Abstract
A set of seven Sm proteins assemble on the Sm-binding site of spliceosomal U snRNAs to form the ring-shaped Sm core. The U7 snRNP involved in histone RNA 3′ processing contains a structurally similar but biochemically unique Sm core in which two of these proteins, Sm D1 and D2, are replaced by Lsm10 and by another as yet unknown component. Here we characterize this factor, termed Lsm11, as a novel Sm-like protein with apparently two distinct functions. In vitro studies suggest that its long N-terminal part mediates an important step in histone mRNA 3′-end cleavage, most likely by recruiting a zinc finger protein previously identified as a processing factor. In contrast, the C-terminal part, which comprises two Sm motifs interrupted by an unusually long spacer, is sufficient to assemble with U7, but not U1, snRNA. Assembly of this U7-specific Sm core depends on the noncanonical Sm-binding site of U7 snRNA. Moreover, it is facilitated by a specialized SMN complex that contains Lsm10 and Lsm11 but lacks Sm D1/D2. Thus, the U7-specific Lsm11 protein not only specifies the assembly of the U7 Sm core but also fulfills an important role in U7 snRNP-mediated histone mRNA processing.
Keywords: Small nuclear ribonucleoprotein, histone pre-mRNA 3′ processing, Sm-like protein, Sm core structure, SMN-Gemin complex
Sm and Sm-like (Lsm) proteins have been found in eukaryotes, archaea, and eubacteria. They are characterized by two closely spaced, conserved Sm motifs 1 and 2 (Hermann et al. 1995; Seraphin 1995) which adopt a fold consisting of an α-helix followed by five β-strands (Kambach et al. 1999b). A common characteristic of Sm/Lsm proteins is their tendency to form oligomers that can close into hepta- or hexameric ring structures which, in turn, control various aspects of RNA metabolism.
The seven prototype Sm proteins B/B′, D1, D2, D3, E, F, and G form the so-called Sm core structure around the conserved Sm-binding site, RAUUU/GUUGR, of the spliceosomal small nuclear RNAs (snRNAs; Lührmann et al. 1990; Raker et al. 1996; Kambach et al. 1999a; Will et al. 1999). The formation of this structure occurs in the cytoplasm and was recently shown to be ATP-dependent and mediated by specific assembly factors. A key player in this process is the “survival of motor neurons” (SMN) protein which is mutated in the neuromuscular disorder spinal muscular atrophy (for review, see Meister et al. 2002; Paushkin et al. 2002). SMN is part of the so-called SMN complex, which is composed of at least 18 distinct proteins, including all Sm proteins. In vitro reconstitution of the assembly reaction revealed that Sm proteins first associate with the SMN complex and are subsequently transferred to the U snRNA (Meister et al. 2001a; Pellizzoni et al. 2002). The functioning of the SMN complex is regulated by the PRMT5 complex. This complex introduces symmetrical dimethylarginines in Sm proteins B/B′, D1, and D3, thereby increasing their affinity for SMN (Brahms et al. 2001; Friesen et al. 2001; Meister et al. 2001b).
In contrast to the canonical Sm proteins found in the Sm core of spliceosomal small nuclear ribonucleoproteins (snRNPs) and the U7 snRNP (see below), the Lsm proteins 1-8 form alternative structures on different classes of RNAs (for review, see He and Parker 2000). Lsm 2-8 associate with the U-rich 3′ end of U6 snRNA in the nucleus, whereas Lsm 1-7 form a cytoplasmic particle involved in mRNA degradation. In addition, recent evidence indicated that Lsm proteins are associated with the U8 small nucleolar RNP (Tomasevic and Peculis 2002) and play a role in pre-tRNA maturation (Kufel et al. 2002).
Although the U7 snRNP shares some structural features with spliceosomal snRNPs, it is not involved in pre-mRNA splicing, but rather in the processing of histone mRNA 3′ ends (for review, see Müller and Schümperli 1997; Dominski and Marzluff 1999). The Sm-binding site of U7 snRNA has a noncanonical sequence, and affinity-purified U7 snRNPs contain the Sm proteins B/B′, D3, E, F, and G but lack Sm D1 and D2 (Pillai et al. 2001). A 14-kD subunit of U7 snRNPs was identified as a new Sm-like protein, termed Lsm10, with high similarity to Sm D1. Based on these findings, we postulated that Lsm10 and an additional Sm-like protein might replace the missing Sm D1 and D2 proteins in a U7-specific Sm core structure. A candidate for this missing subunit was an ∼50-kD protein identified in purified U7 snRNPs (Pillai et al. 2001).
Here we characterize the 50-kD protein as Lsm11, a new, unusual member of the Sm/Lsm protein family, which appears to replace Sm D2. This unique Sm core organization led us to ask whether the U7-specific Lsm proteins functionally contribute to histone RNA 3′ processing, and how the formation of the U7-specific Sm core is specified. We show that Lsm11 is involved in both of these processes. We demonstrate a functional role in histone RNA processing for the long N-terminal extension of Lsm11. In contrast, the C-terminal part of Lsm11 encompassing its two Sm motifs, together with Lsm10 (Pillai et al. 2001) and the special Sm-binding site of U7 snRNA, all participate in specifying the formation of the U7 Sm core. Further, we provide evidence that, like the assembly of spliceosomal U snRNPs, assembly of the U7 snRNP is ATP-dependent and factor-mediated. However, we show that U7 snRNP assembly is not mediated by the SMN complex that mediates spliceosomal U snRNP assembly. Rather it is facilitated by a specialized SMN complex that lacks Sm D1 and D2 but contains the U7-specific proteins Lsm10 and Lsm11. Thus, our data suggest that both the assembly and the function of the U7 snRNP are specified by its unusual set of Sm/Lsm proteins.
Results
Sequence of the U7-specific 50-kD protein Lsm11
To characterize the 50-kD U7 snRNP protein, we isolated it from a preparative SDS-polyacrylamide gel of affinity-purified U7 snRNPs (Pillai et al. 2001) and identified trypticpeptides by nano-electrospray tandem mass spectrometry coupled to the Sequest database search algorithm (Chittum et al. 1998). A total of 11 identified peptides were found to be encoded by related human and mouse expressed sequence tags (ESTs). From these, a complete cDNA sequence was reconstructed using additional overlapping ESTs and the human genomicsequence on chromosome 5 (AC026407). The human gene, represented by UniGene cluster Hs.187117, encodes a protein of 360 amino acids (Fig. 1). The sequence of the corresponding mouse protein (UniGene Mm.45659, 361 amino acids) is 89% identical to the human one. We note that the Xenopus homolog (AF514310) is shorter by 69 amino acids.
Figure 1.
Sequence alignment of Lsm11 proteins of human (Hs; SwissProt P83369; UniGene Hs.187117; genomicsequence AC026407), mouse (Mm; GenBank AF514309; UniGene Mm.45659; LocusLink 72290) and X. laevis (Xl; GenBank AF514310; UniGene Xl.13277). Residues conserved between at least two proteins are shown in inverse print. The conserved Sm motifs 1 and 2 are indicated by shaded boxes. The consensus sequence was adapted from previous sources (Hermann et al. 1995; Achsel et al. 1999). (h) Hydro-phobicamino acids.
The identified protein contains the conserved Sm motifs 1 and 2 (Hermann et al. 1995; Seraphin 1995) and will hence be called Lsm11. However, very unusual for an Sm/Lsm protein, the two Sm motifs are separated by 138 amino acids in human and mouse, and by 99 amino acids in Xenopus Lsm11. Most of this intervening sequence is not conserved between mammals and amphibia. Moreover, Lsm11 has an extensive N-terminal extension, but lacks a C-terminal tail following Sm motif 2.
Lsm11 can be cross-linked to U7 snRNA
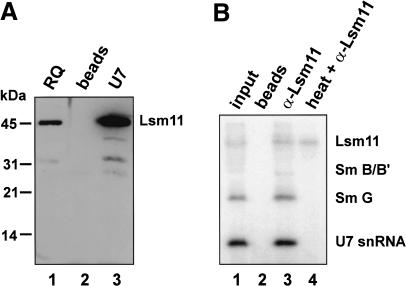
To test whether Lsm11 is an integral U7 snRNP component, affinity-purified polyclonal antibodies against recombinant Lsm11 were used to detect Lsm11 by Western blotting. The antibody reacted with a single protein of 45-50 kD in a Resource Q fraction from HeLa cell nuclear extract enriched for U7 snRNPs (Fig. 2A, lane 1); no signal could be detected in unfractionated whole-cell or nuclear extract, presumably because of the low abundance of Lsm11. The band seen in the Resource Q fraction could be enriched by precipitation with a biotinylated oligonucleotide complementary to the 5′ end of U7 snRNA and magneticstreptavidin beads (Fig. 2A, lane 3), but not by precipitation with beads alone (Fig. 2A, lane 2), indicating that the detected protein is indeed part of the U7 snRNP.
Figure 2.
Lsm11 can be cross-linked to U7 snRNA. (A) Polyclonal antibodies to Lsm11 recognize a specific 45-50-kD polypeptide. Samples of a Resource Q U7 peak fraction (RQ; Pillai et al. 2001) were separated by SDS-PAGE, Western blotted, and analyzed with affinity-purified Lsm11 antibody (lane 1). The same band was also detected by the Lsm11 antibody in affinity-purified U7 snRNPs (U7, lane 3), but not in a control precipitation with beads alone (lane 2). (B) Characterization of a UV-cross-link between Lsm11 and U7 snRNA. (Lane 1) Nuclear extract was subjected to UV irradiation and, after incubation with a radiolabeled oligodeoxynucleotide complementary to the 5′ end of U7 snRNA, subjected to SDS-PAGE and autoradiography. The detected bands correspond to noncross-linked U7 snRNA and UV-adducts to Sm G, B/B′ (very faint), and an ∼50-kD polypeptide, respectively (Mital et al. 1993). (Lane 3) Before oligonucleotide annealing and SDS-PAGE, the extract was subjected to immunoprecipitation by Lsm11 antibody; all UV adducts as well as noncross-linked U7 snRNA were precipitated as parts of intact U7 snRNPs. (Lane 2) Precipitation as in lane 3, but without antibody. (Lane 4) Same as in lane 3, except that the extract was boiled prior to immunoprecipitation to denature snRNPs; only the uppermost UV-adduct was precipitated, indicating that it contains Lsm11.
We showed previously that the Sm proteins G and B/B′, as well as proteins of either 50 kD in mouse nuclear extracts (Mital et al. 1993) or 40 kD in Xenopus oocytes (Stefanovic et al. 1995a), can be cross-linked by UV irradiation to the Sm-binding site of U7 snRNA. Based on its size in mammals and Xenopus, and on its affiliation with the Sm/Lsm protein family (Fig. 1), Lsm11 was a likely candidate for the third cross-linkable protein. To test this hypothesis, we analyzed UV adducts formed in HeLa nuclear extract. U7 snRNA and its photoadducts were detected by annealing a radiolabeled oligonucleotide complementary to the 5′ end of U7 snRNA prior to SDS-PAGE. As expected, we observed the three UV adducts described above for mouse nuclear extracts (Fig. 2B, lane 1). When Lsm11 was immunoprecipitated from the UV-irradiated nuclear extract, uncross-linked U7 snRNA and all three photoadducts were coprecipitated (Fig. 2B, lane 3), but no labeled products were precipitated by beads lacking antibody (Fig. 2B, lane 2). Significantly, after boiling the UV-irradiated nuclear extract, only the 50-kD photoadduct was immunoprecipitated (Fig. 2B, lane 4), demonstrating that this adduct indeed contains the Lsm11 protein.
The N terminus of Lsm 11 is essentialfor histone RNA 3′ end processing
To investigate whether Lsm11 plays a role in histone RNA processing, we first used a special type of assay. We had noticed that a chimeric RNA containing the processing region of histone pre-mRNA physically linked to U7 snRNA sequences (Fig. 3B), when injected into Xenopus oocytes, remained cytoplasmic but was cleaved at the histone processing site (Stefanovic et al. 1995b). This cleavage, like the normal nuclear reaction, was dependent on all conserved sequences of the histone premRNA and U7 snRNA moieties, and these had to be present in cis on the same polynucleotide chain.
Figure 3.
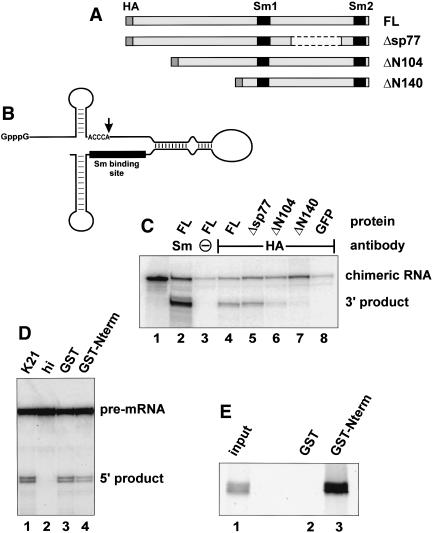
The N terminus of Lsm11 is essential for histone RNA 3′ end processing. (A) Structure of various constructs containing murine Lsm11. An HA tag introduced at the N terminus and the Sm motifs are shown in dark gray and black, respectively. (B) The chimeric histone-U7 RNA (12/12-U7 RNA; Stefanovic et al. 1995b) used in C contains 49 nt of histone pre-mRNA upstream and 36 nt downstream of the cleavage site (vertical arrow), a connector segment of 28 nt, and 65 nt of U7 RNA sequence. The Sm-binding site is indicated by a black bar. (C) Syntheticm RNAs encoding HA-tagged versions of Lsm11 (see A) or of GFP were injected into the cytoplasm of X. laevis oocytes. After overnight incubation to allow for translation of the recombinant proteins, the oocytes were challenged with radiolabeled, chimeric histone-U7 RNA (see B) and further incubated for 3 h. Cytoplasmic extracts were subjected to immunoprecipitation with either anti-Sm or anti-HA antibodies to enrich for total snRNPs or for particles containing HA-tagged Lsm11, respectively, and the radiolabeled RNA was analyzed by denaturing polyacrylamide gel electrophoresis and autoradiography. (Lane 1) Input chimeric RNA. (Lane 3) Precipitation of HA-mLsm11FL extract with beads lacking antibody. (D) Lsm11 N terminus sequesters histone RNA processing factor(s) from K21 mouse mastocytoma cell nuclear extract (Stauber et al. 1990). Note that processing by the extract (lane 1) is strongly reduced by preincubation of the extract with immobilized GST-tagged N terminus of Lsm11, followed by removal of the bound material (lane 4), but is only slightly affected by a similar treatment with immobilized GST (lane 3). (Lane 2) Incubation in extract inactivated by 15 min preincubation at 50°C. (E) Lsm11 N terminus binds to ZFP100 (Dominski et al. 2002) in vitro. Radiolabeled in vitro translated ZFP100 was incubated with glutathione beads preloaded with GST (lane 2) or GST-mLsm11N-term (lane 3), and the bound material was analyzed by SDS-PAGE and autoradiography. Lane 1 shows 1/10 volume of input material.
Importantly, both the input RNA and the 3′ cleavage product contain Sm/Lsm proteins bound to the U7 Sm-binding site, and this can be exploited to analyze by immunoprecipitation whether a complex containing a specific protein is functional in processing. Thus we injected synthetic mRNAs encoding HA-tagged full-length murine Lsm11 cDNA (HA-mLsm11FL) and various HA-tagged deletion mutants (see Fig. 3A) into oocytes and allowed for their translation. The oocytes were then challenged with the radiolabeled, chimeric histone-U7 RNA. After a 3-h incubation, cytoplasmic extracts were prepared and subjected to immunoprecipitation with either anti-Sm or anti-HA antibodies to enrich for total snRNPs or for particles containing HA-tagged Lsm11, respectively. When the labeled RNA from oocytes injected with HA-mLsm11FL mRNA was analyzed by polyacrylamide gel electrophoresis, the anti-Sm immunoprecipitate revealed some uncleaved chimeric RNA, but a large fraction of the RNA was cleaved (Fig. 3C, lane 2). Note that this immunoprecipitate contained RNP complexes assembled with endogenous Lsm11 as well as complexes containing HA-mLsm11FL. Precipitations carried out with beads in the absence of antibody (Fig. 3C, lane 3) or with beads and anti-HA antibodies in oocytes expressing HA-tagged green fluorescence protein (GFP; Fig. 3C, lane 8) contained low amounts of nonspecifically precipitated input RNA, but no cleaved RNA. In the oocytes expressing HA-mLsm11FL, both processed and unprocessed RNAs were precipitated by anti-HA antibody (Fig. 3C, lane 4), indicating that the chimeric snRNPs assembled with HA-mLsm11FL were functional, and providing a reference for the analysis of Lsm11 deletion mutants described below. As expected, the amount of labeled RNA precipitated was lower than with anti-Sm antibodies, suggesting that only a small proportion of the formed complexes contained HA-mLsm11FL.
An internal deletion of 77 amino acids in the spacer between the two Sm motifs did not substantially alter the proportion of cleaved RNA (Fig. 3C, lane 5; Δsp77). In contrast, the cleavage activity was reduced by removing the first 104 amino acids (Fig. 3C, lane 6; ΔN104) and completely abolished by deleting 140 amino acids at the N terminus of Lsm11 (Fig. 3C, lane 7; ΔN140). The amount of input RNA recovered for ΔN104 was somewhat lower than for the full-length protein, but still higher than for the beads and HA-GFP controls. However, such a decrease was not evident for ΔN140 and, moreover, transfection experiments in mammalian cells indicated that all of the truncated proteins were still functional for assembly into U7 snRNPs (see Fig. 4, below). Furthermore, all four HA-tagged Lsm11 variants were expressed in similar amounts and could be coprecipitated by anti-Sm antibodies to a similar extent (data not shown).
Figure 4.
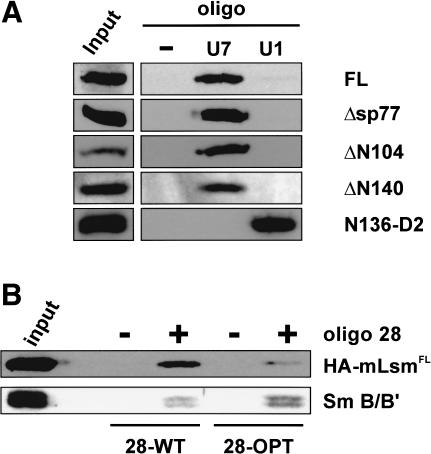
Association of HA-tagged Lsm11 with U7, but not U1, snRNA. (A) Human 293T cells were transiently transfected with plasmids encoding the various HA-tagged Lsm11 proteins shown in Figure 3A or a fusion between HA tag, the first 136 amino acids of Lsm11, and Sm D2 (N136-D2). Nuclear extracts were incubated with biotinylated oligonucleotides complementary to the 5′ ends of U7 or U1 snRNA and precipitated with magnetic streptavidine beads. The samples were subjected to SDS-PAGE and immunoblotted with anti-HA antibody. (-) Precipitation by beads without oligonucleotide; (input) sample of original nuclear extract. (B) Western blots detecting Lsm11 and Sm B/B′ proteins associated in 293T cells with U7 snRNAs containing either the wild-type Sm-binding site or a consensus Sm-binding sequence derived from spliceosomal snRNAs. The cells were cotransfected with plasmids encoding either Sm WT or Sm OPT U7 RNA (Stefanovic et al. 1995a), modified with a 28-nt sequence tag at the 5′ end (Pillai et al. 2001), and with the plasmid encoding HA-mLsm11FL. Nuclear extracts were prepared and processed as described for A, except that precipitations were performed with magnetic streptavidine beads with (+) or without (-) biotinylated oligonucleotide complementary to the 28-nt tag. Anti-HA and Y12 anti-Sm antibodies were used to detect HA-mLsm11FL and Sm B/B′, respectively. (input) Sample of original 28-OPT extract.
These experiments indicated that the N terminus of Lsm11 is essential for histone pre-mRNA cleavage. This was also supported by histone RNA processing reactions carried out in nuclear extract from K21 mouse mastocytoma cells (Stauber et al. 1990); in such a reaction, a histone H4 pre-mRNA substrate was cleaved, resulting in the formation of two 5′ cleavage products differing by two nucleotides (Fig. 3D, lane 1). Incubation of the extract with immobilized GST-tagged N terminus of Lsm11 (the first 136 amino acids), followed by removal of the bound material, led to an approximately twofold reduction in processing activity (Fig. 3D, lane 4), whereas only a minimal reduction was observed with immobilized GST (Fig. 3D, lane 3). This difference was not due to a reduction in U7 snRNA concentration, as was revealed by a primer extension analysis (data not shown). Rather, this result suggested that the N-terminal fragment sequestered an important processing factor.
A good candidate for this factor is the 100-kD zinc finger protein (ZFP100) that has been implicated in stabilizing the interaction between histone pre-mRNA and the U7 snRNP (Dominski et al. 2002; see Discussion). Indeed, in vitro translated ZFP100 was found to bind in vitro to GST-tagged Lsm11 N terminus (Fig. 3E, lane 3) but not to GST alone (Fig. 3E, lane 2). Moreover, the N terminus of Lsm11 and ZFP100 also interacted in a yeast two-hybrid assay (A. Gruber and D. Schümperli, unpubl.). These data therefore suggested that Lsm11 exerts its essential role in histone 3′-end processing at least in part through an interaction with ZFP100.
The C-terminalSm domain of Lsm11 and the special Sm-binding site of U7 snRNA are determinants for forming the U7 Sm core
Although the above experiments indicated that the N terminus and more than half of the sequence between the two Sm motifs are not required for Lsm11 to interact with the chimeric RNA, we wanted to analyze more specifically the ability of the truncated proteins to assemble into U7 snRNPs in mammalian cells and to discriminate between U7 and U1 snRNA. For this purpose, we expressed HA-mLsm11FL and various deletion mutants in human 293T cells by transient transfection of appropriate expression plasmids. U7 or U1 snRNPs were precipitated from nuclear extracts using biotinylated oligonucleotides complementary to the 5′ end of the corresponding snRNA. The HA-tagged proteins were then detected in the precipitates by Western blotting with anti-HA antibody. Neither the two N-terminal deletions nor the internal deletion removing 77 amino acids between the two Sm motifs abolished the incorporation of Lsm11 into U7 snRNPs (Fig. 4A). Moreover, all three truncated proteins associated more efficiently with U7 than with U1 snRNA, similar to the full-length protein. Thus, the C-terminal Sm domain containing the two Sm motifs is not only sufficient for snRNP assembly in general, but also carries the specificity for preferential association with U7 snRNA.
This point was corroborated by an additional finding. As Lsm10 is very similar to Sm D1 and therefore Lsm11 presumably replaces Sm D2 in the U7-specific Sm core, we fused the N-terminal 136 codons of murine Lsm11 to codon 1 of human SmD2 and analyzed the association of the resulting protein with U7 and U1 snRNAs after transfection into 293T cells. In contrast to HA-mLsm11FL and its various deletion mutants, this Sm D2 fusion was preferentially incorporated into U1 snRNPs (N136-D2; Fig. 4A) and, in this respect, behaved similarly to endogenous Sm D2.
Replacing the original Sm-binding site of U7 snRNA (AAUUUGUCUAG; Sm WT) with a consensus sequence derived from spliceosomal snRNAs (AAUUUUUGGAG; Sm OPT) was previously shown to result in U7 snRNPs that did not function in histone RNA 3′ processing (Grimm et al. 1993; Stefanovic et al. 1995a) and in the abrogation of the ∼40-kD cross-link in Xenopus oocytes (Stefanovic et al. 1995a). Most likely, this lack of functionality was due to a failure of U7 Sm OPT RNA to incorporate Lsm11, especially given the above finding that Lsm11 is required for histone RNA processing. We therefore investigated which type of Sm-binding sequence would allow association of U7 RNA with Lsm11. For this purpose, HA-mLsm11FL protein was expressed in 293T cells along with two different versions of U7 snRNA. Both contained an identical 5′-terminal tag of 28 nucleotides, but the Sm-binding site either had the Sm WT (28-WT RNA) or the Sm OPT sequence (28-OPT RNA). After precipitation of the RNAs from nuclear extracts with a biotinylated oligonucleotide complementary to the 28-nucleotide tag, we indeed found HA-mLsm11FL to be present in higher amounts in the affinity-enriched 28-WT snRNPs than in the corresponding sample from cells transfected with 28-OPT (Fig. 4B). When comparing the band intensities, note that the 28-WT snRNA accumulates in the nucleus approximately three times less efficiently than 28-OPT RNA (Grimm et al. 1993; Pillai et al. 2001). This was confirmed by reprobing the Western blot with Y12 anti-Sm antibody, which detects the Sm B/B′ protein present in both the 28-WT and 28-OPT snRNPs (Fig. 4B). Thus, specific incorporation of Lsm11 into U7 snRNPs is to a large extent dictated by the special Sm-binding site of U7 snRNA.
U7 Sm core assembly is active and factor-mediated
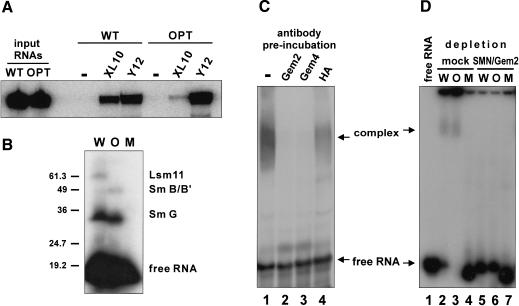
Next, we asked how the unique U7 Sm core domain is formed in the environment of the cell. To address this question, we used an in vitro assembly system based on Xenopus egg extract that faithfully mimics in vivo conditions (Müller et al. 2000). Initially we tested whether incubation of wild-type U7 snRNA in this extract resulted in the formation of the U7-specific Sm core domain. In vitro-transcribed Sm WT and Sm OPT U7 snRNAs were incubated with egg extract and subsequently immunoprecipitated with antibodies directed against Sm core components. The Y12 antibody coprecipitated both the Sm WT and Sm OPT U7 RNAs, indicating that both RNAs are associated with Sm proteins (Fig. 5A). In contrast, precipitation by an antibody against Xenopus Lsm10 was only efficient with Sm WT and very inefficient with Sm OPT RNA, indicating a strong preference of Lsm10 for the special Sm-binding site of U7 snRNA also in this in vitro assembly system. Because antibodies reacting with Xenopus Lsm11 were not available, but Lsm11 can be cross-linked to U7 sn-RNA (Fig. 2B), we performed UV-cross-linking experiments to detect Lsm11 in the assembled complexes. As expected, a UV-adduct corresponding to Sm G could be generated after in vitro assembly of both Sm WT and Sm OPT U7 RNAs (Fig. 5B, lanes W,O, respectively). Note the somewhat higher electrophoretic mobility of this UV-adduct with Sm OPT RNA, which was observed previously after assembly in Xenopus oocytes microinjected with these same U7 snRNAs (Stefanovic et al. 1995a). A band previously identified as a UV-adduct with Sm B was observed for Sm OPT RNA, but was too faint to be detected for Sm WT RNA (this strong difference in intensity also agrees with earlier findings from oocyte injections; Stefanovic et al. 1995a). Importantly, only Sm WT, but not Sm OPT U7 RNA could be cross-linked to a protein of ∼40 kD which, by analogy with Figure 2B and based on our previous mapping of this cross-link to the Sm-binding site of U7 snRNA (Stefanovic et al. 1995a), most likely represents Lsm11. In contrast, an Sm-binding site mutant of U7 snRNA (Sm MUT) that does not assemble into snRNP complexes (Grimm et al. 1993; Stefanovic et al. 1995a; Müller et al. 2000) did not form any UV adducts (Fig. 5B, lane M). As the cross-linking patterns of the three RNAs exactly corresponded to those previously characterized after assembly in microinjected Xenopus oocytes (Stefanovic et al. 1995a), these results are compatible with the notion that the assembly system based on Xenopus egg extract faithfully recapitulates U7 snRNP assembly as it occurs in vivo.
Figure 5.
U7 snRNP assembly in Xenopus egg extract is mediated by an SMN complex. (A) Detection of Xenopus Lsm10 in assembled particles. Radioactively labeled Sm WT or Sm OPT U7 snRNAs (Stefanovic et al. 1995a) were incubated with X. laevis egg extract at 20°C for 30 min. Immunoprecipitations were performed with beads lacking antibody (-), polyclonal antiserum against X. laevis Lsm10 (XL10), or monoclonal anti-Sm antibody Y12. RNA extracted from the precipitates was subjected to denaturing polyacrylamide electrophoresis and autoradiography. (B) Detection of Lsm11 photoadduct in assembled particles. Sm WT (W), Sm OPT (O) U7 snRNAs, or U7 snRNA containing a severely mutated Sm-binding site (Sm MUT; M) were subjected to assembly as in A. The products were UV-irradiated and then subjected to SDS-PAGE and autoradiography. The molecular masses (in kilodaltons) of marker proteins and the positions of the free U7 snRNAs and of photoadducts with the Xenopus Sm G, Sm B/B′, and Lsm11 proteins are indicated. (C) Preincubation of extracts with antibodies against SMN complex components inhibits U7 snRNP assembly. Sm WT U7 snRNA was subjected to assembly as in A. The reaction was subsequently analyzed by electrophoresis on a 5% nondenaturing polyacrylamide gel (lane 1). In the other lanes, the egg extracts were preincubated with anti-Gemin2 (lane 2), anti-Gemin4/GIP (lane 3), and anti-HA (lane 4) antibodies. (D) Immunodepletion of extracts for SMN complex components prevents U7 snRNP assembly. The extracts were either mock-depleted (lanes 2-4) or immunodepleted with antibodies against SMN and Gemin2 (lanes 5-7). Assembly reactions were performed as in A, using Sm WT (W), Sm OPT (O), or Sm MUT (M) U7 snRNAs. (Lane 1) U7 Sm WT RNA in the absence of extract.
Next, we analyzed whether U7 snRNP assembly is an active and factor-mediated process as had been reported for spliceosomal U snRNPs (Meister et al. 2001a; Pellizzoni et al. 2002). When an assembly reaction was analyzed by native gel electrophoresis, a complex migrating significantly slower than the RNA alone was observed (Fig. 5C, lane 1). Control experiments indicated that this complex could be supershifted by Y12 anti-Sm antibody (data not shown) but was not formed on U7 Sm MUT snRNA (Fig. 5D, lane 4), consistent with earlier findings (Müller et al. 2000). Thus, the slowly migrating complex represented the assembled U7-specific Sm core structure that could be immunoprecipitated by Y12 and Lsm10 antibodies (Fig. 5A).
Several lines of evidence suggested that U7 snRNP assembly in the egg extract is energy-dependent and mediated by an SMN complex. First, incubation of the assembly reaction at 4°C instead of 20°C or in the presence of ATP-γS instead of ATP strongly impaired complex formation (data not shown). Previous findings had shown that these conditions block U1 snRNP assembly (Meister et al. 2001a). Second, preincubations with antibodies against Gemin2 and Gemin4/GIP abolished assembly (Fig. 5C, lanes 2,3, respectively), whereas a control antibody directed against the HA tag had only a minimal effect (Fig. 5C, lane 4). Third, and most importantly, immunodepletion of SMN and Gemin2 from the egg extract completely blocked snRNP formation with both Sm WT and Sm OPT U7 snRNAs (Fig. 5D, lanes 5,6), whereas a mock-depleted extract was still active (Fig. 5D, lanes 2,3). As expected, no complexes were formed with U7 Sm MUT RNA (Fig. 5D, lanes 4,7). Taken together, these experiments indicated that the formation of the U7 Sm core is an ATP-dependent process mediated by an SMN complex.
Evidence for separate SMN complexes containing either Sm D1/D2 or Lsm10/11
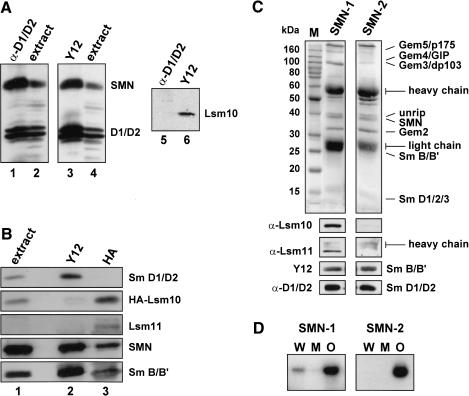
The Sm core domain of the U1 snRNP is generated by an SMN complex that contains all canonical Sm proteins. By analogy, we surmised that assembly of U7 snRNPs would be mediated by a specialized SMN complex containing the Sm proteins B/B′, D3, E, F, G, and, additionally, the U7-specific Lsm proteins 10 and 11. Indeed, Lsm10 and Lsm11 were detectable in purified SMN preparations from HeLa cells by Western blot analysis (Fig. 6C, lane SMN-1). However, compared to the other Sm proteins, Lsm10 and Lsm11 were underrepresented. This finding raised the question of whether an SMN complex containing all Sm proteins was able to bind Lsm10 and Lsm11 additionally, or whether binding of these proteins was mutually exclusive with binding of other Sm proteins such as Sm D1/D2.
Figure 6.
Association of Lsm10 and Lsm11 with a specialized SMN complex. (A) Lsm10 and Sm proteins D1/D2 are associated with separate SMN complexes. Cytoplasmic extract from HeLa cells was immunoprecipitated with an antibody reacting with Sm D1/D2 (lanes 1,5) or with Y12 anti-Sm antibody (lanes 3,6). (Lanes 2,4) Samples of the extracts prior to precipitation. The blots were probed with antibodies specific for SMN, D1/D2, or Lsm10 as indicated. (B) HA-tagged Lsm10 and Sm proteins D1/D2 associate with separate SMN complexes. Cytoplasmic extract from HeLa cells stably expressing HA-tagged human Lsm10 was immunoprecipitated with Y12 antibody (lane 2) or with antibody against the HA peptide (lane 3). (Lane 1) Sample of the extract prior to precipitation. The blots were probed with antibodies against D1/D2, HA, Lsm11, SMN, or with Y12 antibody (revealing Sm B/B′) as indicated. (C) Separate SMN complex preparations differ in their Lsm10 and Lsm11 content. Two different preparations of SMN complex (Meister et al. 2000) were analyzed by SDS-PAGE and Coomassie staining (top panels) and by Western blotting (bottom panels) with antibodies against Lsm10, Lsm11, Y12 antibody (revealing Sm B/B′), or with D1/D2 antibody as indicated. Note that preparation SMN-2 does not contain detectable amounts of Lsm10 or Lsm11. The positions of molecular size markers (M), their sizes in kilodaltons, and the positions of prominent protein bands are indicated. (D) Failure of SMN complex lacking Lsm10/11 to assemble U7 snRNA with its wild-type Sm-binding site. SMN complexes 1 and 2 characterized in C were used in assembly reactions with Sm WT (W), Sm OPT (O), or Sm MUT (M) U7 snRNAs. Assembled complexes were detected by immunoprecipitation with Y12 anti-Sm antibody followed by denaturing polyacrylamide gel electrophoresis and autoradiography of the radiolabeled U7 RNAs.
To address this issue, cytoplasmic HeLa cell extracts were subjected to immunoprecipitation with either Y12 anti-Sm antibody or an antiserum reacting with Sm D1 and D2. Both antibodies precipitated similar amounts of SMN and D1/D2 proteins as detected by Western blotting (Fig. 6A, lanes 1,3). In contrast, Lsm10 was detected only in the Y12 precipitate, not in the anti-D1/D2 immunoprecipitate (Fig. 6A, lanes 5,6). To further verify these results, we stably transfected HeLa cells with HA-tagged Lsm10 and immunoprecipitated cytoplasmic extracts with either Y12 or anti-HA antibody. The immunoprecipitates were then analyzed by Western blotting with various antibodies. Whereas both immunoprecipitates contained Sm B/B′ and SMN, the anti-Sm precipitate contained Sm D1/D2 (which migrated in a single band on this particular gel), but only trace amounts of HA-Lsm10 (Fig. 6B, lanes 2). In contrast, the anti-HA precipitate was devoid of Sm D1/D2 but contained HALsm10 (Fig. 6B, lane 3). Moreover, Lsm11 was barely if at all detectable in the input nuclear extract and in the Y12 precipitate, but clearly visible after anti-HA precipitation. These results strongly indicated that there were two separate SMN entities, one containing Sm D1/D2 and the other containing Lsm10 and Lsm11.
From the above, it seemed likely that the SMN complex containing Lsm10 and Lsm11 was the entity mediating the formation of the U7 Sm core. We were able to elucidate this point due to our observation that some SMN complex preparations lacked detectable Lsm10 and Lsm11 (Fig. 6C, lane SMN-2). Thus, two such SMN complex preparations either lacking or containing Lsm10/11, but containing similar amounts of Sm proteins (as exemplified by Western blotting for B/B′ and D1/D2), were incubated with Sm WT, Sm OPT, and Sm MUT U7 RNAs, and Sm core assembly was subsequently analyzed by immunoprecipitation with Y12 antibody. Strikingly, the SMN complex lacking Lsm10/11 was almost completely inactive in U7 Sm WT assembly but promoted Sm core formation on Sm OPT RNA (Fig. 6D, panel SMN-2). In contrast, the SMN complex containing Lsm10/11 promoted assembly on both RNAs (panel SMN-1). No assembly could be observed in either case for Sm MUT RNA lacking a functional Sm-binding site. Together, these data indicated that a specialized SMN complex containing only the appropriate Sm/Lsm proteins (i.e., Sm B/B′, D3, E, F, and G, together with Lsm10 and 11) is responsible for assembling the U7-specific Sm core.
Discussion
Lsm 11 and the U7 snRNP-specific Sm core
By identifying the ∼50-kD U7 snRNP protein as Lsm11, a new member of the Sm/Lsm protein family (Fig. 1), we have now obtained a complete picture of the unique U7 Sm core structure. Twelve years ago, U7 snRNPs were shown to contain some or all standard Sm proteins, as well as additional proteins of 14 and 50 kD (Smith et al. 1991). Moreover, apart from the Sm proteins G and B/B′, proteins of 50 kD in mammalian cells (Mital et al. 1993) and of 40 kD in Xenopus oocytes (Stefanovic et al. 1995a) could be cross-linked to U7 snRNA. The cross-link to the 40-kD protein in frog oocytes occurred within the Sm-binding site of U7 snRNA, four and two nucleotides downstream from the Sm G and B/B′ cross-links, respectively (Stefanovic et al. 1995a). This was difficult to reconcile with the notion of an snRNP-specific protein that did not belong to the Sm core. Especially when structural models of the Sm core were proposed (Kambach et al. 1999b; Törö et al. 2001), it became clear that the heptameric ring structure would cover a certain stretch of RNA and that close associations of other proteins with the same nucleotide stretch would be highly unlikely. We then demonstrated that purified U7 snRNPs lacked the two standard Sm proteins D1 and D2, but that the 14-kD U7-specific protein was a new member of the Sm/Lsm protein family, which we named Lsm10 (Pillai et al. 2001).
The finding that Lsm11 is also an Sm/Lsm protein identifies the final element of the variant U7 Sm core. Indeed, Lsm11 is the third protein that can be UV-cross-linked to the U7 snRNA Sm-binding site (Fig. 2B). Because Lsm10 resembles Sm D1 (Pillai et al. 2001), Lsm10 and Lsm11 most likely take the positions occupied by D1 and D2, respectively, in standard Sm cores. This is, in fact, supported by the unusual structure of Lsm11. With a predicted MW of ∼40 kD, it is by far the largest Sm/Lsm protein found to date, exceeding Sm B/B′ by ∼16 kD. This extra mass is found in two positions. Lsm11 does not have a C-terminal tail like the Sm proteins B/B′, D1, and D3. Instead, the first Sm motif is preceded by 170 N-terminal amino acids, and 138 amino acids separate the Sm motifs 1 and 2. In most Sm/Lsm proteins, the spacer separating these motifs is very short (1-8 amino acids). Only Sm D2 and its Lsm counterpart, Lsm3, have somewhat longer intervening sequences (21 and 20 residues, respectively), and these proteins also have N-rather than C-terminal extensions flanking the Sm domain, although these extensions are much shorter than in Lsm11. None of our experiments have indicated any role for this intervening sequence, be it in U7 snRNP assembly or in histone RNA processing, and the region between the two Sm motifs is not very well conserved between Lsm11 proteins of different species.
The contribution of Lsm11 to histone RNA 3′ end processing
As shown here, the N-terminal extension of Lsm11 plays an essential role in histone RNA processing (Fig. 3). A functional role for Lsm10 or Lsm11 was not unexpected, because, as already mentioned, U7 Sm OPT RNA was found to be nonfunctional in histone RNA processing (Stefanovic et al. 1995a). Our present data indicate that this defect is due to the formation of a standard Sm core containing Sm D1/D2, as opposed to the U7-specific core containing Lsm10/11, and not to a deficiency of the RNA mutation acting in some other way.
The N-terminal domain contains several patches of conserved residues and one region rich in A, R, and G residues that contains four of the 10 RG dipeptides found in the human protein (mouse 2/6, Xenopus laevis 5/7). It is not yet known whether these residues carry symmetrical dimethylarginine modifications, similar to those present on the RG-rich tails of Sm proteins B/B′, D1, and D3, or of Lsm4 (Brahms et al. 2000, 2001). It will be interesting to see which, if any, of these sequences contribute to histone RNA processing. However, although our experiments demonstrate that the N terminus of Lsm11 plays an essential role, they do not preclude that the C-terminal part, or Lsm10, may also participate in processing. For the C-terminal part (of Lsm11), or Lsm 10, however, it will be more difficult to analyze a possible functional role, because they are also required for U7 snRNP assembly.
Several other Sm/Lsm proteins, such as the Sm core components of the U1 snRNP (Zhang et al. 2001) or the Escherichia coli Hfq protein (Möller et al. 2002; Zhang et al. 2002), have previously been shown to contribute to the functions of their respective RNPs. In these cases, the Sm/Lsm proteins appeared to act by improving the basepairing between the short regulatory RNA and its target mRNA or pre-mRNA. However, Lsm11 seems to stimulate histone RNA processing in a different way, that is, by interacting with at least one essential processing factor. This conclusion is based on our finding that a nuclear processing extract loses part of its activity after adsorption to immobilized N terminus of Lsm11 (Fig. 3D). In spliceosomal snRNPs, such functional roles are more typically associated with the snRNP-specific proteins that bind to other parts of these more complex snRNAs. In this context, it is noteworthy that proteins of the Sm core can stabilize the association of the U1-C and U1-70K proteins with the U1 snRNP (Nelissen et al. 1994). Therefore the spliceosomal Sm core may allow for a modular exchange of various functional proteins, depending on the particular snRNA involved, whereas the U7-specific Sm core directly incorporates specialized functional features via its Lsm11 protein (and possibly also via Lsm10).
An important factor interacting with the N terminus of Lsm11 appears to be ZFP100 (Fig. 3E; A. Gruber and D. Schümperli, unpubl.). This zinc finger protein was originally identified based on its ability to bind to an RNP complex composed of the histone RNA 3′-terminal hairpin and its cognate RNA-binding protein, HBP/SLBP (Dominski et al. 2002). In turn, HBP/SLBP has been shown to contribute to histone RNA processing by stabilizing the interaction between histone pre-mRNA and the U7 snRNP (Streit et al. 1993; Spycher et al. 1994; Dominski et al. 1999), and ZFP100 may be mediating this effect. Thus Lsm11 may contribute to stabilizing the U7-histone pre-mRNA complex after all, but may do so indirectly through the formation of a bridge with ZFP100 and HBP/SLBP. This does not preclude other, more direct roles that Lsm11 or ZFP100 may play in histone RNA processing or additional, yet unknown processing factors that may be tethered to the processing complex via either Lsm11 or ZFP100.
Formation of the Sm core—A specificity problem
The special composition of the U7 Sm core and its functional importance for histone RNA processing pose an important specificity problem for the assembly of U snRNPs. The proven function of Lsm11 (see above) and the finding that U7 Sm OPT snRNPs are not only deficient in histone RNA processing (Stefanovic et al. 1995a) but, when present in oocytes containing endogenous wild-type U7 snRNPs, even exert a dominant negative phenotype (B. Stefanovicand D. Schümperli, unpubl.) strongly suggest that U7 snRNPs incorporating Sm D1 and D2 would be nonfunctional and even inhibitory for histone RNA processing. Conversely, Sm D1 and D2 might have specific functions in spliceosomal snRNPs. Thus the cell should have effective means to prevent such potentially toxic associations of “wrong” Sm/Lsm proteins with a given snRNA. Our data suggest that U7 snRNP assembly is indeed controlled at several levels.
For Lsm11, we have identified which part of the protein is important for this specificity. The N-terminal extension and much of the spacer between the two Sm motifs are dispensable for assembly with U7 snRNA as well as for discriminating against U1 snRNA (Fig. 4A). Moreover, the main feature of U7 snRNA deciding whether or not Lsm11 gets incorporated is the special Sm-binding site (Fig. 4B). Because Lsm10, which consists of a compact Sm domain and a very short C-terminal extension, was shown to have a very similar specificity (Pillai et al. 2001), the main determinants for forming the U7-specific Sm core structure appear to be the Sm domains of Lsm10 and Lsm11 and the sequence of the Sm-binding site.
In the context of a cell, the formation of the U7 Sm core is an ATP-dependent process mediated by an SMN complex (Fig. 5), and SMN must therefore be involved in the specificity of U7 snRNP assembly. This is consistent with a previous report which suggested that the SMN complex serves as a specificity factor for snRNP assembly (Pellizzoni et al. 2002). The SMN complex was found to prevent the spontaneous (potentially promiscuous) association of Sm proteins with RNAs and to allow assembly of Sm proteins on U snRNAs exclusively. Further evidence suggested that the specificity of the SMN-mediated assembly of U1 snRNPs is at least partly due to a direct contact of the SMN complex with hairpin 1 of U1 snRNA (Yong et al. 2002). The picture emerging from this was one of a separate recognition of the U snRNA by the SMN complex using RNA features, at least some of which are distinct from the Sm-binding site.
However, for the U7 snRNP, the problem is more difficult than just preventing non-snRNAs to get assembled, and the strategy used to solve this problem appears to be different. Our results with U7 Sm OPT RNA indicate that a difference of three nucleotides in the Sm-binding site is all that is required to switch from the formation of one type of Sm core to the other (Fig. 4B; Pillai et al. 2001). The other parts of the U7 snRNA, its 5′ end complementary to histone pre-mRNA and the 3′-terminal hairpin, can be replaced without changing the specificity of assembly (Gilmartin et al. 1988; Pillai et al. 2001). Therefore the SMN complex must be able to recognize and specifically combine the U7-specific Lsm10 and Lsm11 proteins and the Sm-binding site of U7 snRNA.
This specificity problem may be somewhat alleviated by the formation of certain Sm/Lsm protein oligomers. The standard Sm proteins form discrete heterodimers and -trimers (B/D3, D1/D2, F/E/G) that act as intermediates of Sm core assembly in spliceosomal snRNPs (Raker et al. 1996, 1999). In this respect, it is noteworthy that the two Sm proteins missing from the U7 snRNP, D1 and D2, are part of the same heterodimer. We have obtained preliminary evidence that Lsm10 and the Sm domain of Lsm11, when coexpressed in E. coli, form a soluble heteromeric complex (R. Pillai and D. Schümperli, unpubl.). However, it is not yet clear whether “illegitimate” heterodimers, for example, between Lsm10 and D2 or between D1 and Lsm11 can be formed. If this were not the case, then the specificity problem would be reduced to the selection of the right combination between only four Sm/Lsm protein subcomplexes.
Even granted this reduction in complexity, the SMN complex must deal with the problem of distinguishing between U snRNAs with only slightly different Sm-binding sites and assigning the right combination of Sm/Lsm proteins to each of them. Important in this respect is our finding that there are at least two separate entities containing SMN and some common Sm proteins, one containing Lsm10 and Lsm11, and the other containing Sm D1/D2 (Fig. 6A-C). Moreover, only Lsm10/11-containing SMN complexes can mediate the assembly of the U7 snRNP, whereas a complex that contains only the seven canonical Sm proteins fails in the same reaction (Fig. 6D). It therefore appears that the Sm/Lsm composition of the SMN complex, together with the Sm-binding site of the RNA, determines what type of Sm core structure is generated. Mismatches, if they occur, must be recognized early during complex formation and eliminated rapidly, as there seems to be hardly any overlap between the two types of complexes. The challenge for the future will be to determine how these two SMN complexes arise and how they prevent a misincorporation of Sm/Lsm proteins into the respective Sm core domains.
Materials and methods
U7 snRNP purification and protein microsequencing
The methods used for the purification of U7 snRNPs from HeLa cell nuclear extracts, including affinity selection with a biotinylated oligonucleotide complementary to U7 snRNA, and the method for microsequencing at the Harvard Microchemistry Facility have been described (Pillai et al. 2001).
Plasmids
IMAGE EST clones were obtained through the Resource Center and Primary Database, Berlin (RZPD). The Lsm11 ORF was amplified by PCR from murine EST AI536381 and subcloned into the pcDNA3-HA vector (Pillai et al. 2001) for expression in mammalian cells as an N-terminal HA tag fusion (HA-mLsm11FL). The murine cDNA was chosen because full-length human EST clones were not available. Deletions of Lsm11 were made by PCR yielding the following HA-tagged constructs: HA-mLsm11ΔN104 (deletes 104 amino acids at the N terminus), HA-mLsm11ΔN140 (deletes first 140 amino acids), and HA-mLsm11Δsp77 (deletes amino acids 246-322, i.e., a major part of the spacer region between the Sm motifs).
For expression in E. coli, a truncated version of the ORF lacking the first 139 amino acid codons was amplified from human EST H16126 and inserted into pET 28a (Novagen) as an N-terminal His-tagged fusion (His-hLsm11ΔN-term). The region encoding the first 136 amino acids of mouse Lsm11 (up to a SmaI restriction site) was amplified by PCR and cloned into the pGex4T3 vector (Pharmacia) resulting in the GST-mLsm11N-term plasmid. The X. laevis Lsm10 ORF was amplified from a Xenopus cDNA library by PCR and cloned into pET28a (HisxLsm10). Details of the constructs are available on request.
The plasmids encoding 28-WT and 28-OPT U7 snRNA derivatives (Pillai et al. 2001) and chimeric histone-U7 RNA (12/12-U7 RNA; Stefanovic et al. 1995b) are as described.
Antibodies
His-hLsm11ΔN-term and His-xLsm10 were produced in E. coli BL21 (Pharmacia). For each, the insoluble protein was recovered from the pellet fraction, resolved by SDS-PAGE, and a gel fragment mixed with adjuvants was used for immunization of rabbits. The polyclonal antibodies were affinity-purified using the respective recombinant proteins blotted onto nitrocellulose membrane essentially as described (Pillai et al. 2001). The antibody against His-hLsm11ΔN-term is usually referred to as antibody against human Lsm11. Antibodies against Sm D1 and D2 were obtained by immunization of rabbits with purified recombinant D1/D2 heterodimer [a generous gift from C. Kambach (Paul Scherrer Institut, Life Sciences, Villigen, Switzerland)] and purified as described (Meister et al. 2001a). Other antibodies used were: polyclonal antibodies to hLsm10 (Pillai et al. 2001) or monoclonal antibodies against Sm proteins B/B′, D1, and D3 (Y12; Lerner et al. 1981), SMN (7B10; Meister et al. 2000), or against the HA tag (BabCo), respectively.
Mammalian cell culture experiments
HeLa cells stably expressing Lsm10 have been described (Pillai et al. 2001). The preparation of nuclear and cytoplasmic extracts, immunoprecipitations, experiments with 28-U7 snRNAs, and Western blots were performed as described (Pillai et al. 2001). Photoadducts between U7 RNA and certain Sm/Lsm proteins were detected by UV-cross-linking of nuclear extracts, followed by annealing of a radiolabeled oligonucleotide complementary to the first 18 nucleotides of U7 snRNA, SDS-PAGE, and autoradiography (Mital et al. 1993; Stefanovic et al. 1995a).
Processing and interaction experiments
Most experimental procedures concerning X. laevis oocytes were done as described (Stefanovic et al. 1995a,b). Capped, unlabeled mRNAs encoding various HA-tagged mLsm11 proteins or HA-tagged green fluorescence protein were transcribed in vitro from linearized plasmids using T7 polymerase and subsequently polyadenylated using yeast poly(A) polymerase according to the manufacturer's instructions (USB Corp./Amersham). The mRNAs were injected into the oocyte cytoplasm, followed by incubation at 18°C for 16 h to allow for protein expression. After a second cytoplasmic injection with radiolabeled chimeric 12/12-U7 RNA (see above), the oocytes were further incubated for 3 h. Cytoplasmic extracts were prepared and subjected to immunoprecipitations with anti-Sm (Y12) or anti-HA antibodies. RNA from the pellet fraction was recovered by phenol extraction and analyzed on a 12% denaturing polyacrylamide gel. As control, the HA-tagged proteins were analyzed for expression and for incorporation into snRNPs by Western blotting with anti-HA antibody without or with prior immunoprecipitation with Y12 antibody, respectively.
Histone RNA 3′ end processing in nuclear extract of K21 mouse mastocytoma cells was analyzed as described (Stauber et al. 1990). For certain experiments, extracts were incubated with glutathione beads preloaded with GST or GST-mLsm11N-term, followed by removal of the beads. These recombinant proteins had been produced in soluble form in E. coli BL21 and purified by glutathione affinity chromatography according to the manufacturer's instructions (Pharmacia). A pSP64 plasmid encoding human ZFP100 (Dominski et al. 2002; a gift from Z. Dominski, UNC Chapel Hill) was used to produce 35S-methionine-labeled protein in rabbit reticulocyte lysate (TnT kit, Promega). The translation products were then mixed with glutathione sepharose beads preloaded with either GST or GST-mLsm11N-term and incubated for 2 h. After washes with PBS, the bound and input material was analyzed by SDS-PAGE and autoradiography.
In vitro assembly of U7 snRNPs
Mouse Sm WT, Sm OPT, and Sm MUT U7 snRNAs (Stefanovic et al. 1995a) were in vitro-transcribed and gel-purified. Next, 20,000 cpm of each RNA was incubated with 1 μg tRNA and 2 μL X. laevis egg extract (Müller et al. 2000). Reactions were incubated for 30 min at 20°C and subsequently loaded on a 5% nondenaturing gel. When required, the egg extracts were preincubated for 10 min with antibodies against Gemin2, Gemin4/GIP (Meister et al. 2001a), or against the HA tag (BabCo) prior to addition of RNA. Alternatively, the extracts were immunodepleted of Xenopus SMN and Gemin2; in fact, the same batches of depleted and mock-depleted extracts were used for which the depletion was previously shown to completely remove SMN and Gemin2, while the mock-depletion had no effect on their levels (Meister et al. 2001a). For immunoprecipitation experiments, in vitro assembly reactions were incubated with antibody-bound Protein G sepharose beads for 2 h, followed by washes and analysis of precipitated RNAs. For UV-cross-linking, samples were irradiated with UV light, and the labeled RNA and photoadducts were analyzed by SDS-PAGE as described (Stefanovic et al. 1995a).
SMN complexes were purified from HeLa cells (Meister et al. 2000), and assembly reactions with these complexes were performed as described (Meister and Fischer 2002).
Acknowledgments
We thank O. Mühlemann and B. Suter for critical comments, and A. Gruber and J. Gygax for technical help. This work was supported by the State of Bern and by Swiss National Science Foundation grant 31-65225.01 to D.S. The DFG-grants SFB 523/A8 and Fi-573/2-3 supported the work in the groups of R.L. and U.F., respectively.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.274403.
References
- Achsel T., Brahms, H., Kastner, B., Bachi, A., Wilm, M., and Lührmann, R. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18: 5789-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms H., Raymackers, J., Union, A., de Keyser, F., Meheus, L., and Lührmann, R. 2000. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J. Biol. Chem. 275: 17122-17129. [DOI] [PubMed] [Google Scholar]
- Brahms H., Meheus, L., de Brabandere, V., Fischer, U., and Lührmann, R. 2001. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7: 1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittum H.S., Lane, W.S., Carlson, B.A., Roller, P.P., Lung, F.D., Lee, B.J., and Hatfield, D.L. 1998. Rabbit β-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry 37: 10866-10870. [DOI] [PubMed] [Google Scholar]
- Dominski Z. and Marzluff, W.F. 1999. Formation of the 3′ end of histone mRNA. Gene 239: 1-14. [DOI] [PubMed] [Google Scholar]
- Dominski Z., Zheng, L.X., Sanchez, R., and Marzluff, W.F. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell Biol. 19: 3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Erkmann, J.A., Yang, X., Sanchez, R., and Marzluff, W.F. 2002. A novel zincfinger protein is associated with U7 snRNP and interacts with the stem-loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes & Dev. 16: 58-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen W.J., Paushkin, S., Wyce, A., Massenet, S., Pesiridis, G.S., Van Duyne, G., Rappsilber, J., Mann, M., and Dreyfuss, G. 2001. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell Biol. 21: 8289-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin G.M., Schaufele, F., Schaffner, G., and Birnstiel, M.L. 1988. Functional analysis of the sea urchin U7 small nuclear RNA. Mol. Cell. Biol. 8: 1076-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Stefanovic, B., and Schümperli, D. 1993. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 12: 1229-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W. and Parker, R. 2000. Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol. 12: 346-350. [DOI] [PubMed] [Google Scholar]
- Hermann H., Fabrizio, P., Raker, V.A., Foulaki, K., Hornig, H., Brahms, H., and Lührmann, R. 1995. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 14: 2076-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C., Walke, S., and Nagai, K. 1999a. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 9: 222-230. [DOI] [PubMed] [Google Scholar]
- Kambach C., Walke, S., Young, R., Avis, J.M., de la Fortelle, E., Raker, V.A., Lührmann, R., Li, J., and Nagai, K. 1999b. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96: 375-387. [DOI] [PubMed] [Google Scholar]
- Kufel J., Allmang, C., Verdone, L., Beggs, J.D., and Tollervey, D. 2002. Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with Lahomologous protein Lhp1p. Mol. Cell Biol. 22: 5248-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner E.A., Lerner, M.R., Janeway, C.A., and Steitz, J.A. 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. 78: 2737-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lührmann R., Kastner, B., and Bach, M. 1990. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim. Biophys. Acta 1087: 265-292. [DOI] [PubMed] [Google Scholar]
- Meister G. and Fischer, U. 2002. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 21: 5853-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Bühler, D., Laggerbauer, B., Zobawa, M., Lottspeich, F., and Fischer, U. 2000. Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum. Mol. Genet. 9: 1977-1986. [DOI] [PubMed] [Google Scholar]
- Meister G., Bühler, D., Pillai, R., Lottspeich, F., and Fischer, U. 2001a. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 3: 945-949. [DOI] [PubMed] [Google Scholar]
- Meister G., Eggert, C., Bühler, D., Brahms, H., Kambach, C., and Fischer, U. 2001b. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 11: 1990-1994. [DOI] [PubMed] [Google Scholar]
- Meister G., Eggert, C., and Fischer, U. 2002. SMN-mediated assembly of RNPs: A complex story. Trends Cell Biol. 12: 472-478. [DOI] [PubMed] [Google Scholar]
- Mital R., Albrecht, U., and Schümperli, D. 1993. Detection of UV-induced RNA: Protein crosslinks in snRNPs by oligonucleotides complementary to the snRNA. Nucleic Acids Res. 21: 1049-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller T., Franch, T., Hojrup, P., Keene, D.R., Bachinger, H.P., Brennan, R.G., and Valentin-Hansen, P. 2002. Hfq: A bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9: 23-30. [DOI] [PubMed] [Google Scholar]
- Müller B. and Schümperli, D. 1997. The U7 snRNP and the hairpin binding protein: Key players in histone mRNA metabolism. Semin. Cell Dev. Biol. 8: 567-576. [DOI] [PubMed] [Google Scholar]
- Müller B., Link, J., and Smythe, C. 2000. Assembly of U7 small nuclear ribonucleoprotein particle and histone RNA 3′ processing in Xenopus egg extracts. J. Biol. Chem. 275: 24284-24293. [DOI] [PubMed] [Google Scholar]
- Nelissen R.L.H., Will, C.L., Van Venrooij, W.J., and Lührmann, R. 1994. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 13: 4113-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S., Gubitz, A.K., Massenet, S., and Dreyfuss, G. 2002. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 14: 305-312. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Yong, J., and Dreyfuss, G. 2002. Essential role for the SMN complex in the specificity of snRNP assembly. Science 298: 1775-1779. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Will, C.L., Lührmann, R., Schümperli, D., and Müller, B. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 20: 5470-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker V.A., Plessel, G., and Lührmann, R. 1996. The snRNP core assembly pathway: Identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 15: 2256-2269. [PMC free article] [PubMed] [Google Scholar]
- Raker V.A., Hartmuth, K., Kastner, B., and Lührmann, R. 1999. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell Biol 19: 6554-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B. 1995. Sm and Sm-like proteins belong to a large family: Identification of proteins of the U6 as well as the U1, U2, U4, and U5 snRNPs. EMBO J. 14: 2089-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H.O., Tabiti, K., Schaffner, G., Soldati, D., Albrecht, U., and Birnstiel, M.L. 1991. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2′-0-methyl oligoribonucleotides. Proc. Natl. Acad. Sci. 88: 9784-9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spycher C., Streit, A., Stefanovic, B., Albrecht, D., Wittop, K., and Schümperli, D. 1994. 3′ end processing of mouse histone pre-mRNA: Evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 22: 4023-4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber C., Soldati, D., Lüscher, B., and Schümperli, D. 1990. Histone-specific RNA 3′ processing in nuclear extracts from mammalian cells. Methods Enzymol. 181: 74-89. [DOI] [PubMed] [Google Scholar]
- Stefanovic B., Hackl, W., Lührmann, R., and Schümperli, D. 1995a. Assembly, nuclear import and function of U7 snRNPs studied by microinjection of synthetic U7 RNA into Xenopus oocytes. Nucleic Acids Res. 23: 3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic B., Wittop, K., and Schümperli, D. 1995b. A synthetic histone pre-mRNA-U7 small nuclear RNA chimera undergoing cis cleavage in the cytoplasm of Xenopus oocytes. Nucleic Acids Res. 23: 3152-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A., Wittop Koning, T.H., Soldati, D., Melin, L., and Schümperli, D. 1993. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone premRNA in vitro. Nucleic Acids Res. 21: 1569-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasevic N. and Peculis, B.A. 2002. Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol. Cell Biol. 22: 4101-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Törö I., Thore, S., Mayer, C., Basquin, J., Seraphin, B., and Suck, D. 2001. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 20: 2293-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L., Schneider, C., Reed, R., and Lührmann, R. 1999. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science 284: 2003-2005. [DOI] [PubMed] [Google Scholar]
- Yong J., Pellizzoni, L., and Dreyfuss, G. 2002. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J. 21: 1188-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Wassarman, K.M., Ortega, J., Steven, A.C., and Storz, G. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9: 11-22. [DOI] [PubMed] [Google Scholar]
- Zhang D., Abovich, N., and Rosbash, M. 2001. A biochemical function for the Sm complex. Mol. Cell 7: 319-329. [DOI] [PubMed] [Google Scholar]