construction of the C termini of the EPA genes is reminiscent of the ALS genes in C. albicans, in which different ALS genes share conserved domains within their C termini (Hoyer 2001). It is not currently known whether genomic localization of the ALS genes in C. albicans might contribute to domain sharing between different ALS family members.
Role of the EPA genes in virulence
HYR1 and EPA1-5 are all predicted by sequence to encode glucan-cross-linked cell wall proteins. HYR1 is a homolog of a cell surface protein in C. albicans which is specifically induced during hyphal growth. C. albicans strains lacking HYR1 have no defect in morphogenesis or in virulence (Bailey et al. 1996). The function of C. albicans HYR1 is not known; whatever the role of HYR1 in C. glabrata, it is not involved in hyphal formation, because C. glabrata does not make true hyphae. EPA1-5 share striking homology within the N-terminal domains, including 11 universally conserved cysteine residues. This overall structural conservation suggests that EPA2-5, like EPA1, might be lectins. We found that a strain deleted for one or both EPA clusters has a modest attenuation in virulence in a murine model of systemic candidiasis. Given that we now know of many EPA genes in addition to the five analyzed here, we hypothesize that the modest attenuation may be due in part to functional redundancy between the cloned and deleted EPA clusters and other EPA genes that we know are still present in the genome. An alternative is that the EPA genes have a more specific role in colonization of mucosal surfaces, a possibility that can be tested with a murine model of C. glabrata vaginitis (Fidel et al. 1996).
In conclusion, we have described that EPA1, an adhesin of C. glabrata, is part of a larger family of highly related genes. EPA1, EPA2, EPA3, EPA4, and EPA5 are present in two regionally silenced subtelomeric clusters. The role of these proteins in virulence is indicated by a modest in vivo mutant phenotype. A better understanding of the precise role of the various EPA family members in virulence must await further functional characterization of individual EPA genes. Lastly, analysis of the transcriptional fate of silenced loci during infection might reveal global transcriptional responses to the host environment.
Materials and methods
Strains
All strains used in this study are summarized in Supplementary Table S1.
Plasmids
All plasmids used in this study are described in Supplementary Table S2.
Primers
All primers used for cloning and sequencing in this study are described in Supplementary Table S3.
Media
Yeast media were prepared as described (Sherman et al. 1986), and 2% agar was added for plates. YPD media contained yeast extract 10g/L, peptone 20g/L, supplemented with 2% glucose. Synthetic complete media (SC) is YNB without NH2SO4 1.7 g/L, NH2SO4 5 g/L, supplemented with 0.6% of casamino acids and 2% glucose and, when needed, supplemented with uracil 25 mg/L and 1.1 g/L 5-FOA for FOA plates. YPD plates were supplemented with either G418 at 200 μg/mL or with Pen/Strep [Penicillin (100 U/mL)/Streptomycin (100 μg/mL)]. Bacterial media was prepared as described (Ausubel et al. 1987), and 1.5% agar was used for plates. Luria-Bertani (LB) media contained yeast extract 5 g/L, bactopeptone 10g/L, and NaCl 10g/L supplemented where needed with either 10μg/mL of chloramphenicol (Cm10), 30μg/mL of kanamycin (Km30), or 100 μg/mL of carbenicillin (Cb100). Phosphate buffer saline (PBS) was 8 g/L NaCl, 0.2 g/L KCl, 1.65 g/L Na2HPO4 7H2O, and 0.2 g/L KH2 PO4.
Transformation
Yeast transformations with linear or supercoiled plasmid DNA were done as described (Castano et al. 2003).
Tn7 mutagenesis
In vitro mutagenesis of plasmids using a modified Tn7 transposon was done as described (Castano et al. 2003).
Identification of the fosmid carrying EPA genes from a fosmid library
We constructed a library of 5280 C. glabrata fosmids, each of which carries a 40-Kb insert (Castano et al. 2003). We arrayed the fosmids in 55 96-well microtiter dishes, and screened for fosmids with primers specific for particular EPA genes. We first screened pools corresponding to each plate of 96 fosmids, and then rows and columns from a particular plate. A PCR screen for EPA1 (primers #498 and #499) yielded one positive fosmid, F4H12, and a screen for EPA4/5 yielded three positive fosmids, F11D8, F23H10, and F29C7 (primers #1124 and #1125).
Sequence
Fosmid sequence was obtained by in vitro mutagenesis with Tn7 as described (Castano et al. 2003). Fosmids F4H12, F11D8, F23H10, and F29C7 were digested with BglII, and the fragments were sublcloned either into pBR322 digested with BamHI or into pACYC184 digested with BstYI (see Supplementary Table S2). The plasmids with Tn7 insertions were isolated using QI mini-preps and then used as templates for sequencing with primers #661 and #662, which anneal at each end of the Tn7 reading away from the Tn7. Because the C termini of EPA2, EPA3, and EPA4/5 include several sequences repeated multiple times and because of the near identity of EPA4 and EPA5, we sequenced the fragments containing these genes by first obtaining Tn7 insertions throughout the sequence. We mapped these insertions by PCR and then obtained sequence with the Tn7 primers #661/#662. The sequence was assembled using the Sequencher program (http://www.genecodes.com/sequencher/index.html), and the amino acid sequence homology analysis was done by ClustalW alignment (Higgins et al. 1996) with the MacVector program (http://www.accelrys.com/products/macvector).
Recombination of Tn7 insertions into C. glabrata
A series of plasmids carrying regions of interest to have Tn7 insertions were mutagenized in vitro with this transposon, and the insertions were mapped and sequenced. The mutant clones were recombined in C. glabrata strains by a one-step replacement procedure: Plasmids were cut with a set of enzymes, and the linear fragments carrying the insertions were gel-purified and used to transform C. glabrata selecting for URA+ on SC-Ura plates. Insertion at the correct locus was verified with locus-specific genomic primers external to the cloned mutagenized fragment. Plasmids used for this procedure are described in Supplementary Table S2. We originally expected that there might be problems in targeting DNA by homologous recombination to subtelomeric regions. However, for the seven fragments (A-G) carrying Tn7-URA3 that were used in monitoring subtelomeric silencing, the frequency of transformation was approximately the same as for fragments targeted elsewhere in the genome (A. De Las Peñas and B. Cormack, unpubl.). For the most telomeric insertion site of the EPA4-5 locus (insertion G), ∼10% of the transformants had integrated the Tn7 correctly at the EPA4 locus, whereas 90% had integrated it elsewhere (perhaps due to the presence of this subtelomeric region multiple times in the genome; see Southern blot in Fig. 3). Insertions that had integrated correctly at this locus, however, were all silenced similarly.
Chromosome walking for sequencing and cloning telomeres
Because the telomeric repeats lack Sau3A restriction sites, our fosmid library contains no fosmids that include the telomeric repeats themselves. Therefore, to clone telomeric genomic DNA from the EPA1-3 or EPA4-5 loci, we exploited Tn7 mutagenesis of fosmid-derived fragments that were already cloned and sequenced. We mutagenized cloned fragments of genomic DNA and recombined these into the chromosomal locus by homologous recombination as detailed above. Genomic DNA was then prepared from strains carrying appropriate Tn7 insertions and digested with restriction enzymes that do not cut in the transposon. The digested DNA was then religated, and the ligation mixture was transformed into strain BW23473, which permits the R6Kγ origin present in the Tn7 to replicate (Castano et al. 2003). Plasmids generated in this way are described in Supplementary Table S2. This methodology permitted recovery of DNA adjacent to the cloned (and mutagenized) fragment. For cloning the terminal telomeric fragment of each locus, the digested DNA was first treated with T4 Polymerase (New England Biolabs) to blunt-end the telomere as well as the other end of the fragment, religated, and used to transform strain BW23473 as above. For these telomeric clones, no transformants were recovered unless the DNA was blunted with T4 polymerase, as expected, because the natural telomeric end will not be compatible with the other restricted end of the genomic fragment. Sequence analysis of these terminal clones also revealed the presence of multiple telomeric repeats, consistent with the telomeric location of these fragments.
Construction of deletion strains
To construct the unmarked deletion strains used in this study, we used the two-step gene replacement procedure. Briefly, we cloned two fragments flanking the area to be deleted in an integrative URA3 plasmid. The plasmid was linearized by digesting it at a unique site in either of the fragments, gel-purified, and then transformed into C. glabrata selecting for Ura+ transformants on SC-Ura plates. Integration of the plasmid at the correct genomic locus was confirmed by PCR analysis using one genomic primer (external to the fragments carried on the plasmid) and one plasmid primer. The integrant colonies were streak-purified on SC-Ura and plated on SC + 5FOA plates. Cells that are Ura+ die on SC + 5FOA plates. 5FOAR colonies could be obtained because the integrated plasmid is able to resolve with subsequent loss of the episome (thus rendering the cells Ura-). The resulting Ura- segregants have either regenerated the wild-type locus or have left the engineered deletion. 5-FOAR colonies were therefore analyzed by PCR and Southern blot to confirm the structure of the deletion. Using this procedure, we made strains BG465 and BG475 (hyr1 epa1 epa2 epa3)Δ, BG570and BG581 (hyr1 epa1 epa2 epa3 epa4 epa5)Δ, BG592 rap1-21, and BG676 sir3Δ (all strains and plasmids used to make them are described in Supplementary Tables S1, S2).
Mouse infections
Eight- to nine-week-old BalbC mice (Taconic) were infected with 2.2 × 107 cells in a volume of 100 μL by tail vein injection. The strains BG462, BG465, and BG475 were grown overnight in YPD, and the cells were washed with 1× PBS and resuspended in 1× PBS to a volume that will give a number of cells of 2.2 × 108/mL. The concentration of cells was determined by reading the O.D.600 nm of the culture (the concentration of BG14 cells at an O.D.600 nm of 1 is 4 × 107/mL), counting the cells in a hemocytometer and by plating serial dilutions and confirming the number of cells the following day. Ten mice per cage were kept until they were sacrificed at day 7 after infection. Kidney, liver, and spleen were retrieved from the mice, and the organs were homogenized. Dilutions of the homogenates were plated on YPD Pen/Strep plates. CFUs were scored the following day; geometric means are reported. We used the Kruskal-Wallis test to examine the significance of the differences in kidney colonization by the two mutant strains (BG465 and BG475) and the wild-type strain (BG462). We made pair comparisons for possible differences in infection outcome across the three strains. The results suggested that the strains BG465 and BG475 were significantly lower in colonization of kidney than the wild-type strain BG462 [χ2(1df) = 7, p = 0.008; χ2(1df) = 4.17, p = 0.04 respectively]. As expected, for the pairwise comparison between the two mutant strains BG465 and BG475, no appreciable difference was found.
Southern blot
Genomic DNA was isolated from wild-type strain BG14 and cut with AgeI and MfeI (New England Biolabs), and the bands separated in a 0.8% agarose gel and transferred to a Hybond-N membrane (Amersham-Pharmacia Biotech). Probe #1 (pAP481) and probe #2 (pAP479) were label using the RmT Random Primer Labeling Kit (Stratagene) with dCTPαP32. The Southern blot was done as described by Castano et al. (2003).
RT-PCR
RNA extraction: First, 2-3 mL overnight cultures were resuspended in 500μL TES (10mM Tris-Cl at pH 7.5, 0.5% SDS, 10 mM EDTA), and 500 μL acid phenol (Acros Organic, catalog no. 32710500) was added. The cells were mixed by vortexing for 1 min followed by 1 h incubation at 65°C with occasional vortexing. The tubes were placed on ice for 5 min after incubation and then centrifuged for 5 min at 4°C at 14,000 rpm. Acid phenol extraction was repeated twice without the incubation at 65°C. Fifty microliters 3 M sodium acetate (pH 5.3) was added before the last extraction, and the RNA was precipitated with 1 mL ice-cold 100% ethanol followed by a wash with 70% ethanol. RNA was resuspended in DEPC-treated water and the concentration was measured.
cDNA synthesis: Reverse transcription (cDNA synthesis) was done according to the protocol of the ThermoScript RT-PCR System (Invitrogen). Briefly, 1 μL of 10μM gene-specific primer, 3 μg RNA, and DEPC-treated water to 10μL were mixed. The RNA and primer were denatured by incubating at 65°C for 5 min, and master reaction mix was added to each tube [4 μL 5× cDNA synthesis buffer, 1 μL 0.1 M DTT, 1 μL RNaseOUT (40 U/μL), 2 μL 10mM dNTP mix, 1 μL of DEPC-treated water, and 1 μL ThermoScript RT (15 U/μL)]. For the no-RT control, 2 μL DEPC-treated water was added instead. The cDNA synthesis reaction was carried out at 55°C for EPA1, EPA3, EPA4, EPA6; 50°C for EPA2 and 60°C for ACT1 for 50min. The reaction was terminated by incubation at 85°C for 5 min. The RNA was degraded by the addition of 1 μL of RNase H and 0.2 μL of RNase cocktail (Ambion, cat. no. 2286; 500 U/mL RNase A and 20,000 U/mL RNase T1) and incubated at 37°C for 30min. Two microliters of the cDNA product was used for the PCR reaction. (primers for RT: EPA1, TAACAGTGTTTTCGTTTGAT; EPA2, GAATGATTTCCTTATTAAAT; EPA3, TAATTTGATCAG TAGCACCG; EPA4, GTCAAATTCTGTAGTGAAAG; and ACT1, GGCTTTCGATTTCTCACC).
PCR: The PCR reaction was performed as follows: Annealing temperatures were 52°C for EPA1, EPA3, EPA4/5, 50°C for EPA2, and 58°C for ACT1. Extension time was 12 sec for all reactions with 26 cycles of amplification for EPA1, EPA2, EPA3, EPA4/5 and 28 cycles for EPA2 and ACT1.
Primers: The reverse primers used were the same as those used in cDNA synthesis. Forward primers used were EPA1, GGGCTCAAAAACAGCTAAAG (143-bp product); EPA2, GG GATCAGATTATGCAAAAG (143-bp product); EPA3, GCAT GTTGATAGTTCCAAAA (143-bp product); EPA4, GCTAACA TTACTGTATTTCT (143-bp product); ACT1, GTGGCAACG GTTTGATGC (124-bp product).
Acknowledgments
We thank Win Cheung for help in identifying EPA4, and Chuan-Yu Chen for help with statistical analyses. We thank Bernard Dujon and Christophe Hennequin for communication of unpublished results. We thank Jeff Corden, Carol Greider, and members of the Cormack lab for careful reading of the manuscript. This work was funded by a UNCF-Pfizer fellowship to A.D.L.P., and by a Burroughs Wellcome Career Award, a Searle Scholars Award, and NIH grants RO1 AI46223 and 2PO1 DK49720to B.P.C.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1121003.
References
- Ai W., Bertram, P.G., Tsang, C.K., Chan, T.F., and Zheng, X.F. 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell. 10: 1295-1305. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Billington, B.L., and Gottschling, D.E. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279-1287. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. 1987. Current Protocols in Molecular Biology. J. Wiley, New York.
- Bailey D.A., Feldmann, P.J., Bovey, M., Gow, N.A., and Brown, A.J. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178: 5353-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P. and Rudenko, G. 1994. Antigenic variation in African trypanosomes. Science 264: 1872-1873. [DOI] [PubMed] [Google Scholar]
- Borst P., Rudenko, G., Taylor, M.C., Blundell, P.A., Van Leeuwen, F., Bitter, W., Cross, M., and McCulloch, R. 1996. Antigenic variation in trypanosomes. Arch. Med. Res. 27: 379-388. [PubMed] [Google Scholar]
- Brul S., King, A., van der Vaart, J.M., Chapman, J., Klis, F., and Verrips, C.T. 1997. The incorporation of mannoproteins in the cell wall of S. cerevisiae and filamentous Ascomycetes. Antonie Van Leeuwenhoek 72: 229-237. [DOI] [PubMed] [Google Scholar]
- Buchman A.R., Kimmerly, W.J., Rine, J., and Kornberg, R.D. 1988. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 210-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone R.A. and Fonzi, W.A. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9: 327-335. [DOI] [PubMed] [Google Scholar]
- Callebaut I., Courvalin, J.C., and Mornon, J.P. 1999. The BAH (bromo-adjacent homology) domain: A link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 446: 189-193. [DOI] [PubMed] [Google Scholar]
- Castano I., Kaur, R., Pan, S., Cregg, R., De Las Penñas, A., Guo, N., Biery, M.C., Craig, N.L., and Cormack, B.P. 2003. Tn7-based genome-wide random insertional mutagenesis of Candida glabrata. Genome Res. 13: 905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M., McEachern, M.J., and Blackburn, E.H. 1998. Telomeric sequence diversity within the genus Saccharomyces. Curr. Genet. 33: 83-91. [DOI] [PubMed] [Google Scholar]
- Cormack B.P., Ghori, N., and Falkow, S. 1999. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 285: 578-582. [DOI] [PubMed] [Google Scholar]
- del Portillo H.A., Fernandez-Becerra, C., Bowman, S., Oliver, K., Preuss, M., Sanchez, C.P., Schneider, N.K., Villalobos, J.M., Rajandream, M.A., Harris, D., et al. 2001. A superfamily of variant genes encoded in the subtelomeric region of Plasmodium vivax. Nature 410: 839-842. [DOI] [PubMed] [Google Scholar]
- Fidel P.L., Cutright, J.L., Tait, L., and Sobel, J.D. 1996. A murine model of Candida glabrata vaginitis. J. Infect. Dis. 173: 425-431. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior L.H., Bottius, E., Pirrit, L.A., Deitsch, K.W., Scheidig, C., Guinet, F., Nehrbass, U., Wellems, T.E., and Scherf, A. 2000. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature 407: 1018-1022. [DOI] [PubMed] [Google Scholar]
- Frieman M.B., McCaffery, J.M., and Cormack, B.P. 2002. Modular domain structure in the Candida glabrata adhesin Epa1p, a β1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 46: 479-492. [DOI] [PubMed] [Google Scholar]
- Gerberding J., Gaynes, R., Horan, T., Abshire, J., Alonso-Echanove, J., and Edwards, J. 1999. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1990-May 1999, issued June 1999. Am. J. Infect. Control 27: 520-532. [DOI] [PubMed] [Google Scholar]
- Gottschling D.E., Aparicio, O.M., Billington, B.L., and Zakian, V.A. 1990. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 63: 751-762. [DOI] [PubMed] [Google Scholar]
- Hackett J.A., Feldser, D.M., and Greider, C.W. 2001. Telomere dysfunction increases mutation rate and genomic instability. Cell 106: 275-286. [DOI] [PubMed] [Google Scholar]
- Haw R., Yarragudi, A.D., and Uemura, H. 2001. Isolation of a Candida glabrata homologue of RAP1, a regulator of transcription and telomere function in Saccharomyces cerevisiae. Yeast 18: 1277-1284. [DOI] [PubMed] [Google Scholar]
- Higgins D.G., Thompson, J.D., and Gibson, T.J. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266: 383-402. [DOI] [PubMed] [Google Scholar]
- Hoyer L.L. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9: 176-180. [DOI] [PubMed] [Google Scholar]
- Johnson L.M., Kayne, P.S., Kahn, E.S., and Grunstein, M. 1990. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 87: 6286-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapteyn J.C., Montijn, R.C., Vink, E., de la Cruz, J., Llobell, A., Douwes, J.E., Shimoi, H., Lipke, P.N., and Klis, F.M. 1996. Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester-linked β-1,3-/β-1,6-glucan hetero-polymer. Glycobiol. 6: 337-345. [DOI] [PubMed] [Google Scholar]
- Kapteyn J.C., Hoyer, L.L., Hecht, J.E., Muller, W.H., Andel, A., Verkleij, A.J., Makarow, M., Van Den Ende, H., and Klis, F.M. 2000. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol. Microbiol. 35: 601-611. [DOI] [PubMed] [Google Scholar]
- Klis F.M., de Groot, P., and Hellingwerf, K. 2001. Molecular organization of the cell wall of Candida albicans. Med. Mycol. 39: 1-8. [PubMed] [Google Scholar]
- Kurtz S. and Shore, D. 1991. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes & Dev. 5: 616-628. [DOI] [PubMed] [Google Scholar]
- Kyrion G., Liu, K., Liu, C., and Lustig, A.J. 1993. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes & Dev. 7: 1146-1159. [DOI] [PubMed] [Google Scholar]
- Lee H.W., Blasco, M.A., Gottlieb, G.J., Horner II, J.W., Greider, C.W., and DePinho, R.A. 1998. Essential role of mouse telomerase in highly proliferative organs. Nature 392: 569-574. [DOI] [PubMed] [Google Scholar]
- Liu C. and Lustig, A.J. 1996. Genetic analysis of Rap1p/Sir3p interactions in telomeric and HML silencing in Saccharomyces cerevisiae. Genetics 143: 81-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Mao, X., and Lustig, A.J. 1994. Mutational analysis defines a C-terminal tail domain of RAP1 essential for Telomeric silencing in Saccharomyces cerevisiae. Genetics 138: 1025-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.F., Kurjan, J., and Lipke, P.N. 1994. A pathway for cell wall anchorage of Saccharomyces cerevisiae α-agglutinin. Mol. Cell. Biol. 14: 4825-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.F., Montijn, R.C., Brown, J.L., Klis, F., Kurjan, J., Bussey, H., and Lipke, P.N. 1995. Glycosyl phosphatidylinositol-dependent cross-linking of α-agglutinin and β 1,6-glucan in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 128: 333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luff B., Pawlowski, L., and Bender, J. 1999. An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol. Cell. 3: 505-511. [DOI] [PubMed] [Google Scholar]
- McEachern M.J. and Blackburn, E.H. 1994. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc. Natl. Acad. Sci. 91: 3453-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P., Freeman, K., Coodly, L., and Shore, D. 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes & Dev. 8: 2257-2269. [DOI] [PubMed] [Google Scholar]
- Pfaller M.A., Messer, S.A., Hollis, R.J., Jones, R.N., Doern, G.V., Brandt, M.E., and Hajjeh, R.A. 1999. Trends in species distribution and susceptibility to fluconazole among blood stream isolates of Candida species in the United States. Diagn. Microbiol. Infect. Dis. 33: 217-222. [DOI] [PubMed] [Google Scholar]
- Pfaller M.A., Diekema, D.J., Jones, R.N., Sader, H.S., Fluit, A.C., Hollis, R.J., and Messer, S.A. 2001. International surveillance of bloodstream infections due to Candida species: Frequency of occurrence and in vitro susceptibilities to fluconazole, ravuconazole, and voriconazole of isolates collected from 1997 through 1999 in the SENTRY antimicrobial surveillance program. J. Clin. Microbiol. 39: 3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde F.E. and Louis, E.J. 1997. Saccharomyces cerevisiae telomeres. A review. Biochemistry (Mosc) 62: 1232-1241. [PubMed] [Google Scholar]
- ____. 1999. Limitations of silencing at native yeast telomeres. EMBO J. 18: 2538-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J. and Herskowitz, I. 1987. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116: 9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaberg D.R., Culver, D.H., and Gaynes, R.P. 1991. Major trends in the microbial etiology of nosocomial infection. Am. J. Med. 91: 72S-75S. [DOI] [PubMed] [Google Scholar]
- Scherf A., Figueiredo, L.M., and Freitas-Junior, L.H. 2001. Plasmodium telomeres: A pathogen's perspective. Curr. Opin. Microbiol. 4: 409-414. [DOI] [PubMed] [Google Scholar]
- Schuman P., Sobel, J.D., Ohmit, S.E., Mayer, K.H., Carpenter, C.C., Rompalo, A., Duerr, A., Smith, D.K., Warren, D., and Klein, R.S. 1998. Mucosal candidal colonization and candidiasis in women with or at risk for human immunodeficiency virus infection. HIV Epidemiology Research Study (HERS) Group [In Process Citation]. Clin. Infect. Dis. 27: 1161-1167. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink, G.R., and Hicks, J.B. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- Stringer J.R. and Keely, S.P. 2001. Genetics of surface antigen expression in Pneumocystis carinii. Infect. Immun. 69: 627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L. and Shore, D. 1991. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: Isolation of viable mutants affecting both silencing and telomere length. Proc. Natl. Acad. Sci. 88: 7749-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick W.E., Fridkin, S.K., Edwards, J.R., Hajjeh, R.A., and Gaynes, R.P. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989-1999. Clin. Infect. Dis. 35: 627-630. [DOI] [PubMed] [Google Scholar]
- Vazquez J.A., Sobel, J.D., Peng, G., Steele-Moore, L., Schuman, P., Holloway, W., and Neaton, J.D. 1999. Evolution of vaginal Candida species recovered from human immunodeficiency virus-infected women receiving fluconazole prophylaxis: The emergence of Candida glabrata? Terry Beirn Community Programs for Clinical Research in AIDS (CPCRA). Clin. Infect. Dis. 28: 1025-1031. [DOI] [PubMed] [Google Scholar]
- Walsh D.W., Wolfe, K.H., and Butler, G. 2002. Genomic differences between Candida glabrata and Saccharomyces cerevisiae around the MRPL28 and GCN3 loci. Yeast 19: 991-994. [DOI] [PubMed] [Google Scholar]
- Wong S., Butler, G., and Wolfe, K.H. 2002. Gene order evolution and paleopolyploidy in hemiascomycete yeasts. Proc. Natl. Acad. Sci. 99: 9272-9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S., Fares, M.A., Zimmermann, W., Butler, G., and Wolfe, K.H. 2003. Evidence from comparative genomics for a complete sexual cycle in the 'asexual' pathogenic yeast Candida glabrata. Genome Biol. 4: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Hayashi, M.K., Merkel, O., Stillman, B., and Xu, R.M. 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 21: 4600-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]