Abstract
To determine which E2F/RB-family members are functionally important at E2F-dependent promoters, we used RNA interference (RNAi) to selectively remove each component of the dE2F/dDP/RBF pathway, and we examined the genome-wide changes in gene expression that occur when each element is missing. The results reveal a remarkable division of labor between family members. Classic E2F targets, encoding functions needed for cell cycle progression, are expressed in cycling cells and are primarily dependent on dE2F1and RBF1for regulation. Unexpectedly, there is a second program of dE2F/RBF-dependent transcription, in which dE2F2/RBF1or dE2F2/RBF2 complexes repress gene expression in actively proliferating cells. These new E2F target genes encode differentiation factors that are transcribed in developmentally regulated and gender-specific patterns and not in a cell cycle-regulated manner. We propose that dE2F/RBF complexes should not be viewed simply as a cell cycle regulator of transcription. Instead, dE2F/RBF-mediated repression is exerted on genes that encode an assortment of cellular functions, and these effects are reversed on sets of functionally related genes in particular developmental contexts. As a result, dE2F/RBF regulation is used to link gene expression with cell cycle progression at some targets while simultaneously providing stable repression at others.
Keywords: E2F, retinoblastoma protein, RBF, Drosophila, cell cycle, differentiation
The E2F and pRB families of transcription factors play a pivotal role in cell division control (for review, see Dyson 1998; Trimarchi and Lees 2002). The six mammalian E2F family members function as heterodimers with one of two DP proteins and can be functionally subdivided into activators (E2F-1, E2F-2, and E2F-3) and repressors (E2F-4, E2F-5, and E2F-6; for review, see Trimarchi and Lees 2002). The activities of E2F1-5 are controlled by their interaction with members of the pRB family of “pocket” proteins, which negatively affect gene expression either by preventing E2F proteins from activating transcription or by recruiting transcriptional corepressor complexes to DNA. E2F/RB proteins provide a module of transcriptional regulation that couples the expression of many genes required for S-phase entry with cell cycle progression. In quiescent cells, complexes containing repressor E2Fs and pRB family members strongly repress transcription. As cells enter the cell cycle, mitogenic signals that act through G1 cyclin-dependent kinases (Cdks) lead to the disruption of E2F/pocket-protein complexes and to the replacement of repressor E2Fs with activator E2Fs at promoters, leading to high levels of expression of E2F-target genes (for review, see DeGregori 2002; Cam and Dynlacht 2003). Consequently, the two E2F groups have opposing effects on cell proliferation. Cells lacking all activator E2Fs (E2F-1, E2F-2, and E2F-3) cannot enter S phase, whereas cells deficient for E2F-4 and E2F-5 are unable to respond to some cell cycle arrest signals (Mann and Jones 1996; Gaubatz et al. 2000; Wu et al. 2001).
The simple view of E2F-dependent transcription has greatly expanded in recent years, and we highlight here three general features. First, “E2F” is a composite activity that is generated by a large number of interconnected and interrelated complexes. This suggests a complexity of function in which individual members may perform distinct tasks. Although it is clear that the collective functions of the RB and E2F families are important, the precise roles played by each of the individual components in this network are not well understood.
Second, the current models for E2F action are based, in large part, on the effects of overexpressing individual E2F proteins or selected combinations of E2F/DP and RB family members. It is not clear how accurately the behavior of overexpressed proteins portrays the normal functions of the endogenous factors. The generation of animals carrying inactivating mutations in the different members of the pathway has given a series of unexpected results (for review, see DeGregori 2002; Trimarchi and Lees 2002; Cam and Dynlacht 2003). Some of these mutant animals have tissue-specific phenotypes and in most cases the cell type-specific changes in E2F-dependent transcription that are responsible for these defects have yet to be identified.
Third, the results of several recent studies suggest that the E2F transcriptional program is far more extensive than initially suspected (for review, see Stevaux and Dyson 2002; Cam and Dynlacht 2003). Microarray analyses of the changes in gene expression caused by the overexpression of activator E2Fs (Ishida et al. 2001; Muller et al. 2001) show that E2F has the potential to affect a large number of genes with functions that extend beyond the “traditional” set of genes with S-phase functions. Similar conclusions are suggested by the analysis of genomic DNA bound by E2F proteins in vivo (Weinmann et al. 2001,2002; Ren et al. 2002). These lists of putative E2F targets contain both positive and negative regulators of cell proliferation, cell differentiation, and apoptosis, as well as a large number of genes with unknown functions, and genes with known functions that had not previously been thought to be regulated by E2F. However, transcription studies with overexpressed proteins do not necessarily define physiological target genes. Moreover, promoter binding assays do not give information about the functional significance of the interactions and cannot distinguish between positive and negative regulation. Most importantly, these studies do not delineate the functional contributions of individual family members to the broad variety of cellular functions that might be linked to E2F regulation.
Here we describe an approach that complements both the overexpression microarray and promoter binding studies that have been used to search for E2F-regulated genes. We have taken advantage of the streamlined dE2F/dDP/RBF pathway present in Drosophila, and of RNA interference (RNAi) techniques to examine the changes in gene expression that occur when each component of the dE2F/dDP/RBF network is removed. These changes in expression identify transcriptional events that depend on the endogenous dE2F/dDP/RBF proteins. The results show how the activities of the various components of the E2F/RB pathway are functionally integrated at the promoters of target genes, and demonstrate redundant, antagonistic, and unique functions for the family members. This study reveals an unexpected aspect of E2F/RB regulation. In addition to the set of “classic” E2F target genes, we found a novel group of E2F-regulated targets, which encode differentiation factors that are repressed by dE2F/RBF complexes in proliferating cells, and are expressed in developmentally regulated and gender-specific patterns.
Results
Microarray analysis of cells depleted of dE2F and RBF proteins
Drosophila contain two E2F genes (de2f1, de2f2), one DP gene (dDP), and two RB family members (rbf1 and rbf2). These have functions and activities that are very similar to their mammalian counterparts. dE2F1 and dE2F2 both dimerize with dDP, but dE2F1 is a potent activator of transcription, whereas dE2F2 is a repressor (Dynlacht et al. 1994; Ohtani and Nevins 1994; Hao et al. 1995; Sawado et al. 1998; Cayirlioglu et al. 2001; Frolov et al. 2001). Like pRB, RBF1 interacts with both repressor and activator E2Fs, whereas RBF2 is only found in a complex with the repressor, dE2F2 (Stevaux et al. 2002).
We used RNAi to determine how gene expression changes when each component of this network is removed. SL2 cells were incubated with double-stranded RNA (dsRNA), and the efficiency of depletion was assessed by Western blotting (Fig. 1A). After 4 d of treatment with the appropriate dsRNA, the levels of dE2F1, RBF1, and RBF2 were greatly reduced, compared to levels in cells treated with a nonspecific control (luciferase). To fully deplete dE2F2 it was necessary to treat SL2 cells for 8 d. To assess functional overlap between the RBF genes, or between the dE2F genes, we examined cells in which the pairs of proteins were inactivated. We were able to codeplete RBF1 and RBF2 by treating cells with both dsRNAs for 8 d. To inactivate all E2F complexes we depleted dDP, the common heterodimeric partner for dE2F1 and dE2F2, because the depletion was more efficient than the codepletion of dE2F1 and dE2F2.
Figure 1.
Specific depletion of dE2F/RBF pathway components by RNA interference. (A) Western blot analysis of whole-cell extracts following treatment with luciferase (Luc), dE2F1 (E1), dE2F2 (E2), dDP (DP), RBF1 (R1), RBF2 (R2), or RBF1 and RBF2 (R1&2). (B) Northern blot analysis of total RNA isolated from depleted cells. (C) FACS profile of cells treated for 4 d with luciferase (Luc), RBF1 (R1), RBF2 (R2), dE2F1 (E1), or dDP (DP) dsRNA; and for 8 d with luciferase (Luc), dE2F2 (E2), or both RBFs (R1&2) dsRNA.
Western blots confirmed that the depletions were specific (Fig. 1A; Frolov et al. 2003; data not shown). Removal of either dE2F1 or dE2F2 did not affect the abundance of the other protein. Consistent with the notion that dE2F1 and dE2F2 form stable complexes with dDP, the levels of both E2F proteins were reduced in dDP-depleted cells (M. Frolov, unpubl.). The levels of RBF2 rose in RBF1-depleted cells, and the Rbf2 mRNA was decreased in dE2F1 and dDP-depleted cells and increased in RBF1-depleted cells (Supplementary Table 1C, bottom panel), a phenomenon previously described for p107 in mammalian cells lacking pRB (Schneider et al. 1994).
The effects of RNAi treatment on cell cycle distribution were assessed by FACS analysis (Fig. 1C). Cells treated with dE2F2, RBF1, RBF2, or RBF1 and RBF2 had profiles similar to control treated cells. As previously reported, the depletion of dE2F1 or dDP reduced the rate of cell proliferation and generated a G1 accumulation by 8 d of treatment (Frolov et al. 2003). After 4 d of treatment with dE2F1 and dDP dsRNA, a significant portion of cells were in S phase, and this was chosen as the optimal timepoint at which protein depletion was maximal but cell cycle effects were minimal.
RNA was isolated from the depleted cells, and changes in expression of known E2F target genes were monitored by Northern blot analysis (Fig. 1B). As expected, the levels of PCNA transcripts increased significantly in RBF1-depleted cells, decreased in dE2F1-depleted cells, and were largely unaffected by the removal of dE2F2. These changes are consistent with previous studies in mutant animals that showed that the patterning of PCNA expression is primarily dependent on dE2F1-mediated activation and RBF1-mediated repression (Duronio et al. 1995; Du and Dyson 1999; Thacker et al. 2003), with de2f2 having a minor role in PCNA expression that becomes more apparent in de2f1 mutant cells (Frolov et al. 2001).
The RNA isolated from depleted cells was used to prepare labeled cRNA for hybridization to Affymetrix GeneChip oligonucleotide arrays containing sequences for ∼13,500 known or predicted Drosophila genes. Cells treated with a nonspecific dsRNA (luciferase) were used as control, and each experiment was performed in triplicate. We identified mRNAs that displayed a significant change in level following the depletion of either dE2F1 or dE2F2. The levels of such transcripts were subsequently examined in cells lacking dDP, RBF1, RBF2, or both RBFs (see Materials and Methods). Because dE2F1 and dE2F2 heterodimerize with dDP and their activities are regulated by interactions with pocket proteins, we focused our initial analysis on transcripts that were changed following the depletion of more than one component of the dE2F/dDP/RBF regulatory network.
The largest cluster of genes affected by the depletion of dE2F1 or dE2F2 contains 157 transcripts that were decreased in dE2F1-depleted cells and/or increased in dE2F2-depleted cells. The changes in this cluster are consistent with previous studies showing that dE2F1 is an activator of transcription and dE2F2, a repressor (Fig. 1B; Frolov et al. 2001). Sixty percent of these transcripts also changed in dDP-depleted cells, and 68% were increased in cells lacking RBF1 and RBF2. This cluster was studied in greater detail as described below. Two additional clusters were observed that were not investigated in this study but are worthy of further consideration. Ninetynine transcripts were found to increase in both dE2F1-depleted cells and dDP-depleted cells. This pattern mirrors changes seen in previous studies of mammalian cells in which the number of genes repressed following the ectopic expression of activator E2Fs was similar to the number of genes induced (Muller et al. 2001). However, fewer than 6% of these transcripts were affected in cells lacking RBF1 and RBF2. This suggests that the vast majority of these genes are either indirectly affected by the loss of dE2F1 or are repressed by a dE2F1/dDP complex in a manner that is independent of RBF proteins. We also observed 170 transcripts that changed in dDP-depleted cells but were not significantly altered in cells lacking either dE2F1 or dE2F2. These targets might be regulated redundantly by dE2F1/dDP and dE2F2/dDP; however, the vast majority of them (>90%) were unchanged following the removal of RBF1 and RBF2 and seem unlikely to be controlled by dE2F/RBF complexes.
dE2F1 and dE2F2 are rate-limiting for the expression of two different sets of genes
The largest cluster of dE2F1/dE2F2-dependent transcripts is displayed in Figure 2; 119 transcripts were decreased in dE2F1-depleted cells, and 47 transcripts were up-regulated in dE2F2-depleted cells. Unexpectedly, the overlap between these two sets of genes is very small.
Figure 2.
Distinct patterns of transcriptional changes upon loss of individual components of the dE2F/RBF network. Fold-change is expressed as a log2 of the difference (C) in expression level between control cells and cells depleted for dE2F1 (E1), dE2F2 (E2), dDP (DP), RBF1 (R1), RBF2 (R2), or RBF1 and RBF2 (R1&2). Color code: log2C ≤ -1.0, dark green; -1.0 ≥ log2C ≥ -0.6, light green; no change, pale yellow; 0.6 ≤ log2C ≤ 1.0, orange; log2C ≥ 1.0, red. Data set is organized by the changes in dE2F1-, dE2F2-, or dDP-treated cells. (A) Down in dE2F1 and in dDP. (B) Down in dE2F1, no change in dDP. (C) Down in dE2F1 and up in dE2F2. (D) Up in dE2F2, no change in dDP. (E) Up in dE2F2 and in dDP.
It is evident from Figure 2 that a continuous gradient exists from genes whose E2F regulation consists mostly of dE2F1-dependent activation to genes where E2F regulation exclusively requires dE2F2-mediated repression. The changes in the levels of these transcripts in cells lacking dDP are largely an integration of the patterns observed in dE2F1- and dE2F2-depleted cells. A careful examination of these patterns suggests that these E2F targets can be subdivided into five groups. At the two extremes, group A and group E, genes are deregulated in the absence of dE2F1 or in the absence of dE2F2 only, and these changes are mirrored in dDP-depleted cells, and in cells depleted of both dE2F1 and dE2F2 (Fig. 3B). Hence, E2F regulation of group A genes appears to be primarily carried out by dE2F1, whereas E-group genes are controlled by dE2F2. The second and fourth groups (B and D) contain transcripts that were changed in the absence of either dE2F1 or dE2F2, but were unaffected in dDP-depleted cells. These may represent promoters where one E2F protein gains activity, or importance, when the other is removed. Similar effects were previously noted in studies of known E2F target genes in SL2 cells and in mutant animals (Frolov et al. 2001,2003). The middle group (C) contains the small number of genes whose expression decreased in dE2F1-depleted cells and increased in dE2F2-depleted cells.
Figure 3.
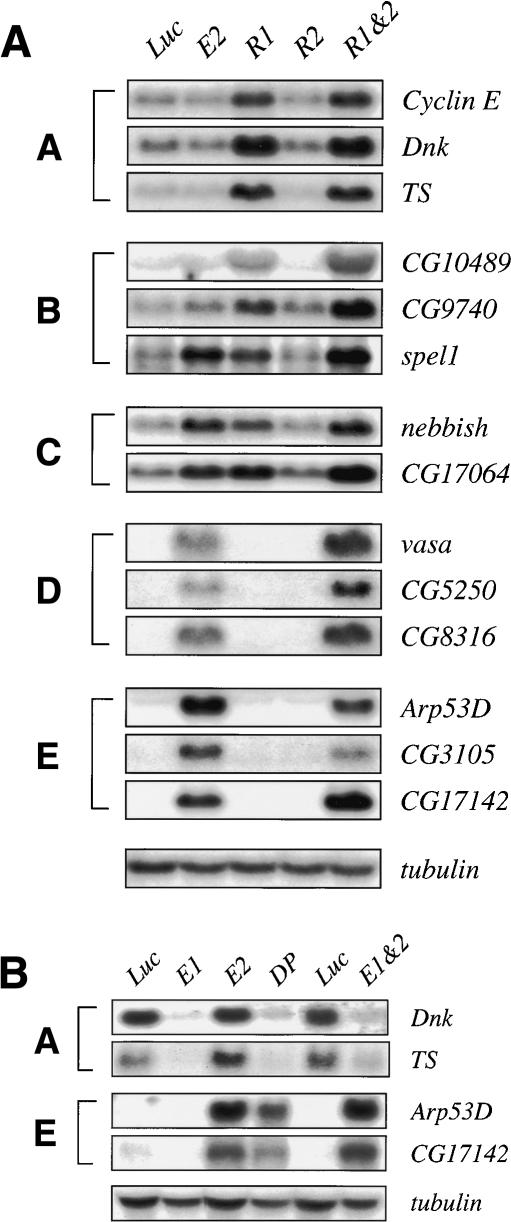
Unique and redundant functions for RBFs, and opposing roles for dE2Fs, at individual E2F-regulated promoters. (A) Northern blot analysis using probes to multiple transcripts from each group of targets illustrates differential requirements for RBF1 and RBF2. Total RNA was isolated from luciferase (Luc), dE2F2 (E2), RBF1 (R1), RBF2 (R2), RBF1 and RBF2 (R1&2). Cells were treated for 8 d with dsRNA. (B) Northern blot analysis using probes to Group A and Group E genes shows two extreme modes of E2F regulation. Total RNA was isolated from cells treated for 4 d with luciferase (Luc), dE2F1 (E1), dE2F2 (E2), dDP (DP); and for 8 d with luciferase (Luc) or with dE2F1 and dE2F2 (E1&2).
Interestingly, the different sets of E2F targets appear to represent different types of cellular functions (Table 1). A striking proportion of the transcripts that decreased in dE2F1-depleted cells provide functions required for cell cycle progression and are homologs of genes that are known or suspected to be E2F-regulated genes in mammalian cells. Groups A and B include genes that encode cell cycle regulators, DNA replication components, DNA repair proteins, proteins with mitotic functions, proteins involved in chromosome condensation and segregation, checkpoint proteins, and proteins with functions in chromatin structure and dynamics. These groups include almost all of the major categories of E2F-regulated genes that have been previously described (for a detailed list, see Supplementary Table 1). Group A also includes several genes that were not known to be E2F-regulated, such as Netrin A and Nf1, as well as numerous genes of unknown function. The composition of Groups A and B supports the idea that E2F proteins are needed to activate the transcription of genes required for G1/S progression, DNA replication (i.e., DNA synthesis and repair), and mitosis.
Table 1.
Functional classification of dE2F-regulated genes
| Function/Group | A | B | C | D | E | T |
|---|---|---|---|---|---|---|
| S phase | 3 | 0 | 0 | 0 | 0 | 3 |
| DNA replication | 16 | 12 | 0 | 0 | 0 | 28 |
| DNA repair | 2 | 7 | 0 | 0 | 0 | 9 |
| Chromatin | 8 | 3 | 0 | 0 | 0 | 11 |
| Mitosis | 9 | 4 | 2 | 0 | 0 | 15 |
| Differentiation | 0 | 0 | 0 | 1 | 7 | 8a |
| Unknown/other | 29 | 17 | 8 | 14 | 14 | 82 |
Number of genes per class of E2F regulated genes encoding various biological functions. (A-E) Classes of E2F regulated genes. (T) Total number of genes for each function.
4/8 function during oogenesis, 1/8 during germ line formation, 3/8 are male-specific as demonstrated in this study (Fig. 6).
In contrast, Groups C, D, and E lack any genes with well studied roles in S-phase entry, DNA replication, or DNA repair. Most of the 47 transcripts up-regulated in dE2F2-depleted cells encode unknown proteins, and none of the known transcripts belonging to these groups were previously thought to be controlled by E2F. Group C contains eight unknown genes and two with mitotic/meiotic functions (Septin and nebbish). Interestingly, groups D and E contain four genes important for oogenesis (vasa, Fcp3, spn-E, and bng; Burke et al. 1987; Gillespie and Berg 1995; Styhler et al. 1998; Tomancak et al. 1998; Lavoie et al. 1999), and the Drosophila homolog of the oocyte-specific Xenopus D7 protein (Smith et al. 1988). These groups also include two transcripts expressed specifically in males (Arp53D and CG8316; Fyrberg et al. 1994; FlyBase Consortium 2003), and a gene, quick-to-court, with a role in male sexual behavior (Gaines et al. 2000).
We conclude that, in SL2 cells, dE2F1 and dE2F2 are rate-limiting for the expression of two different, and nonoverlapping, sets of genes. Based on the distribution of previously studied genes, these two sets of E2F targets likely control different biological functions.
The integration of dE2F and RBF functions at E2F-regulated genes
The biological function of RBF2 was not previously known. The results presented here show that RBF2 acts redundantly with RBF1 at a subset of E2F-dependent promoters to repress transcription.
For most of the genes in Groups A and B, the changes in cells depleted of both RBFs are the same as the changes evident in RBF1-deficient cells (Fig. 3A). None of these genes were affected by the removal of RBF2. In contrast to the genes in Groups A and B, most of the genes that were strongly repressed by dE2F2 (Groups D and E) were unaffected by the removal of either RBF1 or RBF2 alone. As illustrated in Figure 3A, most of these genes were strongly deregulated in the absence of both pocket proteins. Curiously, not all of the transcripts in Groups A and B change in expression when RBF proteins are depleted, whereas every gene in the C, D, and E groups had increased levels of expression in the absence of both pocket proteins (Fig. 2).
This pattern of changes is remarkably consistent with the protein/protein interactions between RBF and E2F proteins. RBF1 and RBF2 can both repress E2F-reporter constructs but RBF2 associates specifically with dE2F2, whereas RBF1 interacts with both dE2F1 and dE2F2 (Stevaux et al. 2002). These results suggest that the functional overlap between RBF1 and RBF2 in the regulation of an E2F-dependent gene is determined, in large part, by the relative functional importance of dE2F1 and dE2F2 in the control of that gene. A unique requirement for RBF1 corresponds strongly with regulation by dE2F1, and a redundant role for RBF1 and RBF2 correlates in most cases with regulation by dE2F2.
Groups A through E contain promoters that are direct targets of E2F/RBF regulation
The gene expression analysis outlined above indicates that the functions of dE2F/RBF proteins are integrated in different ways at different promoters. How are these different patterns achieved? A trivial explanation might be that some of these groups represent direct targets for dE2F/RBF proteins, whereas others are affected indirectly.
We used chromatin immunoprecipitation (ChIP) methods to assay in vivo promoter binding of dE2Fs and RBFs. Analysis of the genomic sequences upstream of the transcriptional start sites revealed that the proximal promoter region (-800bp) of almost all the genes listed in Figure 2 contain potential E2F binding sites (see Supplementary Table 1). We designed primers to amplify the regions containing putative E2F-binding sites for 28 promoters, choosing representatives from each group, and assessed in vivo binding by ChIP. Antibodies specific for each dE2F and RBF protein were used to immunoprecipitate chromatin from SL2 cells, and coprecipitated DNA sequences were analyzed by PCR. To ensure that amplified sequences were enriched relative to a nonspecific sequence, primers for the promoter sequences of rp49, a gene whose expression is not subject to E2F regulation, were included in each PCR reaction. The DNA polymerase α promoter, at which every component of the dE2F/RBF pathway can be found (Frolov et al. 2001; Stevaux et al. 2002), was amplified from every chromatin preparation as a positive control.
In most (22/28) cases, promoter sequences were specifically enriched in dE2F and RBF immunoprecipitations, indicating that these proteins are normally present at the presumptive promoter regions. A representative ChIP analysis is shown in Figure 4A. dDP was detected at all promoters that displayed E2F binding. In six cases, we were unable to detect binding by any dE2F/RBF proteins. It is unclear whether these promoters are not directly controlled by dE2Fs and RBFs, or whether we have yet to identify the critical regulatory region. If we assume that the promoters analyzed above are representative of the groups in general, then most of the genes listed in Figure 2 are likely to be direct targets of dE2F/RBF proteins.
Figure 4.
Identification of the dE2F/RBF family members present at the promoters of each group of E2F target genes. (A) Antibodies against dE2F1, dE2F2, dDP, RBF1, RBF2, and rabbit anti-mouse serum (NS Ab) were used to enrich for the promoter regions of several E2F target genes. The promoters of Group A, B, C, and D genes associate with all proteins. Group E genes and CG8316 are not bound by dE2F1. The asterisks denote genes that were unaffected in RBF depletions. (B) dE2F1 does not bind to the promoter of CG3105, even in the absence of dE2F2. ChIP was performed with chromatin isolated from cells treated for 8 d with control (Luc) or dE2F2 (E2) dsRNA.
Which of the dE2F/RBF family members present at the groups of E2F-regulated promoters are functionally significant?
The differential requirements for dE2F/RBF family members at individual promoters could be explained in two fundamentally different ways. One possibility is that the functional differences reflect binding specificity, with individual dE2F/RBF complexes targeting specific subsets of promoters. This might occur if E2F complexes have differences in their DNA-binding properties, or if their recruitment to DNA depends on selective interactions with adjacent factors. An alternative possibility, based on the idea that most promoters have multiple regulatory elements, is that most E2F-binding sites are associated with the same sets of E2F complexes but the relative importance of dE2F1-mediated activation and dE2F2-mediated repression varies between promoters, depending perhaps on the availability of other transcription factors and the types of coactivators needed for transcription. To distinguish between these models we compared the results of the promoter binding experiments with the expression data. As described below, the results show that both models are true and contribute to the complexity of E2F regulation.
As expected, dE2F1 was found at the promoters of genes that are expressed at reduced levels in the absence of dE2F1 (A, B, and C groups). Similarly, dE2F2 was detected at promoters of C, D, and E group genes that showed elevated expression in dE2F2-depleted cells. Furthermore, dE2F1 and dE2F2 were both detected at the promoters of B and D group genes. This observation is consistent with the idea that the lack of change seen with most B, C, and D group transcripts in dDP-depleted cells reflects the antagonistic effects of dE2F1/dDP and dE2F2/dDP complexes. In each of these cases the presence of dE2F1 or dE2F2 appears to be functionally significant.
However, both dE2F1 and dE2F2 were also detected at each of the Group A promoters tested, even though the transcription patterns point to a specific requirement for dE2F1 (Fig. 3B). Indeed, to date we have been unable to find any promoters that are specifically bound by dE2F1 but not by dE2F2. Thus, dE2F2 binds to a wide spectrum of genes, but the removal of dE2F2 affects the overall level of transcription of only a few of these. In a similar manner, RBF1 and RBF2 could both be detected at almost every promoter tested, even though the microarray and Northern blot data show that some targets are dependent only on RBF1 whereas others were dependent on both RBF proteins (Figs. 2,3). We found two exceptions to this trend: DNK and CG15261, which seemed to be largely RBF1-specific, and CG8316, which gave the strongest signal with RBF2 (Fig. 4A).
This lack of correlation between the promoter binding data and the expression patterns is not due to a lack of specificity in the ChIP assay. The antibodies used are specific for the appropriate antigens. As described below, specific targets do exist, and ChIP signals can be selectively eliminated by RNAi (see below). Hence, at an E2F-regulated promoter, only some of the family members that bind are rate-limiting for the overall level of gene expression, and precisely which family members are important varies from gene to gene.
A further example of this is evident in the subset of Group A genes whose levels of expression are unchanged in RBF1/RBF2-depleted cells. Some of these genes may be indirectly affected by the cell cycle changes caused by the depletion of dE2F1 or dDP (Fig. 1C). However, ChIP analysis of promoter regions revealed dE2F1, dE2F2, dDP, RBF1, and RBF2 binding in two of the four cases examined (Fig. 4A, string and CG14283), suggesting that these are likely to be direct E2F targets. At these promoters dE2F1 is necessary for normal expression, but the release of dE2F1 from the endogenous pool of RBF1 is not sufficient to change the overall level of transcription. string and other genes with this pattern of expression provide functions that are needed for progression through G2/M. This class of targets may represent a wave of E2F-dependent transcription that occurs later in the cell cycle, and expression of these targets may require additional transcription factors.
Group E genes provide an example of a group of targets whose expression patterns correlate with a clear-cut binding specificity. In a manner that directly parallels the changes in gene expression, dE2F2, RBF1, and RBF2 were readily detected at the promoters of Group E genes, but no dE2F1 binding was evident. As an internal positive control, DNA polymerase α sequences were amplified in each PCR reaction. Despite a robust DNA polα signal, no Group E promoter sequences could be detected in dE2F1-associated chromatin. We also failed to detect dE2F1 binding at the promoter of CG8316, a Group D gene (Fig. 4A). We obtained similar results using two additional αE2F1 antibodies (Seum et al. 1996; Royzman et al. 1997). Furthermore, we find that dE2F1 cannot be detected at the CG3105 promoter even in the absence of dE2F2, demonstrating that these genes represent truly dE2F2-specific targets (Fig. 4B).
At Group E genes, dE2F/RBF regulation is uncoupled fromcell cycle position
The generally accepted model of E2F regulation is that the E2F and RB families of proteins are required for the tight coupling between the expression of E2F-regulated genes and cell cycle position. Known E2F-regulated genes are expressed in actively proliferating cells and repressed during quiescence and differentiation. Group E genes do not obviously fit this paradigm. Group E mRNAs were undetectable in actively dividing SL2 cells (Fig. 3). The lack of signal is not due to an inability to detect the transcripts, because they were easily detected when dE2F2 or both RBFs were removed. The differences in expression patterns between well-studied E2F target genes such as PCNA and RNR2 (Group A) and Group E genes are clearly illustrated in Figure 5. The expression of PCNA is restricted to the cells of the first and second mitotic waves in the developing eye disc (Fig. 5A; Frolov et al. 2001), and similarly RNR2 is expressed only in cells synthesizing DNA in stage 13 embryos (Fig. 5B; Duronio et al. 1995). This expression pattern is largely unaltered in de2f2 mutants. In contrast Arp53D, a Group E gene, is not expressed in these cells or in any cells in the eye disc or the embryo. Rather, Arp53D is inappropriately expressed in a diffuse and relatively uniform manner in de2f2 mutant animals. Thus, dE2F2 is necessary for the repression of Arp53D transcription in proliferating and nonproliferating cells in these tissues and at times when other E2F-regulated genes are expressed in a precise cell cycle-dependent and spatiotemporal manner.
Figure 5.
dE2F/RBF regulation of Group E genes is independent of cell cycle position. In situ hybridization of third instar eye discs (A) or stage 13 embryos (B) from w1118 (wild type) and de2f2-/- mutant animals with probes to PCNA or RNR2 (Group A) and Arp53D (Group E). Arp53D does not exhibit the cell cycle-regulated patterns of expression seen with PCNA and RNR2. (C) Binding analysis of dE2F/RBF proteins in S-phase cells at the promoters of DNA polymerase α (Group A) and Arp53D (Group E). dE2F2/RBF repressor complexes are not disrupted in S phase at promoters of Group E genes. (D) dE2F2/RBF complexes repress transcription of Group E genes in S phase. Northern blot analysis of total RNA isolated from S-phase cells treated with control (Luc), dE2F2 (E2), or RBF1 and RBF2 (R1&2) dsRNA.
To understand the mechanism underlying the stable repression of Arp53D we examined the proteins bound to the Arp53D promoter in S phase. Cells were synchronized in early S phase by a double HU/aphidicolin block, then released from the arrest and harvested 3 h after the release. BrdU staining showed that 85% of these cells were in S phase. ChIP analysis revealed that, at this point in the cell cycle, where classic E2F targets are expressed, only dE2F1 could be detected at the DNA polα promoter. In contrast, dE2F2, RBF1, and RBF2 remained present at the Arp53D promoter, indicating that, at Group E promoters, the dE2F/RBF repressor complexes remained intact during S phase (Fig. 5C). To verify that these proteins were actively repressing transcription, we prepared RNA from synchronized populations of S-phase cells after treatment with dE2F2, RBF1 and RBF2, or control dsRNA and analyzed the expression of several Group E genes (Fig. 5D). No mRNA was evident in control S-phase cells, but Group E transcripts were readily detected in dE2F2- or RBF1/RBF2-depleted cells. These results demonstrate that dE2F2/RBF1/RBF2 complexes actively repress transcription of Group E genes in S phase and persist at the promoter, at a time when dE2F1 drives the expression of classic E2F target genes.
dE2F2-mediated repression is required for developmentally regulated patterns of transcription
The expression patterns of several Group D and E genes have been studied previously. For instance, vasa expression is restricted to ovaries (Styhler et al. 1998), quick-to court (qtc) is known to be specific to the olfactory organs, the central nervous system, and the male reproductive tract (Gaines et al. 2000), and Arp53D is expressed in testis (Fyrberg et al. 1994). We asked, therefore, whether other Group D and E genes exhibit gender-specific expression patterns, and whether these patterns require dE2F2.
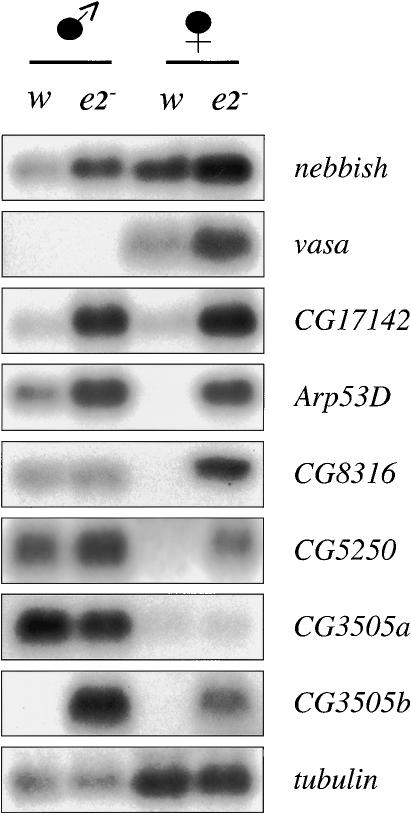
Northern blots were used to compare the levels of some of these transcripts in female and male wild-type (w) and de2f2 mutant (e2-) adults (Fig. 6). nebbish, a Group C gene that is expressed in adult animals only in the reproductive tracts of both males and females, was used as a control (Andrews et al. 2000; FlyBase Consortium 2003). It did not exhibit gender-specific expression, but was deregulated in de2f2 mutant flies. A similar pattern is seen with the Group E gene CG17142. This gene was expressed at low levels in wild-type males and females, and its expression was greatly increased in both male and female de2f2 mutants.
Figure 6.
dE2F2 is required to repress the transcription of genes with tissue- and/or gender-specific expression patterns. Total RNA isolated from w1118 (wild-type) male and female adults (w) and from de2f2 mutant male and female adults (e2-) was analyzed by Northern blot for the expression levels of several Group E and D genes and for the nebbish transcript (a Group C gene).
The most surprising results were obtained with Arp53D, CG8316, and CG5250. These genes are expressed in males, but no transcripts were detected in females (Fig. 6). Strikingly, all three of these genes were misexpressed in de2f2 mutant females. The levels of the Arp53D transcript were also elevated in male de2f2 mutant flies. Thus, dE2F2 is essential for the repression of these male-specific transcripts in wild-type females.
Not all Group D and E genes were deregulated in the same way by the loss of dE2F2. vasa, a gene that is normally only expressed in females, is expressed at higher levels in de2f2 mutant females but remained undetectable in de2f2 mutant males. We detected two different CG3505 transcripts. The faster-migrating transcript (Fig. 6, CG3505a) was expressed at high levels in males, poorly expressed in females, and was unchanged in de2f2 mutants. The slower-migrating form (Fig. 6, CG3505b) was not detected in wild-type flies, but high levels of this transcript appeared in de2f2 mutants, especially in males, and this corresponded to the CG3505 transcript detected in dE2F2-depleted SL2 cells. Taken together, these results show that dE2F2, RBF1, and RBF2 are required for the stable repression of a variety of genes. The loss of dE2F2 causes the misexpression of genes whose expression is normally male-specific (Arp53D, CG8316, CG5250), female-specific (vasa, bng, Fc3), and tissue- or stage-specific (qtc), and of genes that are expressed in both males and females (nebbish, CG17142). Clearly, Groups D and E do not represent a single transcription program. Some of these changes may well explain the defects in both male and female fertility that have been reported for de2f2 mutant animals (Cayirlioglu et al. 2001; Frolov et al. 2001).
Discussion
Here we describe the existence of a program of E2F-dependent transcription that is the very antithesis of the conventional view of E2F/RB action. The analysis of RNAi-treated cells and mutant animals reveals a group of E2F-regulated targets, the Group E genes, that are strongly repressed by dE2F2, RBF1, and RBF2 in actively proliferating cells. These dE2F/RBF-regulated genes are expressed in a variety of developmentally regulated, tissue-specific, and/or gender specific patterns.
These results challenge the dogma that E2F-regulated genes have cell cycle-regulated patterns of expression. At Group E promoters, dE2F2 and RBF proteins provide a repressor activity that is uncoupled from cell cycle progression, and the loss of E2F-mediated repression results in the inappropriate expression of tissue-specific genes and markers of differentiation.
ChIP experiments illustrate two clear-cut differences between the promoters of Group E genes and the more conventional, cell cycle-regulated E2F targets. The first distinction lies in the recruitment of the activator E2F, dE2F1. Whereas dE2F2, dDP, RBF1, and RBF2 were readily detected at most E2F-dependent genes and at each of the different groups of E2F targets uncovered in this study, dE2F1 was conspicuously and specifically absent from Group E promoters. This specificity does not, at first glance, appear to be due to a simple distinction in the types of E2F binding sites. Computer searches revealed multiple E2F-like binding sites upstream of Group E genes, but each of these variants could also be found in Group A and Group B promoters. It seems likely therefore that the specific recruitment of dE2F proteins is influenced by selective interactions with other factors, as was recently demonstrated for mammalian E2F proteins (Schlisio et al. 2002; Giangrande et al. 2003). The absence of dE2F1 at Group E promoters provides a simple mechanism to explain why these promoters escape the cell cycle-regulated burst of dE2F1-mediated activation that occurs during G1/S progression.
A second feature of Group E promoters is that dE2F2/RBF1 and dE2F2/RBF2 complexes appear to be stable and persist in S phase, at times when only dE2F1 is bound at cell cycle-regulated promoters. The implication of this result is that the activation of G1 Cdks is not sufficient to disrupt all dE2F2/RBF repressor complexes; hence, there must be an additional level of control that dictates which repressor complexes remain stable and which repressor complexes are disrupted. It is not yet clear whether dE2F2/RBF compexes remain stable despite being phosphorylated or whether they escape Cdk action. Observations that mammalian E2F4/p107 and E2F4/pRB complexes of unknown function exist in S-phase cells (Schwarz et al. 1993; Moberg et al. 1996) suggest that this type of regulation may not be unique to Drosophila. Moreover, Wells and colleagues (2003) identified human promoters that are bound by E2F1 and pRB during S phase, and found that pRB was poorly phosphorylated at some of these sites. Factors other than Cdks have been found to disrupt E2F/pocket-protein complexes (Wang et al. 1998; Timchenko et al. 1999a,b), and future studies are needed to determine how different signals may distinguish between repressor complexes at different promoters.
The discovery of E2F target genes that are regulated by dE2F and RBF proteins in a manner that is so different from the known pattern illustrates the limitations of current models for E2F action. These models are based in large part on the analysis of overexpressed E2F proteins and the detailed analysis of only a few cell cycle-regulated promoters. The mutation of E2F and RB family members in mice, flies, or worms has given a variety of unexpected and unexplained tissue-specific defects (for review, see DeGregori 2002; Stevaux and Dyson 2002; Trimarchi and Lees 2002; Cam and Dynlacht 2003). It has been suggested that these differentiation phenotypes are caused by defects in cell cycle exit prior to terminal differentiation and as a consequence of changes in the expression of cell cycle regulators. However, the discovery that dE2F/RBF complexes are needed to repress developmentally regulated genes raises the possibility that many of the changes seen in mutant animals may be due, at least in part, to the inappropriate expression of differentiation factors. For example, it seems likely that the misexpression of genes with known functions in gametogenesis and genes normally expressed in gender-specific patterns, which occurs in de2f2 mutants, contributes to the fertility defects seen in these animals (Cayirlioglu et al. 2001; Frolov et al. 2001).
Several workers have pointed out that the key to understanding the biological functions of E2F- and RB-related genes lies in understanding how this network operates over its full range of targets (DeGregori 2002; Cam and Dynlacht 2003). Which E2F proteins are important, at which promoters, and when? And what are the rules that govern how these activities are integrated? By examining the consequences of specifically and systematically removing each dE2F and RBF protein, we are able to identify transcripts that depend on each component of the Drosophila dE2F/RBF network for their normal regulation. The results give the first glimpse of how the functions of these family members are organized. The general conclusions are summarized below.
First, it is clear that there is not one program of E2F-dependent transcription. Instead, different groups of functionally related genes are coordinately regulated by subsets of dE2F/RBF family members. We separate the E2F-regulated genes into two fundamentally different categories (Fig. 7A). The first category consists of genes with functions required for cell cycle progression (DNA replication, DNA repair, chromatin structure, and mitosis) that are highly expressed during cell division. Expression of these targets is mainly dependent on dE2F1 activation. A second category, which includes the Group E genes, contains E2F targets that are not expressed in dividing cells. These genes are strongly repressed by dE2F2, and depend very little if at all on dE2F1 activation. This category includes genes with functions in gametogenesis and markers of differentiation, and these genes have an assortment of dE2F2-restricted expression patterns suggesting that they are normally expressed in distinct developmental programs.
Figure 7.
A novel picture of E2F transcriptional regulation. (A) The genes regulated by dE2F1 activation and dE2F2 repression are required for the regulation of distinct cellular functions. dE2F1 is required to activate the expression of genes involved in cell cycle progression. dE2F2, on the other hand, is required to repress a variety of tissue-specific genes involved in differentiation. The previously reported gain-of-function of dE2F2 at cell cycle promoters in the absence of dE2F1 is represented by the dotted arrow. (B) The mechanism of differential E2F regulation. The expression of dE2F1-dependent transcripts is coupled with a burst of dE2F1 expression at the G1/S transition of the cell cycle and with high G1/S Cdk activity. dE2F2-dependent transcripts are not activated by dE2F1, and dE2F2/RBF complexes present at their promoters are not disrupted by G1/S Cdks. Their expression requires tissue-specific transcription factors (X) and/or developmentally regulated signals.
Second, the functional overlap between RBF1 and RBF2 at a given promoter is tightly connected to the relative roles of dE2F1 and dE2F2. Of the 61 genes up-regulated in RBF1-depleted cells, 57 (93%) were decreased in cells lacking dE2F1. Conversely, of the 52 transcripts increased in cells lacking both RBF1 and RBF2, but not in cells lacking RBF1 alone, 37 (71%) were up-regulated in dE2F2-depleted cells. Hence, genes regulated by dE2F1 activation are mostly dependent on RBF1, whereas genes regulated by dE2F2 repression can be equally repressed by RBF1 or RBF2. These results closely parallel the pattern of protein/protein interactions between dE2F/RBF family members (Stevaux et al. 2002). The observation that RBF2 levels increase in the absence of RBF1 suggests a simple model in which RBF2 can functionally compensate for RBF1 at certain promoters. This situation closely parallels the mammalian pocket-protein network, where p107 is known to compensate for the absence of pRB during myotube formation (Schneider et al. 1994). Because RBF2 is developmentally regulated (Stevaux et al. 2002), its normal function may involve the repression of some dE2F2-regulated genes in cell types or tissues where RBF1 is limiting.
Third, E2F proteins clearly do have different target specificities. However, most E2F-regulated promoters bind all family members, and it is impossible to predict from ChIP experiments precisely which dE2F and RBF family members will be rate-limiting for overall levels of gene expression. This distinction between binding and functional significance is not limited to Drosophila cells. Recent studies in yeast and mammals have found discrepancies between promoter binding assays and experiments that test functional significance (Futcher 2002; Hsu et al. 2003). Studies in mammalian cells show that particular E2F/RB proteins are important at different sets of promoters (DeGregori et al. 1997; Hurford et al. 1997; Muller et al. 2001; DeGregori 2002 and references therein). However, ChIP experiments have failed to find any correlation with selective binding to individual E2F or pRB family members (Takahashi et al. 2000; Wells et al. 2000; Rayman et al. 2002). Thus, in most cases the key issue is not “which E2F or pRB family members bind to a promoter” but “which, of the many E2F and pRB family members that do bind, are important for function.”
Why do the contributions of individual family members vary between promoters that bind the same sets of proteins? We favor an explanation that highlights the combinatorial nature of transcriptional regulation: The contribution of any one component of the dE2F/RBF network to the transcription of any given promoter is determined by both its recruitment to the promoter and by the availability of other activators and repressors. Thus, promoters that are coordinately regulated by dE2F1/RBF1, for example, share a specific requirement for dE2F1/RBF1, rather than being the only promoters where these proteins bind. In addition, differences in affinity and timing of binding may also be important. Although relative differences in ChIP signals are difficult to interpret, we note that the intensity of the dE2F1 and dE2F2 ChIP signals varied between the groups of E2F targets in a manner that paralleled the relative importance of dE2F1 and dE2F2 in the expression patterns. Studies of the mammalian B-Myb promoter have shown that it is bound by activating E2F complexes for only a short window of time at the end of G1, whereas other cell cycle-regulated promoters are occupied by activator E2F throughout S phase (Takahashi et al. 2000; Wells et al. 2000). Such differences in occupancy might affect the strength of E2F-mediated repression and/or activation.
Finally, based on the analysis outlined above, we propose a revised view of E2F regulation in Drosophila (Fig. 7). We suggest that dE2F2-repressor complexes occupy the promoters of a diverse variety of genes. Such dE2F2-mediated repression is relieved at particular subsets of genes in response to cues that may come from developmental signals or from cell cycle signals. At cell cycle-regulated, E2F-controlled promoters, the transcriptional activation is mediated by dE2F1, and this switch from repression to activation is likely to involve Cdk-mediated disruption of the repressor complexes. However, dE2F1 fails to target other dE2F2-repressed genes, and the repressor complexes remain stable. Based on the restricted expression patterns of Group E genes, and our failure to detect dE2F1 at Group E genes even when dE2F2 is removed, we propose that dE2F2/RBF-mediated repression is relieved at these targets by developmentally regulated signals, and that gene expression is driven by factors other than dE2F1 (Fig. 7B). The notion that not all E2F-regulated genes are expressed at any one time raises the question of whether the set of targets that are induced by activator E2Fs in cycling cells is fixed or variable. Recent studies of mammalian E2F proteins show that the recruitment of activator E2Fs to a promoter involves synergistic interactions with adjacent transcription factors (Schlisio et al. 2002; Giangrande et al. 2003). It is therefore easy to imagine how the expression of genes that have the potential to be induced by activator E2Fs might also be tailored in different cellular situations to favor different subsets of targets.
Materials and methods
Cell culture, RNAi, antibodies, and cell cycle analysis
Drosophila SL2 cells were cultured and treated with dsRNA as described (Stevaux et al. 2002 and references therein). To obtain an S-phase population, cells were treated with hydroxyurea/aphidicolin essentially as described (Ina et al. 2001); for details, see the Supplemental Material. Cell cycle analysis using CellQuest (Becton Dickinson) was performed as described (Classon et al. 2000). Antibodies used for Western blot analysis have been described (Stevaux et al. 2002 and references therein).
RNA isolation and Northern blotting
Total RNA was isolated using Trizol (Invitrogen). Northern blots were performed as described (Frolov et al. 2001). Radiolabeled probes were generated by in vitro transcription with T7 RNA polymerase (Promega).
Affymetrix GeneChip and data analysis
Biotin-labeled cRNA was prepared according to the manufacturer's instructions. Total RNA, cRNA, and target (fragmented cRNA) quality were checked using the Agilent BioAnalyser (Agilent Technologies), and by hybridizations to test-chips. All targets used for hybridization to real-chips satisfied the following criteria: 3′/5′ ratios = 1.5-2.5, RawQ < 3, and Background < 80. Hybridization, washes, and staining were performed according to Affymetrix specifications. For data analysis we used the MicroArray Suite version 5.0, MicroDB, and Data Mining Tool software (Affymetrix), and Microsoft Excel. To assess significance of changes in expression, a t-test was performed with a cut-off of 0.025.
Promoter analysis and chromatin immunoprecipitations
DNA sequences were analyzed with Regulatory Sequence Analysis Tools (http://rsat.ulb.ac.be/rsat/). Chromatin immunoprecipitations were performed essentially as described (Frolov et al. 2001; Stevaux et al. 2002). dDP and dE2F1 antibodies for ChIP asasays were raised in rabbits (COCALICO Biologicals) using purified full-length recombinant proteins. Additional dE2F1antibodies were a kind gift from T. Orr-Weaver (Department of Biology, Massachussetts Institute of Technology and Whitehead Institute, Cambridge, MA) and C. Seum (Department of Zoology and Animal Biology, University of Geneva, Geneva, Switzerland).
Fly strains
Wild-type stock: w1118; de2f2 mutant stocks: trans-heterozygous combinations between the two de2f2 mutants described to date: de2f276Q1/CyO (Frolov et al. 2001) and Df(2L)E2f2329/CyO (Cayirlioglu et al. 2001).
In situ hybridization
Hybridizations of eye discs (Du 2000) and stage 13 embryos (Tomancak et al. 2002) were performed as described. Riboprobes were obtained by in vitro transcription with a DIG RNA labeling kit (Roche). Discs were developed for 20 min when labeled with a PCNA probe and for 3.5 h when labeled with Arp53D. Embryos were developed for 2 h when labeled with RNR2 and for 24 h when labeled with Arp53D.
Acknowledgments
We thank Jennifer Couget and the Bauer Center for Genomics Research at Harvard University for training and use of their facilities; Giovanni Bosco, Terry Orr-Weaver, Carole Seum and BobDuronio for providing reagents; Marie Classon for help with FACS analysis; Hristo Houbaviy for advice on RNAi; Patrick Heun and Mike Blower for sharing synchronization protocols; and Toshi Shioda for advice on data analysis. Members of the Dyson and Harlow laboratories are thanked for fruitful discussions. This work was supported in part by an individual NRSA F32 CA93045 to D.D., Rose and Jean Hoguet Foundation and BAEF fellowships to O.S., a Tosteson Postdoctoral Fellowship from MBRC to M.V.F., and by NIH grant GM53203 to N.D. M.V.F. is a Leukemia & Lymphoma Society Special Fellow.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1116703.
References
- Andrews J., Bouffard, G.G., Cheadle, C., Lu, J., Becker, K.G., and Oliver, B. 2000. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 10: 2030-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke T., Waring, G.L., Popodi, E., and Minoo, P. 1987. Characterization and sequence of follicle cell genes selectively expressed during vitelline membrane formation in Drosophila. Dev. Biol. 124: 441-450. [DOI] [PubMed] [Google Scholar]
- Cam H. and Dynlacht, B.D. 2003. Emerging roles for E2F: Beyond the G1/S transition and DNA replication. Cancer Cell 3: 311-316. [DOI] [PubMed] [Google Scholar]
- Cayirlioglu P., Bonnette, P.C., Dickson, M.R., and Duronio, R.J. 2001. Drosophila E2f2 promotes the conversion from genomic DNA replication to gene amplification in ovarian follicle cells. Development 128: 5085-5098. [DOI] [PubMed] [Google Scholar]
- Classon M., Salama, S., Gorka, C., Mulloy, R., Braun, P., and Harlow, E. 2000. Combinatorial roles for pRB, p107, and p130 in E2F-mediated cell cycle control. Proc. Natl. Acad. Sci. 97: 10820-10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J. 2002. The genetics of the E2F family of transcription factors: Shared functions and unique roles. Biochim. Biophys. Acta 1602: 131-150. [DOI] [PubMed] [Google Scholar]
- DeGregori J., Leone, G., Miron, A., Jakoi, L., and Nevins, J.R. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. 94: 7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W. 2000. Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain. Development 127: 367-379. [DOI] [PubMed] [Google Scholar]
- Du W. and Dyson, N. 1999. The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. EMBO J. 18: 916-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio R.J., O'Farrell, P.H., Xie, J.E., Brook, A., and Dyson, N. 1995. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes & Dev. 9: 1445-1455. [DOI] [PubMed] [Google Scholar]
- Dynlacht B.D., Brook, A., Dembski, M., Yenush, L., and Dyson, N. 1994. DNA-binding and transactivation properties of Drosophila E2F and DP proteins. Proc. Natl. Acad. Sci. 91: 6359-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. 1998. The regulation of E2F by pRB-family proteins. Genes & Dev. 12: 2245-2262. [DOI] [PubMed] [Google Scholar]
- FlyBase Consortium. 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31: 172-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov M.V., Huen, D.S., Stevaux, O., Dimova, D., Balczarek-Strang, K., Elsdon, M., and Dyson, N.J. 2001. Functional antagonism between E2F family members. Genes & Dev. 15: 2146-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov M.V., Stevaux, O., Moon, N.S., Dimova, D., Kwon, E.J., Morris, E.J., and Dyson, N.J. 2003. G1 cyclin-dependent kinases are insufficient to reverse dE2F2-mediated repression. Genes & Dev. 17: 723-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B. 2002. Transcriptional regulatory networks and the yeast cell cycle. Curr. Opin. Cell Biol. 14: 676-683. [DOI] [PubMed] [Google Scholar]
- Fyrberg C., Ryan, L., Kenton, M., and Fyrberg, E. 1994. Genes encoding actin-related proteins of Drosophila melanogaster. J. Mol. Biol. 241: 498-503. [DOI] [PubMed] [Google Scholar]
- Gaines P., Tompkins, L., Woodard, C.T., and Carlson, J.R. 2000. quick-to-court, a Drosophila mutant with elevated levels of sexual behavior, is defective in a predicted coiled-coil protein. Genetics 154: 1627-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubatz S., Lindeman, G.J., Ishida, S., Jakoi, L., Nevins, J.R., Livingston, D.M., and Rempel, R.E. 2000. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell 6: 729-735. [DOI] [PubMed] [Google Scholar]
- Giangrande P.H., Hallstrom, T.C., Tunyaplin, C., Calame, K., and Nevins, J.R. 2003. identification of E-box factor TFE3 as a functional partner for the E2F3 transcription factor. Mol. Cell. Biol. 23: 3707-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie D.E. and Berg, C.A. 1995. Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes & Dev. 9: 2495-2508. [DOI] [PubMed] [Google Scholar]
- Hao X.F., Alphey, L., Bandara, L.R., Lam, E.W., Glover, D., and La Thangue, N.B. 1995. Functional conservation of the cell cycle-regulating transcription factor DRTF1/E2F and its pathway of control in Drosophila melanogaster. J. Cell Sci. 108: 2945-2954. [DOI] [PubMed] [Google Scholar]
- Hsu L.-Y., Lauring, J., Liang, H.-E., Greenbaum, S., Cado, D., Zhuang, Y., and Schlissel, M.S. 2003. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity 19: 105-117. [DOI] [PubMed] [Google Scholar]
- Hurford R.K., Cobrinik, D., Lee, M.H., and Dyson, N. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes & Dev. 11: 1447-1463. [DOI] [PubMed] [Google Scholar]
- Ina S., Sasaki, T., Yokota, Y., and Shinomiya, T. 2001. A broad replication origin of Drosophila melanogaster, oriDα, consists of AT-rich multiple discrete initiation sites. Chromosoma 109: 551-564. [DOI] [PubMed] [Google Scholar]
- Ishida S., Huang, E., Zuzan, H., Spang, R., Leone, G., West, M., and Nevins, J.R. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21: 4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie C.A., Ohlstein, B., and McKearin, D.M. 1999. Localization and function of Bam protein require the benign gonial cell neoplasm gene product. Dev. Biol. 212: 405-413. [DOI] [PubMed] [Google Scholar]
- Mann D.J. and Jones, N.C. 1996. E2F-1 but not E2F-4 can overcome p16-induced G1 cell cycle arrest. Curr. Biol. 6: 474-483. [DOI] [PubMed] [Google Scholar]
- Moberg K., Starz, M.A., and Lees, J.A. 1996. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol. Cell. Biol. 16: 1436-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H., Bracken, A.P., Vernell, R., Moroni, M.C., Christians, F., Grassilli, E., Prosperini, E., Vigo, E., Oliner, J.D., and Helin, K. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes & Dev. 15: 267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K. and Nevins, J.R. 1994. Functional properties of a Drosophila homolog of the E2F1 gene. Mol. Cell. Biol. 14: 1603-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman J.B., Takahashi, Y., Indjeian, V.B., Dannenberg, J.H., Catchpole, S., Watson, R.J., te Riele, H., and Dynlacht, B.D. 2002. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes & Dev. 16: 933-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Cam, H., Takahashi, Y., Volkert, T., Terragni, J., Young, R.A., and Dynlacht, B.D. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes & Dev. 16: 245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I., Whittaker, A.J., and Orr-Weaver, T.L. 1997. Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes & Dev. 11: 1999-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawado T., Yamaguchi, M., Nishimoto, Y., Ohno, K., Sakaguchi, K., and Matsukage, A. 1998. dE2F2, a novel E2F-family transcription factor in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 251: 409-415. [DOI] [PubMed] [Google Scholar]
- Schlisio S., Halperin, T., Vidal, M., and Nevins, J.R. 2002. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 21: 5775-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J.W., Gu, W., Zhu, L., Mahdavi, V., and Nadal-Ginard, B. 1994. Reversal of terminal differentiation mediated by p107 in Rb-/- muscle cells. Science 264: 1467-1471. [DOI] [PubMed] [Google Scholar]
- Schwarz J.K., Devoto, S.H., Smith, E.J., Chellappan, S.P., Jakoi, L., and Nevins, J.R. 1993. Interactions of the p107 and Rb proteins with E2F during the cell proliferation response. EMBO J. 12: 1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C., Spierer, A., Pauli, D., Szidonya, J., Reuter, G., and Spierer, P. 1996. Position-effect variegation in Drosophila depends on dose of the gene encoding the E2F transcriptional activator and cell cycle regulator. Development 122: 1949-1956. [DOI] [PubMed] [Google Scholar]
- Smith R.C., Dworkin, M.B., and Dworkin-Rastl, E. 1988. Destruction of a translationally controlled mRNA in Xenopus oocytes delays progesterone-induced maturation. Genes & Dev. 2: 1296-1306. [DOI] [PubMed] [Google Scholar]
- Stevaux O. and Dyson, N.J. 2002. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14: 684-691. [DOI] [PubMed] [Google Scholar]
- Stevaux O., Dimova, D., Frolov, M.V., Taylor-Harding, B., Morris, E., and Dyson, N. 2002. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 21: 4927-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styhler S., Nakamura, A., Swan, A., Suter, B., and Lasko, P. 1998. Vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germ-line cyst development. Development 125: 1569-1578. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Rayman, J.B., and Dynlacht, B.D. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: Distinct E2F proteins mediate activation and repression. Genes & Dev. 14: 804-816. [PMC free article] [PubMed] [Google Scholar]
- Thacker S.A., Bonnette, P.C., and Duronio, R.J. 2003. The contribution of E2F-regulated transcription to Drosophila PCNA gene function. Curr. Biol. 13: 53-58. [DOI] [PubMed] [Google Scholar]
- Timchenko N.A., Wilde, M., and Darlington, G.J. 1999a. C/EBPα regulates formation of S-phase-specific E2F-p107 complexes in livers of newborn mice. Mol. Cell. Biol. 19: 2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko N.A., Wilde, M., Iakova, P., Albrecht, J.H., and Darlington, G.J. 1999b. E2F/p107 and E2F/p130 complexes are regulated by C/EBPα in 3T3-L1 adipocytes. Nucleic Acids Res. 27: 3621-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P., Guichet, A., Zavorszky, P., and Ephrussi, A. 1998. Oocyte polarity depends on regulation of gurken by Vasa. Development 125: 1723-1732. [DOI] [PubMed] [Google Scholar]
- Tomancak P., Beaton, A., Weiszmann, R., Kwan, E., Shu, S., Lewis, S.E., Richards, S., Ashburner, M., Hartenstein, V., Celniker, S.E., et al. 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3: RESEARCH0088.1-RESEARCH0088.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J.M. and Lees, J.A. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 3: 11-20. [DOI] [PubMed] [Google Scholar]
- Wang S., Ghosh, R.N., and Chellappan, S.P. 1998. Raf-1 physically interacts with Rband regulates its function: A link between mitogenic signaling and cell cycle regulation. Mol. Cell. Biol. 18: 7487-7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann A.S., Bartley, S.M., Zhang, T., Zhang, M.Q., and Farnham, P.J. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21: 6820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann A.S., Yan, P.S., Oberley, M.J., Huang, T.H., and Farnham, P.J. 2002. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes & Dev. 16: 235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J., Boyd, K.E., Fry, C.J., Bartley, S.M., and Farnham, P.J. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20: 5797-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J., Yan, P.S., Cechvala, M., Huang, T., and Farnham, P.J. 2003. Identification of novel pRb binding sites using CpG microarrays suggests that E2F recruits pRbto specific genomic sites during S phase. Oncogene 22: 1445-1460. [DOI] [PubMed] [Google Scholar]
- Wu L., Timmers, C., Maiti, B., Saavedra, H.I., Sang, L., Chong, G.T., Nuckolls, F., Giangrande, P., Wright, F.A., Field, S.J., et al. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414: 457-462. [DOI] [PubMed] [Google Scholar]