Abstract
PRC2 is thought to be the histone methyltransferase (HMTase) responsible for H3-K27 trimethylation at Polycomb target genes. Here we report the biochemical purification and characterization of a distinct form of Drosophila PRC2 that contains the Polycomb group protein polycomblike (Pcl). Like PRC2, Pcl-PRC2 is an H3-K27-specific HMTase that mono-, di- and trimethylates H3-K27 in nucleosomes in vitro. Analysis of Drosophila mutants that lack Pcl unexpectedly reveals that Pcl-PRC2 is required to generate high levels of H3-K27 trimethylation at Polycomb target genes but is dispensable for the genome-wide H3-K27 mono- and dimethylation that is generated by PRC2. In Pcl mutants, Polycomb target genes become derepressed even though H3-K27 trimethylation at these genes is only reduced and not abolished, and even though targeting of the Polycomb protein complexes PhoRC and PRC1 to Polycomb response elements is not affected. Pcl-PRC2 is thus the HMTase that generates the high levels of H3-K27 trimethylation in Polycomb target genes that are needed to maintain a Polycomb-repressed chromatin state.
Keywords: Drosophila, gene silencing, histone methylation, PcG, trxG
Introduction
Genetic studies in Drosophila first identified Polycomb group (PcG) genes as regulators that are required for the long-term repression of HOX genes during development (reviewed in Ringrose and Paro, 2004). To date, 17 different genes in Drosophila are classified as PcG members because mutations in these genes cause misexpression of HOX genes (reviewed in Schwartz and Pirrotta, 2007). All Drosophila PcG genes are also conserved in mammals and at least some of them are also conserved in plants (reviewed in Brock and Fisher, 2005; Köhler and Makarevich, 2006; Schwartz and Pirrotta, 2007). In all these organisms, PcG gene products function as repressors of HOX and/or other regulatory genes that control specific developmental programs (reviewed in Sparmann and van Lohuizen, 2006). Moreover, recent studies that analyzed genome-wide binding of PcG proteins in Drosophila and in mammalian cells identified a large number of target sites, and thus a whole new set of genes that potentially is subject to PcG repression (Boyer et al, 2006; Lee et al, 2006; Negre et al, 2006; Schwartz et al, 2006; Tolhuis et al, 2006).
Biochemical purification and characterization of PcG protein complexes has advanced our understanding of the PcG system. To date, three distinct PcG protein complexes have been isolated from Drosophila: PhoRC (Klymenko et al, 2006), PRC1 (Shao et al, 1999; Levine et al, 2002) and PRC2 (Cao et al, 2002; Czermin et al, 2002; Kuzmichev et al, 2002; Müller et al, 2002). The composition and activities of these different complexes and current views on the mechanisms by which these complexes might repress transcription of target genes have been discussed in recent review articles (Müller and Kassis, 2006; Schuettengruber et al, 2007; Schwartz and Pirrotta, 2007).
Biochemically purified Drosophila PRC2 contains the three PcG proteins Enhancer of zeste (E(z)), Suppressor of zeste 12 (Su(z)12) and Extra sex combs (Esc) and, in addition, Nurf55, a protein that is present in many different chromatin complexes (Czermin et al, 2002; Müller et al, 2002). Drosophila PRC2 and the homologue mammalian complex are histone methyltransferases (HMTases) that specifically methylate H3-K27 in nucleosomes (Cao et al, 2002; Czermin et al, 2002; Kuzmichev et al, 2002; Müller et al, 2002). Chromatin immunoprecipitation (X-ChIP) analyses in Drosophila showed that PRC2 binds in a localized manner at Polycomb response elements (PREs) of target genes, but that H3-K27 trimethylation is present across the whole upstream control, promoter and coding region of these genes (Kahn et al, 2006; Mohd-Sarip et al, 2006; Papp and Müller, 2006; Schwartz et al, 2006). Studies that compared the inactive and active state of the HOX gene Ubx in developing Drosophila found that PRC2 is constitutively bound at PREs and, surprisingly, that the whole upstream control region is constitutively trimethylated at H3-K27 (Papp and Müller, 2006). However, presence or absence of H3-K27 trimethylation in the Ubx promoter and coding region correlates tightly with the gene being repressed or active, respectively (Papp and Müller, 2006). H3-K27 trimethylation is thus a distinctive mark of PcG-repressed chromatin.
Analysis of E(z) mutants suggests that E(z) is also responsible for the genome-wide H3-K27 mono- and dimethylation that has been reported to be present on more than 50% of H3 in Drosophila (Ebert et al, 2004). However, biochemical analyses showed that E(z) protein alone does not bind to nucleosomes and is virtually inactive as an enzyme; E(z) needs to associate with Su(z)12 and Nurf55 for nucleosome binding and with Esc for enzymatic activity (Czermin et al, 2002; Müller et al, 2002; Ketel et al, 2005; Nekrasov et al, 2005). This implies that the genome-wide H3-K27 mono- and dimethylation is generated by PRC2 or another E(z)-containing complex that is able to interact in a non-targeted manner with nucleosomes across the whole genome. Conversely, this raises the question whether H3-K27 trimethylation at PcG target genes is simply a consequence of PRC2 being targeted to PREs or whether additional features such as post-translational modifications or associated factors are required.
Previous studies reported that the PcG protein Polycomblike (Pcl) interacts with E(z) in GST pull-down, yeast two-hybrid and co-immunoprecipitation assays (O'Connell et al, 2001; Tie et al, 2003). Like most other PcG proteins, Pcl has also been found to be bound at PREs in Drosophila (Tie et al., 2003; Papp and Müller, 2006). However, to date, no Pcl-containing complexes have been purified and the role of Pcl in PcG repression has remained enigmatic. In this study we report the biochemical purification of Pcl complexes. We show that Pcl exists in a stable complex with PRC2. Our analyses demonstrate that this Pcl complex plays a critical role in generating high levels of repressive H3-K27 trimethylation at PcG target genes.
Results
Biochemical purification identifies Pcl-PRC2 as a distinct PcG protein complex
We used a tandem affinity purification (TAP) strategy (Rigaut et al, 1999) to purify Pcl protein complexes from Drosophila embryos. To this end, we first generated transgenic Drosophila strains that express a TAP-tagged Pcl fusion protein (TAP-Pcl) under the control of the Drosophila a-tubulin promoter. Using a genetic rescue assay, we found that this TAP-Pcl fusion protein is functional and can substitute for endogenous Pcl. Specifically, animals that are homozygous for Pcl21M22, a protein-negative allele of Pcl (see below), die at the end of embryogenesis but are rescued into viable and fertile adults if they carry the transgene expressing TAP-Pcl protein (see Materials and methods).
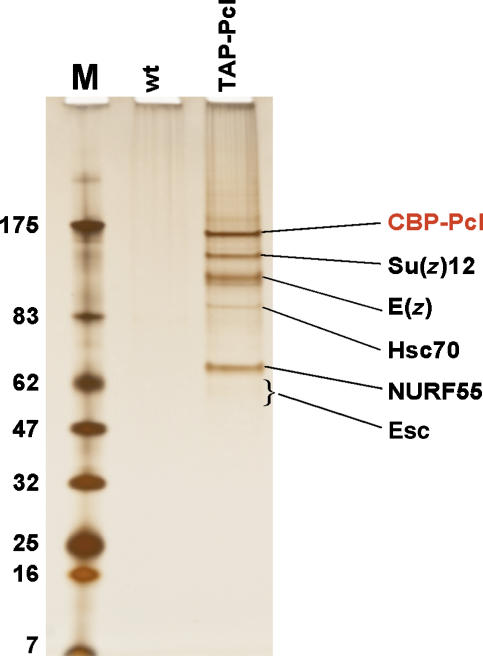
Following the TAP procedure, we purified proteins associated with TAP-Pcl from nuclear extracts that we prepared from TAP-Pcl transformant embryos. The purified material was separated on SDS-polyacrylamide gels and silver staining of the gel revealed five protein bands that consistently co-purified with TAP-Pcl (Figure 1). Sequencing of peptides from these bands by nanoelectrospray tandem mass spectrometry identified these proteins as the PRC2 components E(z), Su(z)12, Nurf-55, Esc and, in addition, heat-shock cognate protein HSC70 (Figure 1; Supplementary Figure S1A). LC-MS/MS analysis of total purified material confirmed that these polypeptides are the main co-purifying proteins (Supplementary Figure S1B). The observation that PRC2 components E(z), Su(z)12, Esc and Nurf-55 co-purify with Pcl suggests that Pcl and PRC2 constitute a specific form of PRC2 that we name Pcl-PRC2.
Figure 1.
TAP of Pcl protein complexes from Drosophila embryonic nuclear extracts. Protein complexes purified from wild-type (wt) and TAP-Pcl; Pcl+/+ embryos. Purified material was separated on a 4–12% polyacrylamide gel and visualized by silver staining; ‘M' indicates molecular weight marker. Input material for mock purification from wild-type embryos, and for purification from transgenic embryos was normalized by protein concentration and equivalent amounts of material eluted from calmodulin affinity resin was loaded. Complexes were eluted with EGTA under non-denaturing conditions. Approximately 0.67 pmol of Pcl–PRC2 complex is loaded, this amount was determined by comparing with mass spectrometry analysis of defined amounts of purified recombinant Pcl, E(z) and Su(z)12 (not shown, see Figure 3). Indicated proteins consistently co-purified with CBP-Pcl in several independent experiments and were identified by microsequencing; CBP in fusion proteins refers to the calmodulin-binding moiety of the TAP-tag. Note presence of PRC2 subunits Su(z)12, E(z), NURF55 and Esc in TAP-Pcl lane; Esc stains poorly and runs as a diffuse band, but is identified by multiple peptides like the other proteins (Supplementary Figure S1). See Supplementary Figure S1 for information on additional proteins identified by mass spectrometry.
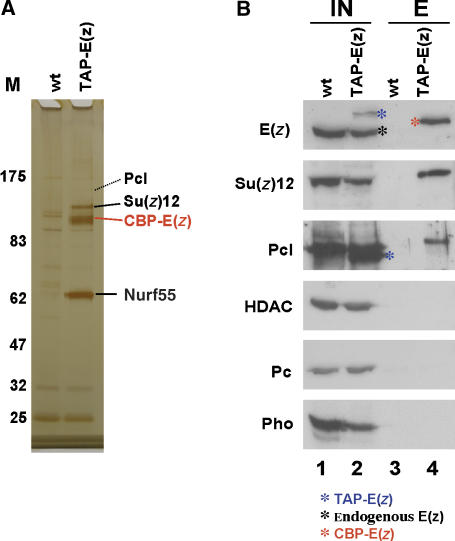
To confirm the association of Pcl with PRC2, we also used the TAP-tag strategy to purify proteins associated with TAP-E(z) protein in Drosophila embryos. Mass spectrometry and western blot analyses revealed that Pcl protein indeed co-purifies with TAP-tagged E(z), albeit clearly at substoichiometric amounts compared to the PRC2 core components (Figure 2; Supplementary Figure S1). In contrast, we note that the material purified either with TAP-Pcl or with TAP-E(z) did not contain detectable amounts of the PhoRC component Pho, the PRC1 component Pc, or HDAC1/RPD3 (Figures 1 and 2; Supplementary Figure S1), three proteins that have been reported to interact with PRC2 subunits in in vitro binding or co-immunoprecipitation assays (Poux et al, 2001; Tie et al, 2001, 2003; Wang et al, 2004). Together, these results further strengthen the view that Pcl-PRC2 is a stable biochemical entity. One possible explanation for the presence of substoichiometric amounts of Pcl in the TAP-E(z) purification would be that Pcl-PRC2 is a distinct form of PRC2 and that only a fraction of PRC2 in the cell is associated with Pcl. An alternative possibility would be that Pcl is a less tightly associated component of PRC2 that tends to dissociate from the complex during biochemical purification. In this context, it is interesting to note that the Hsc70-4 protein is present in the material purified with either TAP-Pcl or TAP-E(z). Hsc70-4 has also been reported to co-purify with both Drosophila and human PRC1 (Saurin et al, 2001; Levine et al, 2002). At present it is not known whether Hsc70-4 is required for assembly, stability or function of Pcl-PRC2, PRC2 and PRC1, and further studies will be needed to address these questions but, intriguingly, Hsc70-4 and Pc also interact in genetic assays (Mollaaghababa et al, 2001).
Figure 2.
TAP of E(z) protein complexes from Drosophila embryonic nuclear extracts. (A) Protein complexes purified from wild-type (wt) and TAP-E(z); E(z)+/+ embryos. Purified material was separated on a 4–12% polyacrylamide gel and visualized by silver staining. Input material for mock purification from wild-type embryos and for purification from transgenic embryos was normalized by protein concentration and equivalent amounts of material eluted from calmodulin affinity resin was loaded; complexes were eluted by boiling of calmodulin resin in SDS buffer. Indicated proteins consistently co-purified with CBP-E(z) in several independent experiments and were identified by microsequencing; CBP in fusion proteins refers to the calmodulin-binding moiety of the TAP-tag. PRC2 subunits Su(z)12, Nurf55 and the weak Pcl band in TAP-E(z) lane were all identified by multiple peptides (Supplementary Figure S1) in excised gel bands; Esc was not detected on the gel. See Supplementary Figure S1 for information on additional proteins identified by mass spectrometry. (B) Western blot analysis of total embryonic nuclear extract input material (IN, lanes 1 and 2) from wild-type (wt) and TAP-E(z) transgenic embryos, and material eluted from calmodulin affinity resin (E, lanes 3 and 4) after purification. All panels come from the same batch of input material, and eluates were all from the same batch of material purified from wild-type and TAP-E(z) embryos, respectively; the same ratio of input versus eluate material was loaded in all cases. CBP-E(z) (red asterisk), TAP-E(z) (blue asterisk) and endogenous E(z) (black asterisk) are indicated; in lane 2, TAP-E(z) is also detected by other antibodies due to protein A tag. Compare the relative enrichment of E(z), Su(z)12 and Pcl in lane 4. For unknown reasons, Su(z)12 and Pcl in lane 4 migrate with slightly higher mobility. Note the lack of signals for HDAC, Pc and Pho in lane 4.
Finally, we explored how Pcl physically interacts with PRC2 subunits. To this end, we used baculovirus expression vectors to coexpress Flag-tagged Pcl with individual PRC2 core components in Sf9 cells, and then used the Flag-epitope for affinity purification from Sf9 cell extracts. We could thus reconstitute a stable dimeric Pcl–E(z) complex (Supplementary Figure S2), consistent with previous studies that reported physical interactions between Pcl and E(z) in GST pull-down assays (O'Connell et al., 2001; Tie et al., 2003). Interestingly, the baculovirus reconstitution assay revealed that Pcl also forms a stable complex with Nurf55 and, albeit less efficiently, also with Su(z)12 (Supplementary Figure S2). In contrast, purification from cells coexpressing Flag-Pcl and Esc resulted in the isolation of Flag-Pcl protein only (Supplementary Figure S2). Together, these data suggest that Pcl associates with PRC2 though interactions with E(z), Nurf55 and Su(z)12.
Pcl-PRC2 and PRC2 are H3-K27-specific HMTases
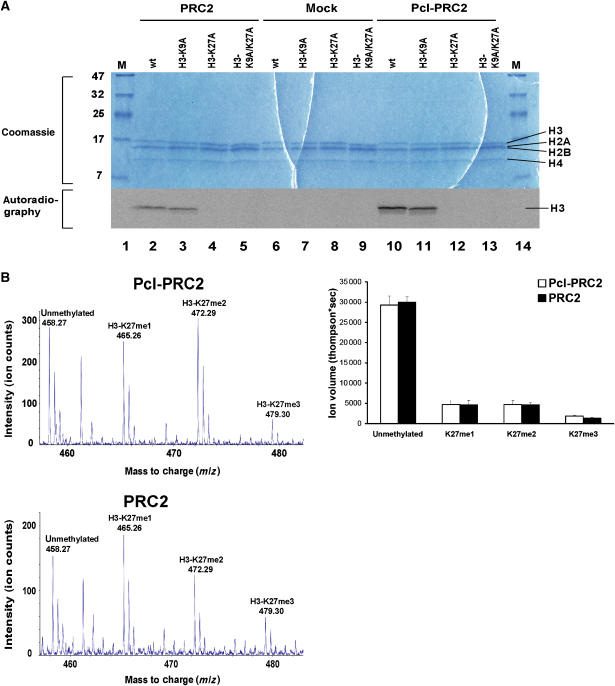
We next compared the HMTase activity of Pcl-PRC2 with the activity of PRC2. For these experiments we used recombinant tetrameric PRC2 (rPRC2) that we reconstituted as previously described (Müller et al, 2002; Nekrasov et al, 2005). Attempts to generate recombinant Pcl-PRC2 of comparable quality with stoichiometric quantities of the five components have been unsuccessful, and we therefore used Pcl-PRC2 purified from Drosophila for these assays. As substrate in the HMTase reactions we used reconstituted recombinant mononucleosomes that contained either wild-type histone H3 or mutant forms of H3, in which lysine 9 (H3K9A) or lysine 27 (H3K27A), or both (H3K9A/K27A) had been mutated to alanine. Pcl-PRC2 and rPRC2 both methylated histone H3 at K27. This specificity is demonstrated by the finding that mononucleosomes containing wild-type H3 or H3K9A are efficiently methylated by either complex, but that no methylation is observed on mononucleosomes containing H3K27A or H3K9A/K27A (Figure 3A). However, purified Pcl-PRC2 appears to be more active than rPRC2 because three-fold molar excess of rPRC2 was needed to obtain comparable H3-K27 methylation signals (Figure 3A). At present it is not known whether this relatively higher HMTase activity of Pcl-PRC2 is due to inclusion of Pcl, because it could also be due to post-translational modifications that are present on other subunits of natively purified Pcl-PRC2 but are missing on rPRC2. Mass spectrometry analysis of the H3 protein band isolated from mononucleosomes after HMTase assays independently confirmed H3-K27 as the site being methylated, and it revealed that Pcl-PRC2 and rPRC2 both trimethylate H3-K27 (Figure 3B). Interestingly, reaction with either complex generated comparable summed intensities of mono-, di- and trimethylated H3-K27, suggesting that both complexes are comparably efficient in generating the different methylated states of H3-K27 (Figure 3B). Taken together, these data suggest that Pcl-PRC2 and PRC2 are both H3-K27-specific HMTases that methylate H3-K27 in nucleosomes in vitro.
Figure 3.
HMTase activity of Pcl-PRC2 and PRC2 in vitro. (A) Molecular weight marker (lanes 1 and 14) and HMTase reactions (lanes 2–13) performed with 2 pmol recombinant PRC2 (lanes 2–5), no complex (‘mock', lanes 6–9) or 0.67 pmol Pcl-PRC2 (lanes 10–13), [14C]-SAM (lanes 2–13) and 2.8 pmol wild-type (wt) or mutant (H3-K9A, H3K27A and H3-K9A/K27A) mononucleosomes as indicated, were resolved on a 20% SDS–polyacrylamide gel. The gel was stained with Coomassie (top) and exposed for autoradiography (below). Note that Pcl-PRC2 and PRC2 both specifically methylate H3-K27 (compare lanes 2–5 with 10–13), no methylation is observed on nucleosomes with H3 containing the K27A mutation. (B) Mass spectrometry analysis of H3 following HMTase reactions with Pcl-PRC2 (top) or PRC2 (bottom). HMTase reactions were performed as in lanes 2 and 10 in panel A except that non-radioactive SAM was used and reactions were allowed to proceed for 12 h; excised H3 bands were digested and analyzed by reverse-phase chromatography and quantitative mass spectrometry. Chromatography separated modified and unmodified H3KSAPATGGVK peptides by only ∼95 s. Therefore a single mass spectrum can show ions of unmodified and all methylation states of H3KSAPATGGVK peptides during their elution. The right lane shows the ion volumes (thompson*sec) of unmodified and modified H3KSAPATGGVK peptides after HMTase reaction. The ion volume is the summed ion intensity of a peptide over its elution time (Fraterman et al, 2007). Note that Pcl-PRC2 and PRC2 generate comparable ion volumes of mono-, di- and trimethylated H3-K27 in this assay. The error bars indicate the standard deviation of a duplicate analysis.
Pcl-PRC2 is needed for high levels of H3-K27 trimethylation at PcG target genes in Drosophila
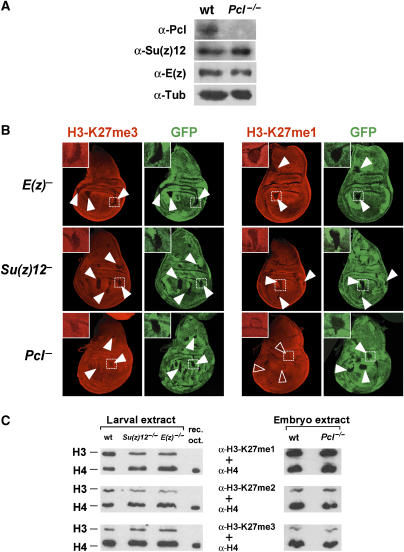
To study the role of Pcl-PRC2 in relation to that of PRC2, we next analyzed Drosophila mutants that lack Pcl protein. For these studies we used the loss-of-function allele Pcl22M21 that we recently isolated (see Materials and methods). Pcl22M21 appears to be a protein-negative allele because no Pcl protein is detected in extracts prepared form 16- to 18-h-old Pcl22M21 homozygous embryos (Figure 4A). Importantly, the levels of Su(z)12 and E(z) protein in such Pcl22M21 homozygous embryos are indistinguishable from those observed in wild-type embryos (Figure 4A); other PRC2 subunits are thus stable in the absence of Pcl protein. This suggests that Pcl22M21 mutants specifically lack the function of Pcl-PRC2 but retain normal levels of PRC2.
Figure 4.
Pcl is needed for high levels of H3-K27 trimethylation in Drosophila. (A) Western blot analysis of extracts from 16- to 18-h-old wild-type (wt) and Pcl22M21 homozygous (Pcl−/−) embryos, probed with antibodies against the indicated proteins; the anti-tubulin (a-Tub) Western blot provides control for loading of equal amounts of extract. Note that in Pcl−/− embryos, no Pcl protein is detected, but that Su(z)12 and E(z) protein levels are comparable to wt embryos. (B) Wing imaginal discs with clones of cells that are homozygous for E(z)731 (E(z)), Su(z)124 (Su(z)12), or Pcl21M22 (Pcl), stained with antibodies against H3-K27me3 (red signal, left column) or H3-K27me1 (red signal, right column). In each case, clones of mutant cells are marked by the absence of GFP signal (green) and discs were analyzed 96 h after clone induction. Su(z)12 and E(z) mutant clones show complete loss of H3-K27me3 and H3-K27me1 signals (arrowheads). Pcl mutant clones also show a clear reduction of H3-K27me3 levels (arrowheads), but H3-K27me1 levels are unaffected (empty arrowheads) and are comparable to those in neighboring wild-type cells. Representative clones (white frame) are shown at higher magnification (C) Western blot analysis of extracts from imaginal disc and CNS tissues from second instar larvae (larval extract, left column) or from 16- to 18-h-old embryos (embryo extract, right column) of the following genotypes: wild-type (wt), Su(z)124/Su(z)124 (Su(z)12−/−), E(z)731/E(z)731 (E(z)−/−), Pcl22M21/Pcl22M21 (Pcl−/−). Recombinant histone octamer (rec. oct.) reconstituted from Xenopus histones expressed in Escherichia coli served as additional control for H3-K27me antibody specificity. In each case, the membrane was simultaneously probed with the antibody against the indicated H3-K27 methylation state and with an antibody against unmodified histone H4, to control for equal extract loading and Western blot processing. Note that in E(z)−/− and Su(z)12−/− larvae, H3-K27me3 Western blot signals are comparable to those observed in wt larvae and only H3-K27me1 and H3-K27me2 signals are detectably reduced. In Pcl−/− embryos, H3-K27me1, H3-K27me2 and H3-K27me3 Western blot signals are comparable to those observed in wt embryos.
We then tested whether Pcl might be required for H3-K27 methylation in vivo. In a first set of experiments we generated clones of Pcl homozygous mutant cells in imaginal discs of developing Drosophila larvae and analyzed the global level of H3-K27 mono- and trimethylation in the mutant cells by immunostaining wing discs with antibodies against H3-K27me1 and H3-K27me3, respectively. Antibodies against H3-K27me2 gave strong staining signals in the cytoplasm of imaginal disc cells and this precluded the analysis of H3-K27 dimethylation in these experiments (data not shown). In parallel to analyzing Pcl mutant clones, we also analyzed wing discs with clones of cells that were homozygous for null mutations in E(z) or Su(z)12, respectively. In all these experiments, discs were analyzed 96 h after clone induction and the clones of mutant cells were identified by absence of a GFP-expressing marker gene. In E(z) or Su(z)12 mutant clones in wing discs, H3-K27me3 and H3-K27me1 signals are reduced to undetectable levels (Figure 4B). The loss of H3-K27me3 and H3-K27me1 signals in cells lacking these two PRC2 core components is consistent with earlier reports that showed that all H3-K27 methylation in Drosophila depends on E(z), the catalytic subunit of PRC2 (Ebert et al, 2004; Ketel et al, 2005; Papp and Müller, 2006). Unexpectedly, we found that H3-K27me3 levels are also reduced in Pcl mutant clones. Although the reduction is not as severe as in E(z) or Su(z)12 mutant clones, it is consistent in all discs examined (Figure 4B). In striking contrast, the level of H3-K27me1 signal in Pcl mutant clones is not diminished and is indistinguishable from that of wild-type cells (Figure 4B). Taken together, these results suggest that, in Drosophila, Pcl-PRC2 is required for H3-K27 trimethylation, but is apparently not required for H3-K27 mono-methylation.
In an independent set of experiments, we compared the levels of total H3-K27me1, H3-K27me2 and H3-K27me3 in wild-type, E(z), Su(z)12 and Pcl mutant animals by Western blot analysis. Like other PcG genes, E(z), Su(z)12 and Pcl are all expressed in the female germline, and wild-type products, deposited into the egg by the mother, rescue the homozygous mutant embryos to a certain extent (Breen and Duncan, 1986; Jones and Gelbart, 1990; Soto et al, 1995; Birve et al, 2001). E(z) and Su(z)12 homozygotes thus develop even into larvae, whereas Pcl homozygotes die at the end of embryogenesis. The imaginal disc tissues in E(z) or Su(z)12 homozygous second instar larvae are only poorly developed, and the cells stop proliferating but, remarkably, they still contain substantial levels of H3-K27me1, H3-K27me2 and H3-K27me3. Compared with wild-type control larvae, we only found H3-K27me1 and H3-K27me2 signals to be detectably reduced in these mutants (Figure 4C). This strongly suggests that H3-K27 methylation and in particular H3-K27 trimethylation that was generated by maternally deposited PRC2 or Pcl-PRC2 in the early embryo persists into the larval stages. Similarly, in 16- to 18-h-old Pcl22M21 homozygous embryos, H3-K27me1, H3-K27me2 and H3-K27me3 Western blot signals are indistinguishable from those in wild-type embryos (Figure 4C), even though maternally deposited Pcl protein is no longer detected in Pcl22M21 homozygotes at this stage (Figure 4A). The inability to detect changes in H3-K27 methylation levels in Pcl mutants in this assay could have different reasons. Like in the case of E(z) or Su(z)12 homozygotes, it is possible that the H3-K27me3 signals present in Pcl mutant embryos represent H3 molecules that were methylated early in embryogenesis by maternally deposited Pcl–PRC2 complexes. Alternatively, it could be that Pcl is not required for H3-K27 methylation in embryos and, finally, it is possible that H3-K27me3 levels are not globally reduced in Pcl mutant embryos but are perhaps only reduced at particular target genes.
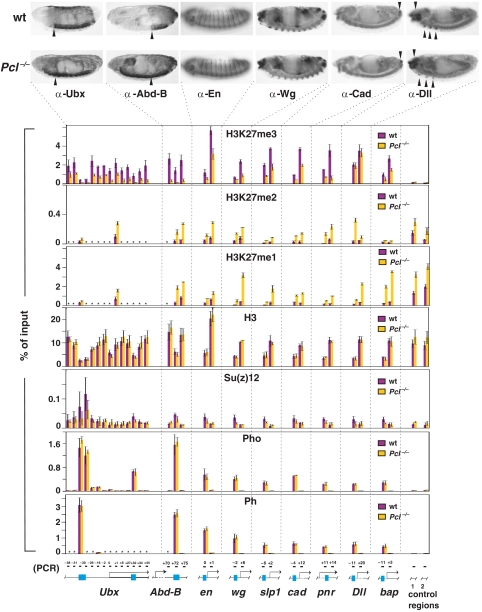
To distinguish between these possibilities, we performed X-ChIP assays to monitor the levels of H3-K27me3, H3-K27me2 and H3-K27me1 at PcG target genes in wild-type and in Pcl22M21 homozygous embryos. Recent X-ChIP on chip studies in tissue culture cells and in Drosophila embryos identified a number of genes to which PRC2, PRC1 and PhoRC components are bound and that also contain chromatin that is trimethylated at H3-K27 (Negre et al, 2006; Schwartz et al, 2006, Tolhuis et al, 2006; K Oktaba and J Müller, unpublished). Among those, we used the Ultrabithorax (Ubx), Abdominal-B (Abd-B), engrailed (en), wingless (wg), sloppy paired 1 (slp1), caudal (cad), pannier (pnr), Distall-less (Dll) and bagpipe (bap) genes for our analysis. We thus prepared chromatin from 16- to 18-h-old wild-type or Pcl22M21 homozygous embryos and performed X-ChIP assays with antibodies against H3-K27me3, H3-K27me2 or H3-K27me1, and also with antibodies against unmodified H3. Unmodified H3 X-ChIP served as a critical control for the ability to detect nucleosomes at the different chromosomal regions and as reference for comparing the levels of H3-K27 methylation in wild-type and in Pcl mutant embryos. In addition we also monitored binding of the three PcG protein complexes PhoRC, PRC1 and PRC2 by performing X-ChIP reactions with antibodies against the PhoRC component Pho, the PRC1 component Ph and the PRC2 component Su(z)12. Real-time quantitative PCR was used to measure the abundance of specific genomic DNA sequences in the immunoprecipitates. At each PcG target gene, we monitored presence of H3, presence of the different methylated states of H3-K27 and binding of the three PcG proteins at (i) the PREs that we had identified by Pho X-ChIP-on-chip in embryos (K Oktaba and J Müller, unpublished) and (ii) at one or more other regions within the transcribed portion of the gene (Figure 5). Two sequences located in two distinct intergenic regions elsewhere in euchromatin served as controls (Figure 5).
Figure 5.
Pcl is required for repression and H3-K27 trimethylation at PcG target genes. (Top) Wild-type (wt, top row) and Pcl22M21 homozygous (Pcl−/−, bottom row) embryos stained with antibodies against Ubx, Abd-B, Engrailed (En), Wingless (Wg), Caudal (Cad) and Distal-less (Dll) proteins. In Pcl−/− embryos, Ubx and Abd-B are misexpressed outside of their normal expression domains; arrowheads mark anterior margin of ps5 and ps10 that correspond to the anterior boundary of the Ubx and Abd-B expression domain in wt animals, respectively. Expression of En is essentially normal in Pcl−/− animals and only very few En-positive cells that are not visible here are present in addition to the wild-type En expression pattern in the posterior compartment of every segment (cf. Moazed and O'Farrell, 1992). The complex expression pattern of Wg, expression of Cad at the posterior end of the embryo (arrowhead) and Dll expression in the head and thorax (arrowheads) are all normal in Pcl−/− animals and no misexpression is detected. The slightly reduced levels of Dll expression in the imaginal disc primordia in thoracic segments (arrowheads) in Pcl−/− animals is likely due to downregulation by misexpressed BX-C proteins (Vachon et al, 1992). (Bottom). X-ChIP analysis in wild-type (wt, purple bars) and Pcl mutant (Pcl−/−, yellow bars) embryos, respectively. Each bar shows the result from at least three independent immunoprecipitation reactions on independently prepared batches of chromatin, performed with the indicated antibodies against H3-K27me3, H3-K27me2, H3-K27me1, unmodified H3, Su(z)12, Pho or Ph; X-ChIP signal levels are presented as percentage of input chromatin precipitated for each region. The location of PREs (blue boxes) and other regions with respect to the transcription start sites are indicated in kilobases, see Supplementary Figure S4 for information on exact location of PCR primer pairs. Note that at each region, H3 X-ChIP signals are comparably high in wt and Pcl mutant embryos. The comparably lower H3-X-ChIP signal at each PREs in both wt and Pcl mutant embryos suggests inefficient detection of nucleosomes or, more likely, reduced nucleosome occupancy at PREs, as previously observed at Ubx (Kahn et al, 2006; Mohd-Sarip et al, 2006; Papp and Müller, 2006). In wt animals, H3-K27me3 X-ChIP signals are enriched at all target genes compared to control regions 1 and 2. Note that in Pcl mutant embryos, H3-K27me3 signals are at least two-fold lower in each region; only at Dll, H3-K27me3 signals are unaltered. In wild-type embryos, H3-K27me1 and H3-K27me2 signals at target genes are overall lower than in control regions 1 and 2. Note that in Pcl mutants H3-K27me1 and H3-K27me2 signals are strongly increased at target genes and that they are also increased at control regions 1 and 2 (see text for details). Note the specific enrichment of Su(z)12, Pho and Ph at PREs. In Pcl mutant embryos, binding of Pho and Ph is undiminished compared to wild-type embryos but Su(z)12 binding is reduced. Note that Su(z)12 X-ChIP signals at the bxd PRE in Pcl mutants are still higher than in control regions 1 and 2. Compared to X-ChIP assays in imaginal discs (Papp and Müller, 2006), we find that in embryos a smaller fraction of input material is precipitated with Pho, Ph and, in particular, with Su(z)12 antibodies. It is possible that the intrinsically different fixation procedure is responsible for a lower crosslinking efficiency in embryos compared with discs. Asterisks (*) indicate regions where X-ChIP signals were not measured.
Comparison of the H3-K27me3, H3-K27me2 and H3-K27me1 profiles between wild-type and Pcl mutant embryos revealed that the levels of these three modifications are very differently affected in the absence of Pcl. In particular, Pcl mutants show a 2- to 3-fold reduction of H3-K27me3 signals in most regions of the nine target genes and this reduction is accompanied by a 2- to 4-fold increase of H3-K27me1 and H3-K27me2 signals at these regions (Figure 5). Several specific aspects of these observations should be noted. First, trimethylation of H3-K27 in target gene chromatin is only reduced but not abolished in Pcl mutants, and although the reduction is seven-fold in the region of the iab-7 PRE in Abd-B or in the coding region of pnr, H3-K27me3 levels are not detectably reduced at the Dll gene (Figure 5). Together with the observation that global H3-K27me3 levels are not detectably reduced in Pcl mutant embryos (Figure 4C), but that they are clearly reduced in Pcl mutant clones in imaginal discs (Figure 4B), this suggests that Pcl-PRC2 is required to generate high levels of H3-K27 trimethylation at many but not all PcG target genes in embryos, but probably becomes required for this methylation at most target genes during larval development. Second, in wild-type animals, H3-K27me1 and H3-K27me2 X-ChIP signals at target genes are lower than in the control regions 1 and 2 but they reach comparable levels in Pcl mutant embryos (Figure 5). This suggests that in wild-type animals, nucleosomes in PcG target gene chromatin are extensively trimethylated at H3-K27 and that in the absence of Pcl, these nucleosomes are mono- and dimethylated similar to the rest of the genome. Third, in Pcl mutants, H3-K27me1 and H3-K27me2 X-ChIP signals are also two-fold increased at control regions 1 and 2 (Figure 5). One possible explanation for this effect could be that in the absence of Pcl, a larger proportion of ‘free' PRC2 becomes available for the untargeted genome-wide mono- and dimethylation of H3-K27.
One possible explanation for the reduced H3-K27 trimethylation at PcG target genes in Pcl mutants could be that Pcl is required for anchoring of PRC2 at PREs. In X-ChIP assays with antibodies against E(z), we were unable to detect E(z) at PREs in wild-type embryos, but antibodies against Su(z)12 revealed enrichment of Su(z)12 at the PREs of all nine genes in wild-type embryos (Figure 5). We note that the Su(z)12 X-ChIP signals at HOX gene PREs in embryos are substantially lower than in imaginal discs (Papp and Müller, 2006), and, at all PREs, the signals were lower than the Pho or Ph X-ChIP signals in embryos (Figure 5). Nevertheless, we found that Su(z)12 X-ChIP signals at most PREs are slightly reduced in Pcl mutant embryos compared with wild-type embryos even though at most PREs the reduction was within the experimental error and binding of Su(z)12 was still higher than in the control regions 1 and 2 (Figure 5). It thus appears that PRC2 binding at PREs is reduced but not abolished in the absence of Pcl. In contrast, Pho and Ph X-ChIP signals are indistinguishable in Pcl mutant and wild-type embryos (Figure 5). Binding of PhoRC and PRC1 to PREs thus seems to be unaffected in the absence of Pcl protein. This observation is consistent with earlier findings that suggest that Pho directly or indirectly targets PRC1 to PREs, independently of H3-K27 trimethylation (Mohd-Sarip et al, 2005, 2006; Klymenko et al, 2006).
Finally, we asked how the lack of Pcl protein and the concomitant reduction of H3-K27 trimethylation affect repression of these target genes. To this end we compared the expression patterns of Ubx, Abd-B, en, wg, cad and Dll in wild-type and in Pcl mutant embryos by staining embryos with antibodies against their protein products. Pcl mutant embryos show widespread misexpression of Ubx and Abd-B, whereas en is only misexpressed in a few rare cells, and we have been unable to detect misexpression of cad, wg or Dll in these mutant embryos (Figure 5). Similar misexpression phenotypes were observed in embryos homozygous for other PcG mutations: only Ubx and Abd-B show widespread misexpression, en and cad are only misexpressed in a few rare cells, and wg and Dll show no detectable misexpression (McKeon and Brock, 1991; Moazed and O'Farrell, 1992; Simon et al, 1992; J Müller, unpublished observations). Intriguingly, this was also true for embryos that were homozygous for the temperature-sensitive allele E(z)61 and had been reared at the restrictive temperature and therefore lacked detectable levels of K27 di- and trimethylation (data not shown; Cao et al, 2002; Ketel et al, 2005). Taken together, these observations suggest the following: first, in Pcl mutants, H3-K27 trimethylation levels are evidently also reduced in the chromatin of target genes that are not becoming widely misexpressed in the embryo and it also occurs in upstream control regions. This suggests that the reduction of H3-K27me3 levels is probably directly due to the lack of Pcl and/or the reduction of PRC2 binding at target genes, and is not a secondary consequence of these target genes becoming transcriptionally active. Second, in Pcl and other PcG mutants, not all target genes show the widespread misexpression that is observed in the case of HOX genes. This suggests that removal of PcG function alone does not result in transcriptional activation of these target genes, but that specific transcriptional activators are needed in order for these genes to become activated in cells outside of their normal expression domains. Consistent with this, we find that even though en is only subtly misexpressed in Pcl mutant embryos, during larval stages, en shows widespread misexpression in Pcl mutant clones in imaginal discs (Supplementary Figure S3). Similarly, we find that cad, Dll and wg become misexpressed in PcG mutant clones in imaginal discs (Beuchle et al, 2001, and data not shown). This suggests that although high levels of H3-K27 trimethylation may not yet be critical during embryonic development, they appear to become critical for repression of these target genes during larval development.
Discussion
In this study, we show that Pcl-PRC2 is a distinct form of the PRC2 HMTase, with a critical role in H3-K27 trimethylation at PcG target genes. In the following sections we shall discuss in turn the main conclusions that can be drawn from the data reported here.
Biochemically purified Pcl complexes contain Pcl together with the four core subunits of PRC2. In contrast, our biochemically purified E(z) complexes contain only substoichiometric amounts of Pcl and the previously described purifications of PRC2 failed to reveal Pcl in the purified material (Cao et al, 2002; Czermin et al, 2002; Müller et al, 2002; Kuzmichev et al, 2002). Moreover, fractionation of crude nuclear extracts by gel filtration indicated that Pcl and PRC2 components Esc, E(z) and Su(z)12 co-fractionate in high-molecular-weight assemblies, but that the bulk of these other PRC2 components is present in lower-molecular-weight fractions that do not contain Pcl (O'Connell et al, 2001; Tie et al, 2003). Taken together, these observations suggest that only a fraction of PRC2 is associated with Pcl and that Pcl-PRC2 is a distinct complex.
Previous X-ChIP studies showed that Pcl and Su(z)12 colocalize at Ubx and Abd-B PREs (Papp and Müller, 2006). This suggests that Pcl-PRC2 is bound at these PREs. Here, the analysis of Drosophila mutants that lack Pcl protein and therefore lack Pcl-PRC2, allowed us to obtain insight into the function of this complex. Our results provide strong evidence that Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation in the chromatin of PcG target genes. Unlike in E(z) or Su(z)12 mutants, removal of Pcl in embryos or in imaginal discs only reduces but does not eliminate H3-K27 trimethylation. Nevertheless, repression of several PcG target genes is abolished in Pcl mutants. This suggests that not only the mere presence of H3-K27me3, but presence of high levels of H3-K27me3 is crucial for maintaining these PcG target genes in the repressed state. Previous studies on the Ubx gene suggested that presence of H3-K27 trimethylation in the promoter and coding region is critical for PcG repression (Papp and Müller, 2006). One possibility would be that it is the overall density of H3-K27me3-marked nucleosomes across the promoter and coding region that determines whether a PcG target gene is repressed. Another possibility would be that even though a whole chromatin domain becomes trimethylated at H3-K27, only a few H3-K27me3-marked nucleosomes at a particular position (e.g., around the transcription start site) are actually required for repression, and failure to maintain this trimethylation results in loss of repression.
The observation that Su(z)12 binding and H3-K27 trimethylation are reduced but not lost in the absence of Pcl is consistent with the idea that Pcl might help anchoring PRC2 to PREs, but it also suggests that at least some PRC2 must be targeted to PREs independently of Pcl. It seems likely that the residual H3-K27 trimethylation present in Pcl mutant embryos and in Pcl mutant clones in imaginal discs is generated by PRC2 that is bound at PREs independently of Pcl. In this context it is important to note that we found that not only Pcl-PRC2 but also PRC2 is able to trimethylate H3-K27 in recombinant nucleosomes in vitro. Apart from the suggested role in tethering of PRC2 to PREs, it is possible that Pcl also functions in a post-recruitment step to help PRC2 generate high levels of H3-K27 trimethylation at target genes. For example, the tudor domain and PHD fingers of PRE-bound Pcl might interact with modified nucleosomes in the promoter and coding region of target genes to ensure that they become trimethylated at H3-K27 by the associated PRE-tethered PRC2.
Finally, we found no evidence that Pcl-PRC2 would be required for the genome-wide H3-K27 mono- and dimethylation. Our X-ChIP analyses suggest that H3-K27 mono- and dimethylation across the genome might even slightly increase in the absence of Pcl (Figure 5). In contrast, there is a loss of all H3-K27 methylation in either E(z) or Suz)12 mutants (Ebert et al, 2004; Figure 4B). This suggests that PRC2 or another E(z)-containing complex generates the genome-wide H3-K27 mono- and dimethylation. The experiments in Pcl mutants thus allowed us to dissect the role of different H3-K27 methylation states in Drosophila. The selective reduction of H3-K27me3 levels, and the concomitant loss of repression of PcG target genes in Pcl mutants, provides compelling evidence that only the trimethylated state of H3-K27 is functional in PcG repression in Drosophila. Pcl-PRC2 is evidently critically needed to generate the high levels of H3-K27 trimethylation that are required to maintain a Polycomb-repressed chromatin state.
Materials and methods
Tandem affinity purification
The a-tubulin-TAP-Pcl and a-tubulin-TAP-E(z) transgenes in the Drosophila transformation vector CaSpeR have the following structure: a 2.6-kb fragment of the a-tubulin 1 gene, containing promoter and 5′ untranslated region sequences (Struhl and Basler, 1993) linked to the N-terminal TAP tag (Rigaut et al, 1999), followed by Pcl or E(z) cDNA fragments that contain the ORF of Pcl (Pcl1−1043) or E(z) (E(z)1−760), respectively (plasmid maps are available on request). Rescue function of the a-tubulin-TAP-Pcl and a-tubulin-TAP-E(z) transgenes was tested by introducing the transgene into a Pcl22M21/Pcl22M21 or E(z)731/E(z)63 mutant background, respectively. Specifically, in the case of Pcl, we recombined a copy of the a-tubulin-TAP-Pcl transgene onto an FRT40 FRT42D y+Pcl22M21 chromosome and found that yw; a-tubulin-TAP-Pcl FRT40 FRT42D y+Pcl22M21/a-tubulin-TAP-Pcl FRT40 FRT42D y+Pcl22M21 animals are wild type in appearance, viable and fertile. Similarly, we found that the a-tubulin-TAP-E(z) transgene rescues animals that are trans-heterozygous for two protein-negative E(z)-null alleles (Müller et al, 2002); w; a-tubulin-TAP-E(z); E(z)731FRT2A/E(z)63FRT2A animals are wild type in appearance, viable and fertile. TAP was performed from embryonic nuclear extracts as previously described (Klymenko et al, 2006); see Supplementary Figure S6 for detailed information.
Mass spectrometry
A detailed list of peptide sequences obtained from mass spectrometry analysis of the protein bands shown in Figure 1 is available in Supplementary Figure S1. For quantitative mass spectrometry of modified peptides, the samples were separated on a nano-flow 1D-plus Eksigent (Eksigent, Dublin, CA) HPLC system coupled to a qStar Pulsar i quadrupole time-of-flight MS (Applied Biosystems, Darmstadt, Germany). The resulting MASCOT search result file and the generic mass spectrometry data of each sample were parsed using MSQuant in a no-label setting (Kratchmarova et al, 2005). The quantitation result (peptide ion volumes in thompson·sec) for each peptide was presented in a diagram (Figure 3B). The presented data was an average of a duplicate analysis. To minimize differences between the samples, the ion volume of all modified and unmodified peptides was summed and normalized (Fraterman et al, 2007).
Immunostaining of discs and embryos
Staining of imaginal discs and embryos was performed as described (Beuchle et al, 2001).
X-ChIP assays
X-ChIP on wt and Pcl−/− embryos was performed as described in Klymenko et al (2006). Pcl22M21/Pcl22M21 embryos were collected from an FRT40 FRT42D y+Pcl22M21/CyO ubi-nGFP strain by selecting for GFP-negative embryos using an embryo sorter (COPAS™ SELECT, Union Biometrica). Primers used for amplification are listed in Supplementary Figure S4.
Antibodies
All antibodies used in this study are listed in Supplementary Figure S5.
Protein expression and purification
Recombinant PRC2 complex was expressed and purified as described in Nekrasov et al (2005).
HMTase assay and mononucleosome assembly
Assays were performed as described (Nekrasov et al, 2005).
Embryonic extract preparation
For Western blot experiments shown in Figure 4, embryos were dechorionated and taken up in ice-cold PBS buffer containing 0.01% Triton. Embryos were homogenized with a glass dounce homogenizer, lysate was cleared at 400 g for to pellet the embryonic carcasses. Supernatant was centrifuged at 1100 g and the pellet was resuspended in SDS–Laemmli buffer. For Western blots on larval extracts, imaginal disc and CNS tissues were dissected from wild-type or mutant larvae and resuspended in SDS–Laemmli buffer.
Fly strains and alleles
The following strains were used in this study:
w; FRT40 FRT42D y+Pcl22M21/SM6B
w; E(z)731FRT2A/TM6C
w; E(z)63FRT2A/TM6C
w; Su(z)1214FRT2A/TM6C
y w hs-flp; hs-nGFP FRT2A
y w hs-flp; FRT42D hs-nGFP
Pcl22M21 was isolated in a screen for new PcG mutation; details for the screen will be described elsewhere (Gaytán de Ayala Alonso et al, 2007). In brief, Pcl22M21 was induced with EMS on an FRT40 FRT42D y+ chromosome; complementation tests with PclD5 revealed that Pcl22M21 is an allele of Pcl. Sequence analysis revealed a single nucleotide exchange in Pcl22M21, changing the codon E701 from GAG into a premature stop codon (TAG).
Supplementary Material
Supplementary Figures
Acknowledgments
We are grateful to Thomas Jenuwein and Susanne Opravil for the generous gift of antibodies. We thank Brigitte Wild for molecular characterization of the Pcl22M21 allele, Ann-Mari Voie for help with fly injections, Vladimir Benes and the GeneCore for excellent support with qPCR analysis and Andrew Riddell for help with the embryo-sorter, and also thank Danny Reinberg and Kavitha Sharma for discussions. Comments from Eileen Furlong helped to improve the paper. MN is supported by an ‘E-STAR' fellowship funded by the EC's FP6 Marie Curie Host fellowship for Early Stage Research Training under contract number MEST-CT-2004-504640. TK and BP are supported by grants from the Deutsche Forschungsgemeinschaft. Work in the lab of HS is supported by funding from the 6th Research Framework Programme of the European Union Project Heroic (LSHG-CT-2005-018883).
References
- Beuchle D, Struhl G, Müller J (2001) Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128: 993–1004 [DOI] [PubMed] [Google Scholar]
- Birve A, Sengupta A, Beuchle D, Larsson J, Kennison J, Rasmuson-Lestander A, Müller J (2001) Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development 128: 3371–3379 [DOI] [PubMed] [Google Scholar]
- Boyer L, Plath K, Zeitlinger J, Brambrink T, Medeiros L, Lee T, Levine S, Wernig M, Tajonar A, Ray M, Bell G, Otte A, Vidal M, Gifford D, Young R, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Breen TR, Duncan IM (1986) Maternal expression of genes that regulate the bithorax complex of Drosophila melanogaster. Dev Biol 118: 442–456 [DOI] [PubMed] [Google Scholar]
- Brock HW, Fisher CL (2005) Maintenance of gene expression patterns. Dev Dyn 232: 633–655 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G (2004) Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev 18: 2973–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraterman S, Zeiger U, Khurana TS, Wilm M, Rubinstein NA (2007) Quantitative proteomic profiling of sarcomere associated proteins in limb and extraocular muscle allotypes. Mol Cell Proteomics 6: 728–737 [DOI] [PubMed] [Google Scholar]
- Gaytán de Ayala Alonso A, Gutiérrez L, Fritsch C, Papp B, Beuchle D, Müller J (2007) A genetic screen identifies Novel polycomb group genes in Drosophila. Genetics (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R, Gelbart W (1990) Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics 126: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn TG, Schwartz YB, Dellino GI, Pirrotta V (2006) Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J Biol Chem 281: 29064–29075 [DOI] [PubMed] [Google Scholar]
- Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA (2005) Subunit contributions to histone methyltransferase activities of fly and worm Polycomb group complexes. Mol Cell Biol 25: 6857–6868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Köcher T, Schelder M, Fritsch C, Wild B, Wilm M, Müller J (2006) A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev 20: 1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Makarevich G (2006) Epigenetic mechanisms governing seed development in plants. EMBO Rep 7: 1223–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M (2005) Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science 308: 1472–1477 [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Jenner R, Boyer L, Guenther M, Levine S, Kumar R, Chevalier B, Johnstone S, Cole M, Isono K, Koseki H, Fuchikami T, Abe K, Murray H, Zucker J, Yuan B, Bell G, Herbolsheimer E, Hannett N, Sun K et al. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE (2002) The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Cell 17: 6070–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon K, Brock H (1991) Interactions of the Polycomb group of genes with homeotic loci of Drosophila. Rouxs Arch Dev Biol 199: 387–396 [DOI] [PubMed] [Google Scholar]
- Moazed D, O'Farrell PH (1992) Maintenance of the engrailed expression pattern by Polycomb group genes in Drosophila. Development 116: 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, Cleard F, Mishra R, Karch F, Verrijzer P (2005) Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes Dev 19: 1755–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, van der Knaap JA, Wyman C, Kanaar R, Schedl P, Verrijzer P (2006) Architecture of a polycomb nucleoprotein complex. Mol Cell 24: 91–100 [DOI] [PubMed] [Google Scholar]
- Mollaaghababa R, Sipos L, Tiong SY, Papoulas O, Armstrong JA, Tamkun JW, Bender W (2001) Mutations in Drosophila heat shock cognate 4 are enhancers of Polycomb. Proc Natl Acad Sci USA 98: 3958–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Müller J, Kassis JA (2006) Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr Opin Genet Dev 16: 476–484 [DOI] [PubMed] [Google Scholar]
- Negre N, Hennetin J, Sun L, Lavrov S, Bellis M, White K, Cavalli G (2006) Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol 4: 0918–0932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M, Wild B, Müller J (2005) Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep 6: 348–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell S, Wang L, Robert S, Jones CA, Saint R, Jones RS (2001) Polycomblike PHD fingers mediate conserved interaction with Enhancer of zeste protein. J Biol Chem 276: 43065–43073 [DOI] [PubMed] [Google Scholar]
- Papp B, Müller J (2006) Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev 20: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux S, Melfi R, Pirrotta V (2001) Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev 15: 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R (2004) Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet 38: 413–443 [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE (2001) A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412: 655–660 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G (2007) Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Schwartz Y, Kahn T, Nix D, Li X, Bourgon R, Biggin M, Pirrotta V (2006) Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Schwartz Y, Pirrotta V (2007) Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE (1999) Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37–46 [DOI] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W (1992) Ten different Polycomb group genes are required for spatial control of the abd-A and Abd-B homeotic products. Development 114: 493–505 [DOI] [PubMed] [Google Scholar]
- Soto MC, Chou TB, Bender W (1995) Comparison of germline mosaics of genes in the Polycomb group of Drosophila melanogaster. Genetics 140: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M (2006) Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 6: 846–856 [DOI] [PubMed] [Google Scholar]
- Struhl G, Basler K (1993) Organizing activity of wingless protein in Drosophila. Cell 72: 527–540 [DOI] [PubMed] [Google Scholar]
- Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ (2001) The Drosophila Polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128: 275–286 [DOI] [PubMed] [Google Scholar]
- Tie F, Prasad-Sinha J, Birve A, Rasmuson-Lestander A, Harte PJ (2003) A 1-megadalton ESC/E(Z) complex from Drosophila that contains polycomblike and RPD3. Mol Cell Biol 23: 3352–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M (2006) Genome-wide profiling of PRC1 and PRC2. Polycomb chromatin binding in Drosophila melanogaster. Nat Genet 38: 694–699 [DOI] [PubMed] [Google Scholar]
- Vachon G, Cohen B, Pfeifle C, McGuffin ME, Botas J, Cohen SM (1992) Homeotic genes of the bithorax complex repress limb development in the abdomen of the Drosophila embryo through the target gene Distal-less. Cell 71: 437–450 [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS (2004) Hierarchical recruitment of polycomb group silencing complexes Mol. Cell 14: 637–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures