Abstract
Background
Honey bee (Apis mellifera) workers are characterized by complex social behavior. Their life-history is dominated by a period of within-nest activity followed by a phase of long-distance flights and foraging. General insights into insect metabolism imply that foraging onset is associated with fundamental metabolic changes, and theory on social evolution suggests metabolic adaptations that are advantageous for the colony as a whole.
Results
Here we address the life-history characteristics of workers with LC-MS/MS based relative quantification of major proteins. Our approach includes: i. Calculation of a false positive rate for the identifications, ii. Support of relative protein quantification results obtained from spectral count by non-parametric statistics, and iii. Correction for Type 1 error inflation using a bootstrap iteration analysis. Our data are consistent with the use of glucose as the main fuel for honey bee flight. Moreover, the data delivers information on the expression of ATPsynthases/ATPases, and provide new insights into nurse- and forager-specific patterns of protection against oxidative stress.
Conclusion
The results show the suitability of this approach to investigate fundamental biochemical changes in an insect, and provide new evidence for metabolic specializations that occur during the social ontogeny of worker honey bees.
Background
Studies of the biochemistry of life-history progression provide insights into the dynamic properties of biological systems, including metabolic change [1]. A fundamental step in evolution was the emergence of social life, as exemplified by social insects [2]. The best-studied insect model in sociobiology is the honey bee, which has proven useful for understanding regulatory changes that may have occurred during social evolution [3-7]. Work on honey bee life-history progression has focused on the transition from nest-tasks to flight activity and foraging, a behavioral shift characteristic of the caste of facultative sterile worker females (reviewed by [8]). Several studies have demonstrated changes in brain mRNA levels when worker bees shift from working in the nest to foraging flights approximately 2–3 weeks after adult emergence [9-12]. Differences are found for genes involved in signal transduction and primary metabolism [13], but mRNA expression does not always correlate with protein concentrations and metabolic state [14-17].
Enzyme activity measurements and flux analyses on worker bees, including studies of hexokinase, phosphofructokinase, glyceraldehyde-3-phosphate-dehydrogenase, citrate synthase and cytochrome oxidase, indicate that oxidation of carbohydrates is the primary energy source used for foraging flights (e.g. [18-20]. Also, recent use of a 2d-gel based proteomic approach in combination with enzyme assays point to metabolic changes in honey bee thorax muscle that can increase flight ability after foraging onset (increase in troponin T 10a and citrate synthase, [21]). We address this important life-history transition by using an electrospray tandem mass spectrometry approach for relative quantification of the whole-body proteome of nest workers and foragers. A common argument against whole-body analyses is that the contributions of different organs or tissue types to the overall pattern are unclear. Yet whole-body analyses have provided fundamental insights into key biological processes like aging, development, and immunity (e.g. [22-24]: They focus quantitatively on the most important changes in the organism as a whole, and complement more targeted tissue- or organ-specific profiling methods.
Although not routine, recent descriptions of some honey bee subproteomes (subset of the whole proteome of an organism, i.e. royal jelly proteome, bee venome proteome, hypopharyngeal gland proteome, haemolymph proteome), prove that proteomic research on honey bees using mass spectrometry is feasible [25-28]. Our approach included an estimate of the false positive rate, non-parametric statistics, and iterative correction against Type 1 error inflation. In total, 113 proteins were identified and 47 of them quantified by our approach. Levels of 15 proteins diverged significantly between nest workers and foragers. Our data are consistent with the use of glucose as the main fuel for honey bee flight. Moreover, the data delivers information on the expression of ATPsynthases/ATPases, and provide new insights into nurse- and forager-specific patters of protection against oxidative stress.
Results and discussion
Methodology setup and validation
Experimental manipulation and data analysis at a systemic, whole body level can readily increase our knowledge about living organisms. This principle is apparent e.g. when looking at the outcome of systemic RNA interference approaches (e.g. [29,30]). Here, tissue specific effects are not well defined but global response can be monitored and understood. If differences can be observed on the whole-organism level they are likely to be substantial, as they are robust to deflation by combining body compartments. Further, although separating an organism into its parts can help to clarify how contributions of different tissues translate into global patterns, the data cannot easily be added up to the overall pattern of any protein. This is because background matrix will vary from tissue to tissue and might impair quantification itself. Therefore, whole body analyses can contribute in a unique way to increase our knowledge about biochemical changes during honey bee ontogeny.
Our approach consists of a protein extraction step, followed by protein digestion, separation of peptides via HPLC and characterization of the proteins by mass spectrometry. We were able to identify 113 proteins and several hundred peptides with this approach [see Additional files 1 and 2]. Relative quantification is accomplished by spectrum count where the spectra that are identified for any given protein in one sample are counted and this value is compared to the value obtained for the same protein in another sample [31-35]. Thus, every quantified protein was identified several times leading to a high reliability of identification.
In our experiment, actin showed the most stable protein quantity over all samples and was used for standardization. Use of honey bee actin for relative quantification is also common in mRNA expression studies (e.g., [3]). We further validated our approach to data handling by standardization toward the overall spectral count obtained for each sample. Statistics were in good agreement between these two methods of correction [see Additional file 1], and even without standardization more than 75% of the observed differences in protein expression remained (data not shown). Our analysis, therefore, is both reproducible and robust, and in the following we report the results based on correction toward actin. Of the 47 quantifiable proteins, 15 were expressed at significantly different levels in nest bees and foragers [as determined by Mann-Whitney U-test, P < 0.05, n = 12, see Additional file 1]. A hierarchical clustering analysis computed on the quantifiable proteins revealed distinct clustering of behavioral groups with one exception [Figure 1]. Surprisingly, one forager clusters much closer to the nest bees than to the other forager bees. This could be because the respective bee was on the verge of changing its tasks (bees in this study were of unknown age) what would imply that some physiological features of the nest bee stage are retained in young foragers and that there is a gradual rather than an abrupt change in the expression pattern of some proteins when bees change from in-hive tasks to foraging.
Figure 1.
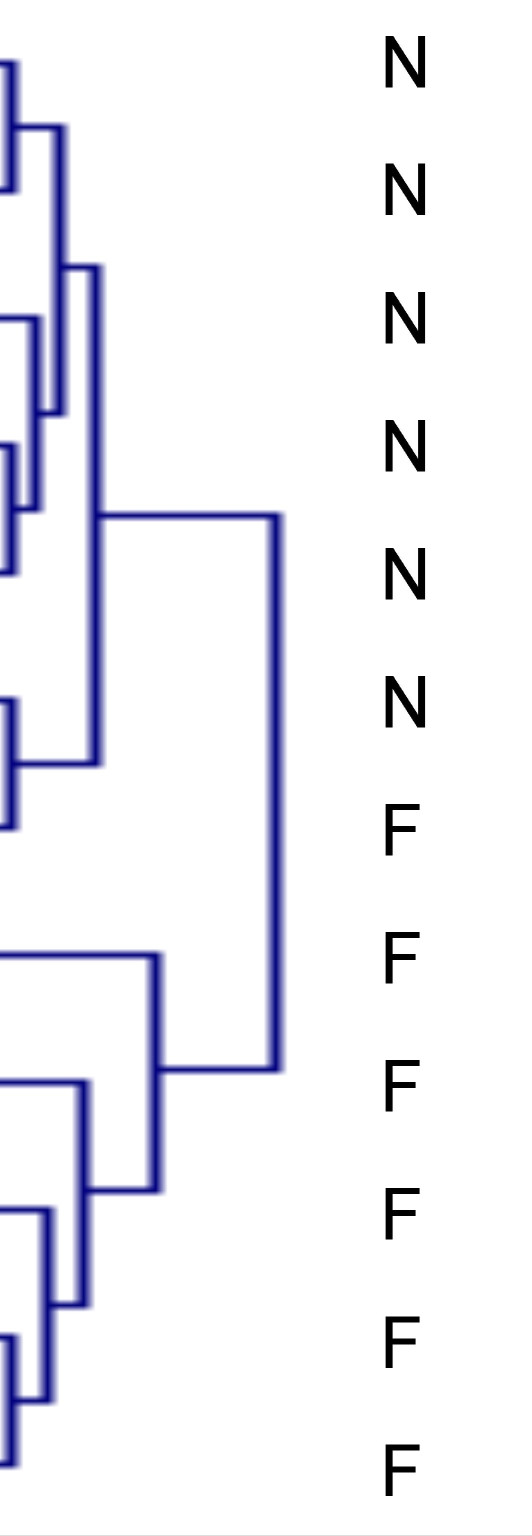
Result of a hierarchical clustering analysis computed on quantifiable proteins.
Glucose consumption
The main task of foragers is to provide the colony with nectar and pollen. Consequently, one of their major behavioral characteristics is long-distance flights for the purpose of food collection. There is current agreement that honey bee flight is predominately fueled through hexose sugars [20,36]. Thereby, primarily glucose is used to produce ATP and NADH necessary for the exhaustive demands of the flight process. The bee thorax is largely dominated by flight muscle tissue [37], and proteins of the thorax make up a major fraction of total bee protein [38]. In addition, metabolic rate during flight is much higher than during non-flight activities [39], and activities of glycolytic enzymes increase from the nest bee stage to the forager stage [40]. These observations led us to expect that in honey bees the concentrations of proteins in the major pathways of ATP production and use can reflect flight muscle contents, and that they should be higher in foragers than in nest bees. This higher level would indicate a major physiological adaptation associated with the shift in social behavior from nest tasks to intense flight activity.
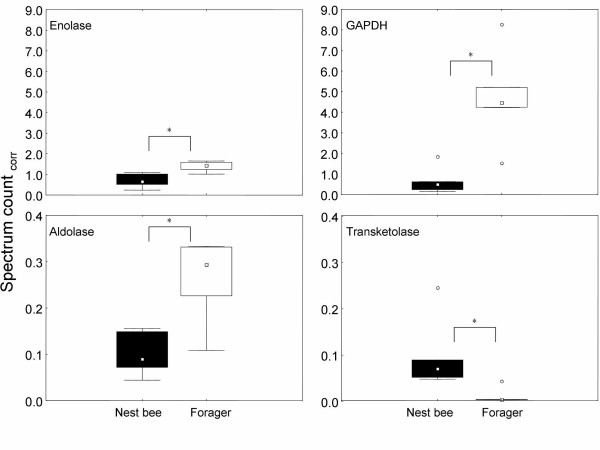
In accordance with this prediction, we found that proteins similar to the glycolytic enzymes fructose-1,6-bisphosphate aldolase, glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) and enolase are significantly elevated in foragers [Fig. 2].
Figure 2.
Box-and-whisker-plots of selected proteins associated with glucose processing Boxes represent 25–75% percentiles of the data, outliers are marked as open circles. * denotes significant differences between nest bees and foragers, for p-values see Additional file 1. Aldolase: fructose-1-6-bisphosphate aldolase, GAPDH: glyceraldehyde-3-phosphate-dehydrogenase. Spectrum countcorr: spectrum count corrected for actin.
Food synthesis and processing
Nest bees and foragers have different provisioning functions. Nest bees care for the brood and queen, whereas foragers bring in the resources needed for colony growth and reproduction. Unlike foragers, nest bees synthesize proteinaceous jelly in a set of paired acinous head glands (hypopharyngeal glands), and these secretions are fed to the brood and to other adult bees including foragers [41]. A family of proteins specific to jelly are the major royal jelly proteins [42,43]. In general, the bee's biology of jelly synthesis and transfer matches well with our finding of higher values for a major royal jelly protein in nest bees compared to foragers [see Additional file 1]. We also observed that a protein similar to a transketolase, which is an important enzyme in the pentosephosphate-pathway, is downregulated in foragers [Fig. 2]. One of the major functions of this pathway is the production of NADPH and building blocks that can be used in biosynthetic reactions [44]. This may point to a more important role of reactions involving NADPH in nest bees compared to foragers.
To fuel flight, foragers must utilize high energy containing food. Their capacity of glycolysis appears to be upregulated compared to nest bees (described above). However, we reasoned that enzymes involved in sugar uptake and processing would also be upregulated. Interestingly, a protein highly similar to an alpha-glucosidase, a protein that is involved in the processing of disaccharides to glucose, shows higher levels in foragers [see Additional file 1]. This result agrees well with previous reports by Kubo et al. on elevated levels of alpha-glucosidase mRNA and protein in foragers [45-47]. We conclude that our observations generally agree with a higher capacity of sugar processing and NADH production in foraging honey bees.
Citric acid cycle enzymes
Pyruvate produced by glycolysis is converted to acetyl-CoA and enters the citric acid cycle, which serves further production of reduction equivalents and biosynthetic precursors. On an organismal level, our data suggest that the dynamics of the honey bee citric acid cycle enzymes are complex. While a malate dehydrogenase-like protein is upregulated in foragers, the amounts of other enzymes of the cycle including an isocitrate dehydrogenase-like protein and a protein similar to oxoglutarate dehydrogenase remained seemingly unchanged [see Additional file 1]. These results suggest that some enzymes of the citric acid cycle can be regulated by metabolites or posttranslational modifications rather than on the level of enzyme amount [48], that higher expression levels in one tissue are accompanied by lower expression levels in another tissue, that differences were too subtle to be detected by the method we used, and/or that malate dehydrogenase has an additional role besides its involvement in the citric acid cycle, e.g. its function in an aspartate/malate shuttle [49]. Interestingly, Sacktor et al. reported a sharp increase of malate concentration at the beginning of flight in blowfly while levels of citrate, oxaloacetate and α-ketoglutarate remained seemingly unchanged [50]. However, no explanatory framework was provided with their observation.
One major consumer of NADH generated by glycolysis and the citric acid cycle is the mitochondrial respiratory chain. While electrons are transported from NADH to oxygen, an electrochemical gradient is build up over the inner mitochondrial membrane. This gradient in turn is used to drive the generation of ATP by ATPsynthases. These proteins, accordingly, play a key role in production of ATP. As muscle activity depends largely on the availability of ATP, and as an increase in mitochondria and cytochromes from young nest bees to older foragers was reported by Herold and coworkers [51,52], we expected higher levels of respiratory chain enzymes in foragers. The lack of differences in cytochrome c-levels (a protein of the mitochondrial respiratory chain) was thus surprising. Notably, the overall spectrum count for cytochrome c was low, and therefore the lack of differences between nest bees and foragers could to be due to methodological limitations. A change in the protein extraction technique could enable improved insight into these dynamics in future analyses.
Enzymes related to ATP generation and consumption
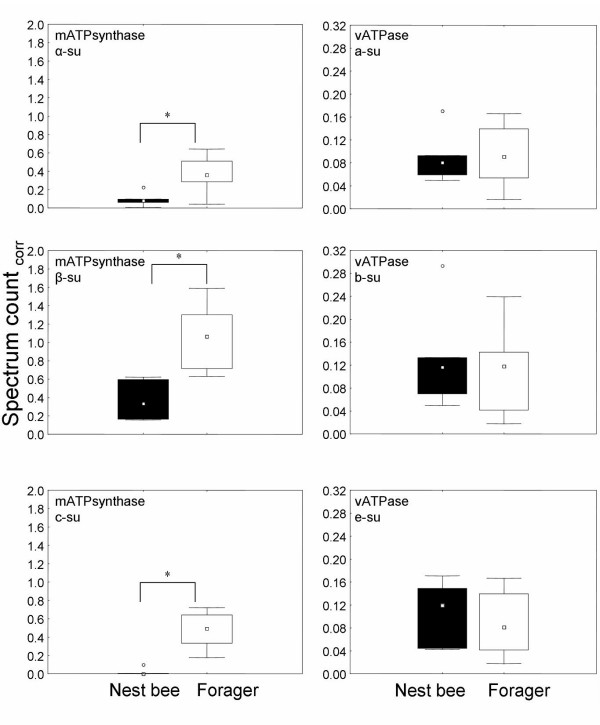
ATPsynthase subunit homologs were more abundant in foragers, in accordance with foragers having larger ATP production capacities than nest bees [Fig. 3].
Figure 3.
Box-and-whisker-plots of proteins homologues to ATPases/synthases. Boxes represent 25–75% percentiles of the data, outliers are marked as open circles. * denotes significant differences between nest bees and foragers, for p-values see Additional file 1. Left: subunits (su) of the mitochondrial ATPsynthase. Right: subunits (su) of a v-type ATPase. Spectrum countcorr: spectrum count corrected for actin.
In contrast, levels of proteins similar to the subunits of a v-type ATPase remained unchanged [Fig. 3]. V-type ATPases are structurally similar to mitochondrial ATP synthases but serve other functions [53]. Instead of generating ATP, they establish and maintain an electrochemical gradient over plasma membranes under the consumption of ATP [54]. Our data indicate that v-type ATPases are equally important for both nest and forager bees and may illustrate how two structurally similar proteins (mATPsynthase and vATPase) can differ profoundly in their roles in life-history transitions.
The data discussed so far suggest that in foragers a whole pathway involved in the processing of glucose and the generation of ATP and NADH is optimized to meet the demands of the energy consuming flight process. However, we also observed differences that can not be explained by this line of argument. Arginine kinase, for example, converts arginine plus ATP into phosphoargine plus ADP and vice versa [55], and can be used to control the ATP resources and rapidly release ATP when necessary. Indeed, a highly related protein (creatine kinase) is one of the major players in muscle contraction and maintenance of ATP/ADP balance in mammals [56,57]. Arginine kinase activity has been reported in insect flight muscle and its role in maintaining high levels of ATP at the sites of muscle contraction has been discussed [58-60]. Thus, higher arginine kinase activities are often associated with greater locomotory performance. Based on these findings, one could assume that foragers – in constant demand for rapidly released energy, for maintenance of adequate ATP/ADP ratios during flight, and with greater locomotory performance – have higher levels of arginine kinase. Yet we found that arginine kinase levels are significantly higher in nest bees [see Additional file 1]. A possible explanation is that nest workers, which perform defecation flights and also use flight muscle contraction to maintain optimal nest temperature (fanning, warming), utilize more energy phosphates than foragers as their oxidative catabolic pathways may not yet be optimized for flight. However, arginine kinase mRNA is highly expressed in honey bee tissues other than the thorax (head, abdomen) [61], and thus the contribution of flight muscle arginine kinase to whole-body patterns are likely confounded by other sources. Ubiquitous distribution of arginine kinase might suggest its involvement in a synthetic redox/shuttle mechanism that is common to many tissue types.
Reactive oxygen defense
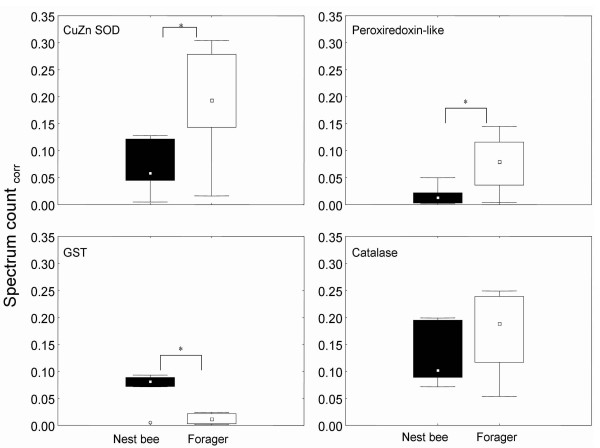
One class of proteins that in honey bees appears to deviate from general biochemical reasoning [7,62] is involved in protection against reactive oxygen species (ROS). A major threat for organisms that depend on oxygen for metabolism is production of ROS that cause oxidative damage to cellular components. Due to a high demand for oxygen during flight and, thereby, an assumed increase in ROS and a documented increase in oxidative damage [63]; one might reason that foragers generally invest in higher levels of proteins involved in ROS protection. Interestingly, at the whole-organism level we found no general bias of the relative abundance of proteins putatively involved in the protection against oxidative stress in foragers compared to nest bees [Fig. 4].
Figure 4.
Box-and-whisker-plots of proteins homologues to proteins putatively involved in oxygen stress response. Boxes represent 25–75% percentiles of the data, outliers are marked as open circles. * denotes significant differences between nest bees and foragers, for p-values see Additional file 1. CuZn SOD: CuZn superoxide dismutase. GST: glutathione-s-transferase. Spectrum countcorr: spectrum count corrected for actin.
Levels of CuZn superoxide dismutase and of a peroxiredoxin-like protein were observed to be at least 3-fold higher in foragers. Homologs of these proteins have been shown to protect mammalian muscle from oxidative damage and this might also be true in honey bees [64]. Other enzymes commonly associated with ROS protection are catalase and thioredoxin reductase, but their levels remained unchanged in foragers [Fig. 4 and Additional file 1]. In sharp contrast, glutathione-s-transferase (GST) levels, another enzyme involved in the response against oxidative stress and in detoxification but also in synthetic pathways [65], were at least 7-fold higher in nest bees. For some insects age-related decrease of GST activities and levels have been reported previously [66-68]. Studies on GSTs in honey bees revealed a complex influence of several parameters on GST activity including colony size, brood/adult ratio [69], insecticide treatment, temperature, and starvation [70]. Smirle and Winston reported that the total detoxification capacity related to GST and mixed function oxidases was lower in forager midguts compared to nest bee midguts [71]. However, since the enzyme activity per milligram midgut protein was higher in foragers, they stated that foragers account for loss of GST levels to some extent by increasing GST activity.
Proteins involved in the defense against ROS have major effects on longevity in many if not all organisms [72,73]. It has been suggested that lack of sufficient ROS defense mechanisms in foragers reflects reduced somatic maintenance, that this reduced maintenance conserves energetic resources at the colony level, and that it in part is responsible for rapid somatic decline in foraging bees [63]. In a recent study, our group showed that nest bees in comparison with foragers are more tolerant to the ROS inducing agent paraquat [7]. This effect was directly and positively linked to the circulating vitellogenin protein levels in the respective bees, and vitellogenin was shown to exhibit characteristic antioxidant function by being preferentially carbonylated by ROS. In the present study we were unable to quantify vitellogenin, although a possible peptide from this protein was detected [see Additional file 2]. This inability to quantify vitellogenin is likely due to relatively low whole-body abundance in the sampled bees. However, our proteomic data suggest that other compounds like GST might contribute to the higher ROS tolerance of nest bees, and that protection mechanisms in foragers may be limited to proteins indispensable for their foraging performance, as we have suggested previously [74-76].
Conclusion
In this study we compare the overall protein profiles of honey bee nest workers and foragers. The resulting data provide new biochemical insights into the major adult life-history transition of this social insect, although they do not permit the separation of behavioral and age effects. Our findings should encourage future studies on tissue-specific profiles that ultimately can reveal how different parts of the body contribute to the overall proteomic profile, and also show how each tissue contributes to major biochemical processes.
Methods
Chemicals
Unless indicated otherwise, chemicals were obtained from Sigma-Aldrich (St. Louis, USA) and Roche (Indianapolis, USA).
Bee sampling and protein extraction
Nest workers and foragers were collected from 2 wild type (unselected) honey bee colonies in the apiary of Arizona State University. Bees were sampled based on established behavioral assays: nest workers were collected in the brood nest while inserting their head into cells containing larvae (performing nursing behavior) [77], and foragers were collected at the hive entrances when returning from foraging flights [78]. Nectar and/or pollen loads were determined to confirm forager identity. Bees where then frozen in liquid nitrogen, and stored at -80°C until further use. Protein extraction was essentially performed as described in [79]. Briefly, a protein extraction mixture containing 50 mM hepes pH 7.5, 6 M urea, 40% sucrose, 1% β-mercaptoethanol, and 60 mM sodium fluoride (300 μl/bee) as well as tris buffered phenol (900 μl/bee) was added and bees were ground in this mixture at room temperature followed by incubation at 4°C for 15 min on a sample rotator. Subsequently, samples were centrifuged at 12.000 rpm for 5 min, and 300 μl of the upper phase were precipitated with ice-cold acetone at -20°C over night. On the following day the samples were centrifuged for 5 min at 10.000 rpm and the pellet was washed twice with 300 μl ice-cold methanol each. Finally, pellets were dried at RT for 20 min.
Protein digestion and sample preparation for HPLC
For tryptic digestion protein pellets were first redissolved in 50 μl of a dissolvation buffer containing 50 mM ammoniumbicarbonate, 8 M urea, and 1 mM CaCl2. Then, 150 μl of digestion buffer (50 mM ammoniumbicarbonate, 1 mM CaCl2) and 2 μl of a 0.5 μg/μl trypsin solution (trypsin in 1 mM HCl) were added and proteins were incubated over night at 37°C. Samples were desalted on the next day using C18 extraction plates (3 M empore, St. Paul, USA) following the instructions provided in the manual. Desalted peptides were dried in a speed vacuum device and stored at -20°C until further use.
HPLC and mass spectrometry
Dried peptides were redissolved in 8 μl of 5% acetonitrile, 2% TFA of which 6 μl were used for analyses. Peptides were separated on a monolithic column (100 μm ID, Merck, Darmstadt, Germany) using a 95 min gradient ranging from 95% A (0.1% formic acid, 99.9% H2O) to 80% B (0.1% formic acid, 99.9% acetonitrile) followed by a 15 min equilibration step. Peptides were eluted from the reversed phase μLC column directly into an LTQ mass spectrometer (Thermo, San Diego, USA). The isolation window was set to 3 m/z, collision energy to 35, and the activation time to 30 ms. MS2 was triggered for the three most abundant peaks in each MS spectrum. Using the open source search tool OMSSA [80] the spectra were matched against an A. mellifera sequence database retrieved from the National Center for Biotechnology Information (NCBI, Bethesda, MD, USA) containing additional trypsin and keratin sequences. The following criteria were used: 0.8 Da fragment tolerance, 2.0 Da precursor tolerance [81], maximum of 2 missed cleavages, only tryptic sequences allowed, only one hit per peptide reported (only the best hit for a given spectrum with a given scan number was shown), variable modifications: methionine oxidation.
To assess the rate of false positive identifications and to obtain an adequate e-value cutoff, peptides were also searched against a database containing the same sequences as the original database but in reversed order. False positive spectra were determined to make up less than 1% at an e-value of 1 (evalue -he 1), which was used in all analyses. In addition, a protein hit was only accepted if at least three spectra were obtained representing two distinct peptides or one single peptide if it represented more than 20% of the protein sequence and displayed an e-value of at least 0.1. Using these additional criteria no false positive protein hits were identified in a reverse database search. When hits with ambiguous identifications were obtained due to the presently unfinished annotation of the A. mellifera genome, affiliation of the identified peptides to certain proteins was ascertained using BLAST [82]. Proteins were quantified using spectrum count, which delivers an estimate for relative quantification [31,32,35]. For standardization purposes it was determined which of the proteins displayed the lowest variation throughout all samples. The individual values for this protein were then used to correct protein values by dividing each protein value by the "standard protein" value for the respective sample. In addition, we calculated ratios of individual spectral count of every quantified protein divided by the overall spectral count for every sample. This leads to normalized values that are corrected for overall protein amount. Proteins were only quantified, if accepted (criteria for acceptance as described) in at least four of the six measured samples belonging to one sample group (hive bee or forager). In the remaining samples where no peptides were found for the respective proteins, an arbitrary value of 0.1 was used for quantification calculations.
Statistics
Of 113 proteins identified, 47 met the criteria for quantification (n = 12, 6 nest bees and 6 foragers, one run each). Of these 47, data for 4 proteins did not conform to assumptions of ANOVA, as determined by Hartley-Cochran-Bartlett and the Levene's test. Therefore, the non-parametric Mann-Whitney U-test was used to test for significant differences between nest bees and foragers. To control the Type 1 error rate, 1000 bootstrap iterations were run for each protein using values corrected for actin amounts (see Results and Discussion). During one iteration, 6 expression values were randomly assigned to each of two groups, and a P-value calculated using the Mann-Whitney U-test. The 1000 P-values were subsequently sorted in ascending order, and the bootstrap cut-off value of the 5% lower tail was determined for the P-value distribution. This cut-off was > 0.13 for all the examined proteins, which implies that our report of significance at an alpha level of 0.05 is not associated with inflation of Type 1 error. The analysis was conducted with Statistica 6.0. The bootstrap algorithm was written in MatLab 6.5.
The hierarchical clustering analysis was conducted on the actin normalized values of the quantifiable proteins. Euclidean distance and average linkage clustering was used to visualize sample groups with common features (TIGR Multiexperiment Viewer version 4.0 b [83]).
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
FW designed the study and carried out the experimental work. GVA supervised the collection of bees for analysis, guided the non-parametric statistical analysis, and performed the bootstrap correction. FW and GVA wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Proteins quantified by spectrum count. Table displaying the results from a non-parametric Mann-Whitney-U-test (rank sums and p-level) for proteins quantified by spectrum count (threshold level for significant differences p < 0.05).
Peptides identified by LC-MS/MS. Table displaying all peptides identified by OMSSA with an e-value ≤ 1.0E-01. Lower case letter m: oxidized methionine. For details on OMSSA see [80].
Acknowledgments
Acknowledgements
We thank Nick Baker and M. Kim Fondrk for help with bees and Jon F. Harrison and Zhengping Yi for helpful comments on the manuscript. GVA was supported by NFR #171958, #175413, NSF #0615502 and The PEW Foundation and FW by a Feodor Lynen stipend provided by the Humboldt foundation.
Contributor Information
Florian Wolschin, Email: f.wolschin@gmail.com.
Gro V Amdam, Email: gro.amdam@asu.edu.
References
- Voit E, Neves AR, Santos H. The intricate side of systems biology. Proc Natl Acad Sci USA. 2006;103:9452–9457. doi: 10.1073/pnas.0603337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EO. The Sociogenesis of Insect Colonies. Science. 1985;228 doi: 10.1126/science.228.4707.1489. [DOI] [PubMed] [Google Scholar]
- Amdam GV, Norberg K, Fondrk MK, Page Jr. RE. Reproductive ground plan may mediate colony-level selection effects on individual foraging behavior in honey bees. PNAS. 2004;101:11350–11355. doi: 10.1073/pnas.0403073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch G, Wheeler D, Robinson GE. Endocrine Influences on the Organization of Insect Societies. In: Pfaff D, Arnold A, Etgen A, Fahrbach S, Moss R, Rubin R, editor. Hormones, Brain and Behavior. Vol. 3. San Diego , Academic Press; 2002. pp. 195–235. [Google Scholar]
- Robinson GE, Ben-Shahar Y. Social behavior and comparative genomics: new genes or new gene regulation? Genes Brain Behav. 2002;1:197–203. doi: 10.1034/j.1601-183x.2002.10401.x. [DOI] [PubMed] [Google Scholar]
- Robinson GE, Vargo EL. Juvenile hormone in adult eusocial Hymenoptera: gonadotropin and behavioral pacemaker. Arch Insect Biochem Physiol. 1997;35:559–583. doi: 10.1002/(SICI)1520-6327(1997)35:4<559::AID-ARCH13>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Seehuus SC, Norberg K, Gimsa U, Krekling T, Amdam GV. Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc Natl Acad Sci U S A. 2006;103:962–967. doi: 10.1073/pnas.0502681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GE, Grozinger CM, Whitfield CW. Sociogenomics: Social life in molecular terms. Nat Rev Gen. 2005;6:257–270. doi: 10.1038/nrg1575. [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Cziko AM, Robinson GE. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302:296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Band MR, Bonaldo MF, Kumar CG, Liu L, Pardinas JR, Robertson HM, Soares MB, Robinson GE. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res. 2002;12:555–566. doi: 10.1101/gr.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto M, Aoki M, Takada M, Kanou Y, Sasagawa H, Kitagawa Y, Kadowaki T. The changes of gene expression in honeybee (Apis mellifera) brains associated with ages. Zoolog Sci. 2004;21:23–28. doi: 10.2108/0289-0003(2004)21[23:TCOGEI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Kucharski R, Maleszka R. Evaluation of differential gene expression during behavioral development in the honeybee using microarrays and northern blots. Genome Biol. 2002;3:RESEARCH0007. doi: 10.1186/gb-2002-3-2-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield CW, Cziko AM, Robinson GE. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302:296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- Lehman HK, Schulz DJ, Barron AB, Wraight L, Hardison C, Whitney S, Takeuchi H, Paul RK, Robinson GE. Division of labor in the honey bee (Apis mellifera): the role of tyramine beta-hydroxylase. J Exp Biol. 2006;209:2774–2784. doi: 10.1242/jeb.02296. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie L, Wu G, Zhang W. Correlation between mRNA and protein abundance in Desulfovibrio vulgaris: a multiple regression to identify sources of variations. Biochem Biophys Res Commun. 2006;339:603–610. doi: 10.1016/j.bbrc.2005.11.055. [DOI] [PubMed] [Google Scholar]
- Hack CJ. Integrated transcriptome and proteome data: the challenges ahead. Brief Funct Genomic Proteomic. 2004;3:212–219. doi: 10.1093/bfgp/3.3.212. [DOI] [PubMed] [Google Scholar]
- Suarez RK, Lighton JR, Joos B, Roberts SP, Harrison JF. Energy metabolism, enzymatic flux capacities, and metabolic flux rates in flying honeybees. Proc Natl Acad Sci U S A. 1996;93:12616–12620. doi: 10.1073/pnas.93.22.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez RK, Darveau CA, Welch KC, Jr., O'Brien DM, Roubik DW, Hochachka PW. Energy metabolism in orchid bee flight muscles: carbohydrate fuels all. J Exp Biol. 2005;208:3573–3579. doi: 10.1242/jeb.01775. [DOI] [PubMed] [Google Scholar]
- Gmeinbauer R, Crailsheim K. Glucose-Utilization during Flight of Honeybee (Apis-Mellifera) Workers, Drones and Queens. J Insect Physiol. 1993;39:959–967. [Google Scholar]
- Schippers MP, Dukas R, Smith RW, Wang J, Smolen K, McClelland GB. Lifetime performance in foraging honeybees: behaviour and physiology. J Exp Biol. 2006;209:3828–3836. doi: 10.1242/jeb.02450. [DOI] [PubMed] [Google Scholar]
- Irving P, Troxler L, Heuer TS, Belvin M, Kopczynski C, Reichhart JM, Hoffmann JA, Hetru C. A genome-wide analysis of immune responses in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15119–15124. doi: 10.1073/pnas.261573998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Scarselli R, Donadio E, Giuffrida MG, Fortunato D, Conti A, Balestreri E, Felicioli R, Pinzauti M, Sabatini AG, Felicioli A. Towards royal jelly proteome. Proteomics. 2005;5:769–776. doi: 10.1002/pmic.200401149. [DOI] [PubMed] [Google Scholar]
- Santos KS, dos Santos LD, Mendes MA, de Souza BM, Malaspina O, Palma MS. Profiling the proteome complement of the secretion from hypopharyngeal gland of Africanized nurse-honeybees (Apis mellifera L.) Insect Biochem Mol Biol. 2005;35:85–91. doi: 10.1016/j.ibmb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Peiren N, Vanrobaeys F, de Graaf DC, Devreese B, Van Beeumen J, Jacobs FJ. The protein composition of honeybee venom reconsidered by a proteomic approach. Biochim Biophys Acta. 2005;1752:1–5. doi: 10.1016/j.bbapap.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Chan QW, Howes CG, Foster LJ. Quantitative comparison of caste differences in honeybee hemolymph. Mol Cell Proteomics. 2006;5:2252–2262. doi: 10.1074/mcp.M600197-MCP200. [DOI] [PubMed] [Google Scholar]
- Patel A, Fondrk MK, Kaftanoglu O, Emore C, Hunt G, Frederick K, Amdam GV. The Making of a Queen: TOR Pathway Is a Key Player in Diphenic Caste Development. PLoS ONE. 2007;2:e509. doi: 10.1371/journal.pone.0000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May RC, Plasterk RH. RNA interference spreading in C. elegans. Methods Enzymol. 2005;392:308–315. doi: 10.1016/S0076-6879(04)92018-6. [DOI] [PubMed] [Google Scholar]
- Cox B, Kislinger T, Emili A. Integrating gene and protein expression data: pattern analysis and profile mining. Methods. 2005;35:303–314. doi: 10.1016/j.ymeth.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Wienkoop S, Larrainzar E, Niemann M, Gonzalez EM, Lehmann U, Weckwerth W. Stable isotope-free quantitative shotgun proteomics combined with sample pattern recognition for rapid diagnostics. J Sep Sci. 2006;29:2793–2801. doi: 10.1002/jssc.200600290. [DOI] [PubMed] [Google Scholar]
- Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M, Paiement J, Kearney RE, Nilsson T, Bergeron JJ. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- Blatt J, Roces F. Haemolymph sugar levels in foraging honeybees (Apis mellifera carnica): dependence on metabolic rate and in vivo measurement of maximal rates of trehalose synthesis. J Exp Biol. 2001;204:2709–2716. doi: 10.1242/jeb.204.15.2709. [DOI] [PubMed] [Google Scholar]
- Harrison JF, Fewell JH, Anderson KE, Loper GM. Environmental physiology of the invasion of the Americas by Africanized honeybees. Integr Comp Biol. 2006;46:1110–1122. doi: 10.1093/icb/icl046. [DOI] [PubMed] [Google Scholar]
- Haydak MH. Changes in total nitrogen content during the life of the imago of the worker honeybee. Journal of Agricultural Research. 1934;49:21–28. [Google Scholar]
- Harrison JM. Caste-Specific Changes in Honeybee Flight Capacity. Physiol Zool. 1986;59:175–187. [Google Scholar]
- Hersch MI, Crewe RM, Hepburn HR, Thompson PR, Savage N. Sequential Development of Glycolytic Competence in Muscles of Worker Honeybees. Comp Biochem Phys B. 1978;61:427–431. [Google Scholar]
- Crailsheim K. Interadult feeding of jelly in honeybee (Apis mellifera L.) colonies. J Comp Physiol B. 1991;161:55–60. [Google Scholar]
- Schmitzová J, Klaudiny J, Albert S, Schröder W, Schreckengost W, Hanes J, Júdová J, Simúth J. A family of major royal jelly proteins of the honeybee Apis mellifera L. CMLS Cell Mol Life Sci. 1998;54:1020–1030. doi: 10.1007/s000180050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crailsheim K. The flow of jelly within a honeybee colony. J Comp Physiol B. 1992;162:681–689. [Google Scholar]
- Wood T. Physiological functions of the pentose phosphate pathway. Cell Biochem Funct. 1986;4:241–247. doi: 10.1002/cbf.290040403. [DOI] [PubMed] [Google Scholar]
- Kubo T, Sasaki M, Nakamura J, Sasagawa H, Ohashi K, Takeuchi H, Natori S. Change in the expression of hypopharyngeal-gland proteins of the worker honeybees (Apis mellifera L) with age and/or role. J Biochem-Tokyo. 1996;119:291–295. doi: 10.1093/oxfordjournals.jbchem.a021237. [DOI] [PubMed] [Google Scholar]
- Ohashi K, Natori S, Kubo T. Change in the mode of gene expression of the hypopharyngeal gland cells with an age-dependent role change of the worker honeybee Apis mellifera L. Eur J Biochem. 1997;249:797–802. doi: 10.1111/j.1432-1033.1997.t01-1-00797.x. [DOI] [PubMed] [Google Scholar]
- Ohashi K, Sawata M, Takeuchi H, Natori S, Kubo T. Molecular cloning of cDNA and analysis of expression of the gene for alpha-glucosidase from the hypopharyngeal gland of the honeybee Apis mellifera L. Biochem Biophys Res Commun. 1996;221:380–385. doi: 10.1006/bbrc.1996.0604. [DOI] [PubMed] [Google Scholar]
- Krebs HA. Rate control of the tricarboxylic acid cycle. Adv Enzyme Regul. 1970;8:335–353. doi: 10.1016/0065-2571(70)90028-2. [DOI] [PubMed] [Google Scholar]
- Marden JH. Variability in the size, composition, and function of insect flight muscles. Annu Rev Physiol. 2000;62:157–178. doi: 10.1146/annurev.physiol.62.1.157. [DOI] [PubMed] [Google Scholar]
- Sacktor B, Wormser-Shavit E. Regulation of metabolism in working muscle in vivo. I. Concentrations of some glycolytic, tricarboxylic acid cycle, and amino acid intermediates in insect flight muscle during flight. J Biol Chem. 1966;241:624–631. [PubMed] [Google Scholar]
- Herold RC, Borei H. Cytochrome Changes During Honey Bee Flight Muscle Development. Dev Biol. 1963;53:67–79. doi: 10.1016/0012-1606(63)90026-5. [DOI] [PubMed] [Google Scholar]
- Herold RC. Development and ultrastructural changes of sarcosomes during honey bee flight muscle development. Dev Biol. 1965;12:269–286. doi: 10.1016/0012-1606(65)90031-x. [DOI] [PubMed] [Google Scholar]
- Arechaga I, Jones PC. The rotor in the membrane of the ATP synthase and relatives. FEBS Lett. 2001;494:1–5. doi: 10.1016/s0014-5793(01)02300-6. [DOI] [PubMed] [Google Scholar]
- Beyenbach KW, Wieczorek H. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol J Exp Biol. 2006;209:577–589. doi: 10.1242/jeb.02014. [DOI] [PubMed] [Google Scholar]
- Cheung AC. Kinetic properties of arginine phosphokinase from honeybees, Apis mellifera L. (Hymenoptera, Apidae) Arch Biochem Biophys. 1973;154:28–39. doi: 10.1016/0003-9861(73)90031-3. [DOI] [PubMed] [Google Scholar]
- Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiological Reviews. 2000;80:1107–1213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
- Cain DF, Davies RE. Breakdown of adenosine triphosphate during a single contraction of working muscle. Biochem Biophys Res Commun. 1962;8:361–366. doi: 10.1016/0006-291x(62)90008-6. [DOI] [PubMed] [Google Scholar]
- Schneider A, Wiesner RJ, Grieshaber MK. On the Role of Arginine Kinase in Insect Flight-Muscle. Insect Biochemistry. 1989;19:471–&. [Google Scholar]
- Newsholme EA, Beis I, Leech AR, Zammit VA. Role of Creatine Kinase and Arginine Kinase in Muscle. Biochem J. 1978;172:533–537. doi: 10.1042/bj1720533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacktor B, Hurlbut EC. Regulation of metabolism in working muscle in vivo. II. Concentrations of adenine nucleotides, arginine phosphate, and inorganic phosphate in insect flight muscle during flight. J Biol Chem. 1966;241:632–634. [PubMed] [Google Scholar]
- Kucharski R, Maleszka R. Arginine kinase is highly expressed in the compound eye of the honey bee, Apis mellifera. Gene. 1998;211:343–349. doi: 10.1016/s0378-1119(98)00114-0. [DOI] [PubMed] [Google Scholar]
- Corona M, Hughes KA, Weaver DB, Robinson GE. Gene expression patterns associated with queen honey bee longevity. Mech Ageing Dev. 2005;126:1230–1238. doi: 10.1016/j.mad.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Seehuus SC, Krekling T, Amdam GV. Cellular senescence in honey bee brain is largely independent of chronological age. Exp Gerontol. 2006 doi: 10.1016/j.exger.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers SK, Ji LL, Leeuwenburgh C. Exercise training-induced alterations in skeletal muscle antioxidant capacity: a brief review. Med Sci Sports Exerc. 1999;31:987–997. doi: 10.1097/00005768-199907000-00011. [DOI] [PubMed] [Google Scholar]
- Enayati AA, Ranson H, Hemingway J. Insect glutathione transferases and insecticide resistance. Insect Molecular Biology. 2005;14:3–8. doi: 10.1111/j.1365-2583.2004.00529.x. [DOI] [PubMed] [Google Scholar]
- Hazelton GA, Lang CA. Glutathione S-Transferase Activities in the Yellow-Fever Mosquito [Aedes-Aegypti (Louisville)] during Growth and Aging. Biochem J. 1983;210:281–287. doi: 10.1042/bj2100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotze AC, Rose HA. Glutathione-S-Transferase in the Australian Sheep Blowfly, Lucilia-Cuprina (Wiedemann) Pesticide Biochemistry and Physiology. 1987;29:77–86. [Google Scholar]
- Kostaropoulos I, Mantzari AE, Papadopoulos AI. Alterations of some glutathione S-transferase characteristics during the development of Tenebrio molitor (Insecta:Coleoptera) Insect Biochem Molec. 1996;26:963–969. [Google Scholar]
- Smirle MJ. The influence of colony population and brood rearing intensity on the activity of detoxifying enzymes in worker honey bees. Physiological Entomology. 1993;18:420–424. [Google Scholar]
- Papadopoulos AI, Polemitou I, Laifi P, Yiangou A, Tananaki C. Glutathione S-transferase in the insect Apis mellifera macedonica kinetic characteristics and effect of stress on the expression of GST isoenzymes in the adult worker bee. Comp Biochem Physiol C Toxicol Pharmacol. 2004;139:93–97. doi: 10.1016/j.cca.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Smirle MJ, Winston ML. Detoxifying Enzyme-Activity in Worker Honey Bees - an Adaptation for Foraging in Contaminated Ecosystems. Can J Zool. 1988;66:1938–1942. [Google Scholar]
- Nystrom T. Role of oxidative carbonylation in protein quality control and senescence. Embo J. 2005;24:1311–1317. doi: 10.1038/sj.emboj.7600599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Amdam GV, Seehuus SC. Order, disorder, death: Lessons from a superorganism. Adv Cancer Res. 2006;95:31–60. doi: 10.1016/S0065-230X(06)95002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam GV, Page RE. Intergenerational transfers may have decoupled physiological and chronological age in a eusocial insect. Aging Res Rev. 2005;4:398–408. doi: 10.1016/j.arr.2005.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omholt SW, Amdam GV. Epigenic regulation of aging in honeybee workers. Sci Aging Knowl Environ. 2004;26:pe28. doi: 10.1126/sageke.2004.26.pe28. [DOI] [PubMed] [Google Scholar]
- Huang ZY, Robinson GE. Regulation of honey bee division of labor by colony age demography. Behav Ecol Sociobiol. 1996;39:147–158. [Google Scholar]
- Amdam GV, Aase ALTO, Seehuus SC, Norberg K, Hartfelder K, Fondrk MK. Social reversal of immunosenescence in honey bee workers. Exp Gerontol. 2005;40:939–947. doi: 10.1016/j.exger.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolschin F, Weckwerth W. Combining metal oxide affinity chromatography (MOAC) and selective mass spectrometry for robust identification of in vivo protein phosphorylation sites. Plant Methods. 2005;1:9. doi: 10.1186/1746-4811-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteins quantified by spectrum count. Table displaying the results from a non-parametric Mann-Whitney-U-test (rank sums and p-level) for proteins quantified by spectrum count (threshold level for significant differences p < 0.05).
Peptides identified by LC-MS/MS. Table displaying all peptides identified by OMSSA with an e-value ≤ 1.0E-01. Lower case letter m: oxidized methionine. For details on OMSSA see [80].