Abstract
The GTPase RhoA is essential for the development of pre-T cells in the thymus. To investigate the mechanisms used by RhoA to control thymocyte development we have used Affymetrix gene profiling to identify RhoA regulated genes in T cell progenitors. The data show that RhoA plays a specific and essential role in pre-T cells because it is required for the expression of transcription factors of the Egr-1 and AP-1 families that have critical functions in thymocyte development. Loss of RhoA function in T cell progenitors causes a developmental block that pheno-copies the consequence of losing pre-TCR expression in Recombinase gene 2 (Rag2) null mice. Transcriptional profiling reveals both common and unique gene targets for RhoA and the pre-TCR indicating that RhoA participates in the pre-TCR induced transcriptional program but also mediates pre-TCR independent gene transcription.
Keywords: Pre-TCR, RhoA, AP-1, Egr, Thymus
1. Introduction
The T cell antigen receptor (TCR) complex comprises variable α/β subunits that recognise peptide/major histocompatibility (MHC) complexes and invariant signal transduction subunits of the CD3 antigen. A key stage in T cell development in the thymus is the selection of cells that have successfully rearranged their TCR-β locus. This occurs in T cell precursors, which do not express of the MHC receptors, CD4 and CD8 (double negative (DN) thymocytes) [1]. The DN stages of intrathymic differentiation can be followed by the sequential pattern of expression of CD44 and CD25. T cell progenitors enter the thymus as CD44+CD25− cells (termed DN1); these then express CD25 (DN2) and begin to rearrange T-cell receptor β loci. Cells then lose CD44 expression and continue β chain rearrangements to completion (DN3). Cells that successfully rearrange their TCR-β locus will express a functional receptor complex known as the pre-TCR comprising a TCR-β chain, the p-Tα subunit and the signalling subunits of the CD3 antigen [2–4]. When the pre-TCR is expressed at the cell membrane it promotes cell survival and entry into the cell cycle [5]; cells downregulate CD25 and transit to the DN4 pre-T cell subset. DN4 T cells undergo proliferative expansion and differentiate to CD4+CD8+ double positive (DP) cells. These cells then undergo TCR α-chain gene rearrangements and upon expression of a functional α/β TCR complex are subjected to positive and negative selection to generate CD4+ or CD8+ single positive (SP) thymocytes [6–8].
Normal pre-T cell development requires the coordination of a complex program of gene transcription by signal transduction networks mediated by the pre-TCR complex and cytokine and stromal signals [9,5]. Mice deficient for the recombinase activating (RAG) genes which are unable to rearrange TCR-β subunits and express a pre-TCR are unable to progress beyond the DN3 stage of thymocyte development [10]. Similarly, the absence of pre-Tα or CD3 subunits blocks thymocyte development at the DN3 stage [11,3]. The transition of thymocytes beyond the pre-T cell stage of thymocyte differentiation is also dependent on signal transduction networks mediated by tyrosine and serine kinases [3,12,13]. In this context, crucial responses are mediated by Rho family guanine nucleotide binding proteins such as RhoA, Rac-1 and CDC42 [14–17].
The importance of RhoA for thymocyte development has been demonstrated by studies of transgenic mice that express Clostridium botulinum C3-transferase under the control of T cell specific promoters such as the p56lck and CD2 promoters [17,15,18]. This toxin selectively ADP-ribosylates RhoA within its effector-binding domain and abolishes its biological function. Transgenic mice that express C3-transferase under the control of the p56Lck promoter have a small thymus and severely reduced numbers of peripheral T cells [15,17]. This phenotype is caused by survival defects in DN2 and DN3 thymocytes that lack Rho function [15]. During embryogenesis no cells progress beyond the DN1 stage but in adult mice a few T cell progenitors survive and develop to DN4s and beyond [15]. This ‘leakiness’ either reflects selection of cells that compensate for loss of Rho function in DN2/3 thymocytes or reflects heterogeneous and asynchronous expression of the lck promoter in adult DN thymocytes. The few DP thymocytes and mature T cells found in adult Lck-C3 transgenic mice have numerous defects including reduced survival, proliferation, integrin mediated cell adhesion and defective cell motility [15,19]. A complementary strategy to probe Rho function in the thymus used the CD2 locus control region (LCR) to target C3 transferase to T cell progenitors (CD2–C3 mice) [18]. The inhibition of Rho function at the DN2/3 stage in CD2–C3 mice is not leaky and causes T cells to become blocked in differentiation at the DN3 stage of thymus development. CD2–C3 mice thus have a thymic phenotype indistinguishable from the phenotype of Recombinase gene null mice or mice lacking key structural or signaling components of preTCR complex [10,18,20,21]: thymocyte development is blocked at the pre-T cell/DN3 stage [18].
The basis for the failure of pre-T cell differentiation in CD2–C3 transferase mice is not known. One way to address this issue is to use microarray gene expression profiling to determine the impact of losing Rho function on transcriptional responses in pre-T cells. Previous studies in transformed cell lines have identified a role for RhoA in regulating activating protein-1 (AP1) family of transcription factors [22–24] but transcriptional targets for RhoA signal transduction in primary non-transformed cells have not been explored. The present data show that RhoA regulates expression of genes encoding members of the Fos/Jun and early growth response (Egr) family of transcription factors but also has an impact on expression of genes regulating diverse biological functions including serine/threonine kinases, protein phosphatases, enzymes that regulate protein biosynthesis and proteins that regulate nuclear structure and function.
2. Materials and methods
2.1. Mice
Mice were bred and maintained under specific pathogen-free conditions in the transgenic animal unit. RAG2−/− and C3 transgenic mice, which selectively express the bacterial toxin C3-transferase under the control of CD2 promoter and locus control region in the thymus, have been described in detail elsewhere [10,18].
2.2. Flow cytometric analysis
Fluorescein isothiocyanate (FITC), phycoerythrin (PE), allophycocyanin (APC), and biotin-conjugated antibodies were obtained from Pharmingen (San Diego, CA). Tricolour (PE/Cy5 and APC/Alexa750) conjugated antibodies and fluorophore-streptavidin conjugates were from Caltag (Burlingame, CA). Thymocytes were stained for cell surface markers and analysed on a FACS Calibur (Becton Dickinson Immunocytometry Systems Franklin Lakes CA). Data were analysed using CellQuest BDIS or FlowJo (Treestar Inc, Ashland OR) software. CD4−CD8− double negative thymocyte subsets were analysed for CD44 and CD25 expression following lineage exclusion of mature DP and SP cells as well as non-T cell lineage cells using a cocktail of biotinylated antibodies (CD4, CD8, CD3 B220, Mac-1, NK, Gr-1, and γδ) revealed with streptavidin-tricolour and costained with CD25-FITC, CD44-PE and Thy1.2-APC. Intracellular phospho-S6 and TCR-β intracellular staining was carried out on thymocytes pre stained to identify DN3 and DN4 subpopulations. Cells were subsequently fixed in 1% paraformaldehyde for 10 min at room temperature, washed in PBS and permeabilized with saponin buffer (0.5% saponin, 5% FBS, 10 mM HEPES [pH 7.4] in PBS) for 10 min at room temperature. Permeabilized cells were incubated with PE-conjugated TCRβ antibody and phospho S6 antibody for 45 min at room temperature, washed in saponin buffer and subsequently stained with FITC conjugated donkey anti rabbit IgG (Jackson Immunoresearch, West Grove PA). Cells were analysed on an LSR Flow Cytometer (BDIS).
2.3. RNA extraction
Thymocytes were isolated from 4- to 6-week-old RAG2−/−, CD2–C3 and C57BL/6 control mice. DN3 pre-T cells were purified firstly by removing CD4, CD8 DP cells using MACS CD4+ and -MACS CD8+ T cell isolation kits (Miltenyi Biotec, Ltd.) followed by magnetic autoMACS separation. CD4/CD8 depleted thymocytes were then labelled with Thy1.2-APC, PE conjugated CD4, CD8, CD44 and TCRγδ antibodies, FITC conjugated CD25 antibodies and the DN3 subpopulation (Thy1+CD4−/CD8−/CD44−/CD25+) was purified by cell sorting on a FACS Vantage (BDIS) using Cell Quest software (BDIS). All sorted cells were analysed by flow cytometry and only used for experiments if the purity was 95–98%. Total RNA was extracted from sorted cells using the Qiagen RNeasy kit according to manufacturers instructions. Total RNA was quantified using the RNA 6000 Nano LabChip® kit and analyzed on an Agilent 2100 Bioanalyzer (Agilent Technologies). Total RNA from up to 10 thymi were pooled for microarray analysis and quantitative PCR.
2.4. Microarray
Microarray analysis was carried out by the Finnish DNA Microarray Centre at the Centre for Biotechnology, Turku, Finland. For Affymetrix sample preparation, 100–1000 ng of total RNA was used as the starting material to synthesise target cRNA using the GeneChip® Eukaryotic Small Sample Target Labeling Assay Version II according to manufacturers’ instructions (Affymetrix, Santa Clara, CA). The cRNA target sample was hybridised to the Affymetrix Mouse Genome 430 2.0 Array (45,101 probes sets and expressed sequence tag (EST) clusters). Data was analysed with Affymetrix Gene Chip Operating Software (GCOS) version 1.1 and filtered according to recommendations of the manufacturer. Comparison analysis was used to compare the expression profiles from two arrays – results were obtained by analyzing the experimental array in comparison to the baseline array. The global method of scaling was used for normalizing hybridization intensity between arrays. The statistical algorithm used by GCOS defined the expression status of each gene-specific probe set as not changed, increased, marginally increased, decreased or marginally decreased. Genes that were absent or unchanged between comparisons were excluded from the results. Comparison analysis defines a gene as up-regulated if the signal log ratio between the baseline sample and the experimental sample is larger than 1 (2-fold) and the experimental sample is present. Similarly changed genes were further analyzed/classified using the gene ontology tool (www.Affymetrix.com) and represented by biological function. Gene lists were complied and sorted by genes that were either commonly or uniquely regulated in CD2-C3 and RAG2−/− thymocytes.
2.5. Real-time RT-PCR analysis
Purified total RNA (200 ng) was reverse transcribed using the iScript™ cDNA synthesis kit (BioRad). Real time RT-PCR was performed in a 96-well plate using iQ™ SYBR Green based detection (BioRad) on a BioRad iCycler in 20 μl reaction volume containing 1 μl cDNA (20 ng), 0.8 μl 10μM sense and antisense primers, 10 μl iQ™ SYBR Green supermix, and 4 μl nuclease free water. Each reaction was performed in duplicate and each experiment repeated in triplicate. 18S rRNA levels were used to normalise RNA concentrations between samples and the relative mRNA levels were calculated using the equation:
where E is the efficiency of PCR, ct is the threshold cycle, u is the mRNA of interest, r is the reference gene (18S rRNA), s is the sample and c is the control sample. Primers used for RT-PCR were designed using Beacon Designer 2 software. Primer sequences are: Egr1 forward 5′-ACAGAAGGACAAGAAAGCAGAC-3, reverse 5-CCAGGAGAGGAGTAGGAAGTG-3′, Fos forward 5′-CTACTGTGTTCCTGGCAATAGC-3′, reverse 5′-AACATTGACGCTGAAGGACTAC-3′, Egr3 forward 5′-TGACCAACGAGAAGCCCAATC-3′, reverse 5′-GCTAATGATGTTGTCCTGGCAC-3′, Jun forward 5′-CGCCTCGTTCCTCCAGTC-3′ reverse 5′-ACGTGAGAAGGTCCGAGTTC-3′, JunB forward 5′-CTTCTACGACGATGCCCTCAAC-3′, reverse 5′-GTTCAAGGTCATGCTCTGTTTTAGG-3′, Nur77 forward 5′-CCTGTTGCTAGAGTCTGCCTTC-3′, reverse 5′-CAATCCAACACCAAAGCCACG-3′, 18S RNA forward 5′-GTAACCCGTTGAACCCCATT-3′, reverse 5′-CCATCCAATCGTAGTA-3′.
3. Results and discussion
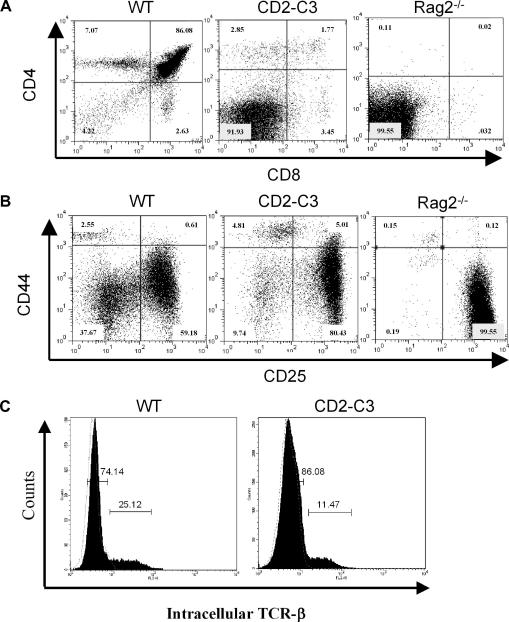
Mice expressing the RhoA inhibitor Clostridium Botulinum C3 transferase under the control of the CD2 locus control region (LCR) have been described previously [18]. Loss of RhoA function in CD2-C3 mice results in loss of CD4/CD8 double positive thymocytes (Fig. 1A) with a block in thymocyte development at the DN3 stage (Fig. 1B). Rag2−/− mice also block thymus development at the DN3 stage (Figs. 1A and B) due to failed expression of the pre-TCR complex. Despite this superficial similarity, loss of Rho function does not prevent TCR beta locus rearrangements although there is a reduced frequency of DN3 cells expressing intracellular TCR-β subunits (Fig. 1C). Ectopic expression of transgenic TCR complexes cannot reverse the developmental block in CD2–C3 thymocytes demonstrating that failed expression of TCR complexes does not explain the developmental defects caused by loss of Rho function [18]. Rho thus appears necessary for pre-TCR function although these results do not discriminate between a direct role for RhoA in pre-TCR signalling or a role for this GTPase in the cytokine/stromal signalling pathways that synergise with pre-TCR signals to control pre-T cell differentiation [25–27].
Fig. 1.
Expression of C3 transferase in the thymus of adult mice prevents up regulation of thymocyte developmental markers. (A) Data show flow cytometry analysis of CD4 and CD8 expression on Thy1+ thymocytes from CD2–C3, RAG2−/− and C57BL/6 control mice. (B) Data show flow cytometry analysis of CD25 and CD44 expression on Thy1+ CD4−CD8− thymocytes from CD2-C3, RAG2−/− and C57BL/6 control mice. (C) Histograms show intracellular TCR-β staining in DN3 cells (CD44−CD25+ Thy1+ CD4−CD8−) from wild type or CD2–C3 transferase mice. Isotype-matched control antibody shows negative staining (dotted line). In 5 experiments, TCR-β positive cells in wild type DN3s averaged 24% (range 20–25%) whereas in CD2–C3 DN3s averaged 10% (range 7.35–11.47%).
CD2–C3 transferase mice provide a good model system to probe the immediate transcriptional consequences of losing RhoA function in pre-T cells since the CD2-LCR initiates expression of transgenes in T cell progenitors as they transit from the DN2 to DN3 stage of thymocyte development [18]. Accordingly, it is possible to isolate DN3 thymocytes ex vivo that have only just switched on expression of C3 transferase and prepare RNA for DNA microarray analysis. The Affymetrix Mouse Genome 430 2.0 array, representing 39 000 murine gene transcripts, was used to transcriptionally profile DN3 pre-T cells purified from the thymi of CD3–C3 transgenic mice. In these experiments DN3 thymocytes from wild type control mice were transcriptionally profiled and used for comparisons.
The microarray data were analyzed with the Affymetrix Gene Chip Operating Software (GCOS, version 1.1) by comparing the gene expression profiles of CD2–C3 pre-T cells to the profile of normal cells to reveal changes above or below wild type levels. DN3 pre-T cells expressed approximately 18 000 genes and there were only small changes in the transcriptional profile of CD2–C3 DN3 thymocytes compared to control wild type DN3s. We focused our analysis on ⩾2-fold changes, which were further analyzed using the Affymetrix gene ontology tool. A full list of the genes regulated 2-fold or greater by loss of RhoA function in DN3 thymocytes is shown in Supplementary Fig. 1. There were approximately 18 000 genes expressed in wild type DN3 thymocytes: loss of RhoA function caused a ⩾2-fold decrease in expression of 383 genes and a ⩾2-fold increase in expression of 190 genes. RhoA regulated genes in DN3 thymocytes encode proteins with diverse biological functions including serine/threonine kinases, protein phosphatases, enzymes that regulate protein biosynthesis.
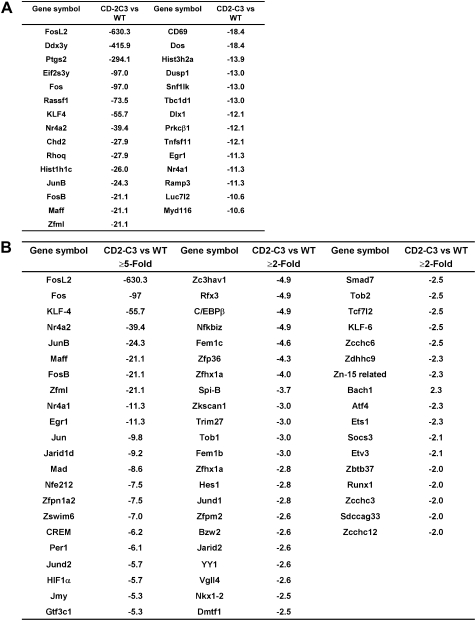
This point is illustrated in Fig. 2A, which shows the genes whose expression is regulated more than 10-fold. The largest decrease was in expression of the gene encoding FosL2, a Fos family transcription factor (630-fold decrease) but other large changes included a subunit of eukaryotic translation initiation factor 2, Protein Kinase C beta and the membrane receptor CD69. There was also a loss of expression of genes encoding proteins that regulate nuclear structure and function such as genes encoding histones that are involved in nucleosome assembly. In a similar category there was reduced expression of mRNA encoding special AT-rich sequence binding protein 1 (SATB1) in CD2-C3 DN3s (Supplementary Fig. 1). SATB1 acts as a scaffold for chromatin modifiers and is known to regulate higher order chromatin structure and to regulate gene expression by acting as a “docking site” for several chromatin remodeling enzymes [28–30].
Fig. 2.
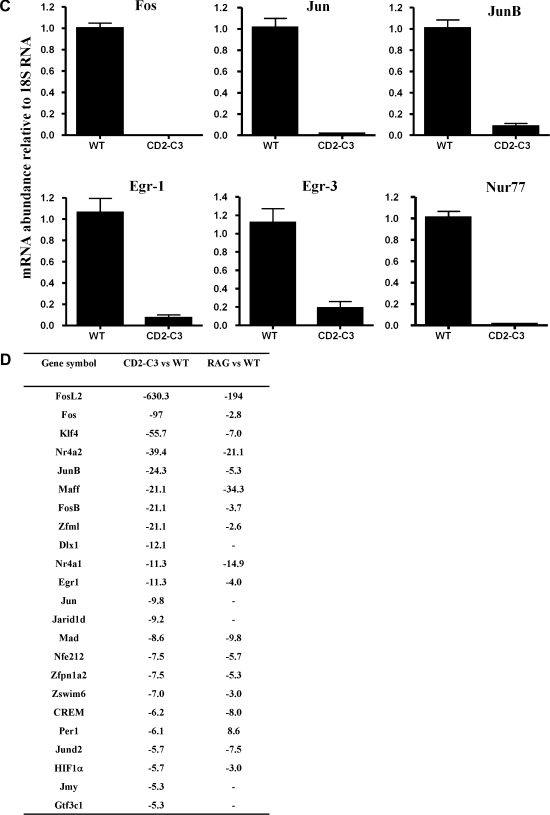
Transcriptional changes caused by loss of endogenous RhoA function in DN3 pre-T cells. (A) Table showing the diverse number of genes that display a 10-fold or greater reduction in expression in CD2–C3 DN3 cells compared to wild type DN3 cells. The symbol (–) preceding the numerical value indicates a fold decrease. (B) The table displays total number of transcription factors that display a 2-fold or greater reduction in expression in CD2–C3 DN3 thymocytes compared to wild type DN3 cells. (C) Real time PCR analysis of several transcription factor genes downregulated in CD2–C3 DN3 cells compared to wild type DN3 cells. Error bars indicate the standard deviation obtained from three independent experiments. (D) Table showing comparison of transcription factors downregulated in RAG2−/− DN3 cells with CD2–C3 DN3 cells.
Of 55 genes whose expression was decreased ⩾5-fold, 23 encoded transcription factors; of 384 genes decreased ⩾2-fold, 63 were transcription factors (Fig. 2B). Transcription factors downregulated in CD2-C3 DN3 thymocytes included Kruppel family transcription factors KLF4, KLF6 and KLF9. There was also loss of expression of members of the immediate early gene family Egr1 and Egr3, AP-1 family members Fos, FosL2, FosB, Jun, JunB and downregulated expression of genes encoding Ets1 and Nurr77. To validate the microarray analysis, quantitative real-time PCR was used to compare expression of a selection of these different transcription factors in wild type versus CD2-C3 thymocytes (Fig. 2C). The experiments confirmed the reduction in Fos, Jun, JunB, Egr1 and Egr3 and NURR77 in CD2–C3 DN3s compared to wild type controls.
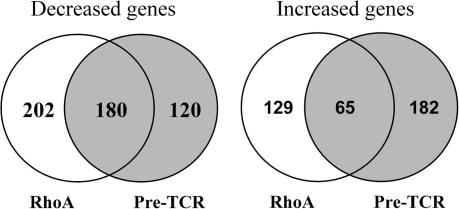
A number of gene targets for Egr transcription factors have been described [31] and in this context, RhoA inactivation caused loss of expression of multiple members of the MAP kinase phosphatase family DUSP 1, 2, 4, 6, 16. The latter are induced in negative feedback mechanisms that modulate MAP kinase activity and have been described as targets for Egr family transcription factors [31–33]. The loss of DUSP expression is thus almost certainly a secondary consequence of failed expression of Egr molecules. Egr1 and Egr3 are well characterised pre-TCR induced genes [34–38]. To verify this we compared the transcriptional profile of CD2-C3 DN3s with Recombinase gene 2−/− (RAG2−/−) DN3s which lack expression of the pre-TCR complex because they fail to rearrange their TCR beta locus. Of the 23 transcription factors downregulated ⩾5-fold in CD2–C3 thymocytes 18 were also downregulated in Rag2−/− DN3s (Fig. 2D) and these included Egr1, Egr3, Fos, Fosl2, FosB, Ets1, Nurr77. These microarray analyses were confirmed by quantitative real-time PCR analysis of wild type versus Rag2−/− thymocytes (data not shown). The common loss of expression of members of the Egr family in CD2–C3 and Rag2−/− pre-T cells is thus consistent with a RhoA requirement for the pre-TCR induced gene transcription. However, comparisons of the total number of genes regulated uniquely in CD2–C3 and Rag2−/− DN3s (Fig. 3) (VENN diagrams) indicate that there are a number of gene changes in CD2–C3 cells that are not seen in Rag2−/− DN3s. The full list of genes that were commonly and uniquely regulated 2-fold and greater in CD2 C3 versus Rag2−/− pre-T cells is shown in Supplementary Fig. 2. In terms of ⩾ 2-fold, gene repression, CD2-C3 and Rag2−/− DN3s had 180 gene targets in common with 202 genes being downregulated uniquely in CD2–C3 pre-T cells and 120 genes downregulated uniquely in Rag2−/− cells. In terms of ⩾2-fold gene increases, CD2–C3 and Rag2−/− DN3s had 65 gene targets in common, 129 genes that were increased uniquely in CD2-C3 DN3s and 182 increased uniquely in Rag2−/− cells (Fig. 3).
Fig. 3.
Differential gene comparison in DN3 pre-T cells lack either endogenous RhoA function or TCR-β signalling. Venn diagrams showing the number of genes that are uniquely and commonly regulated in CD2–C3 DN3s (lacking RhoA function) versus RAG2−/− DN3s (lacking TCR-β signalling). The region of overlap regulates genes that are commonly regulated. Gene changes 2-fold and above are shown.
The common transcriptional changes caused by loss of the pre-TCR in Rag2−/− DN3s or loss of Rho function in CD2–C3 mice indicates that Rho function is necessary for some pre-TCR induced transcriptional responses. However, there are genes downregulated in DN3s lacking RhoA function that are unchanged in Rag2−/− DN3s (Supplementary Fig. 3) indicating that RhoA is not just required to mediate the preTCR induced gene program but may also participate in cytokine/stromal cell-initiated signalling pathways that control pre-T cell differentiation. The genes uniquely lost in CD3–C3 DN3 thymocytes include serine kinases (Protein Kinase C beta and Map3k14); transcription factors (SpiB and Dlx1) and regulators of protein biosynthesis (Eif2s3y) (Supplementary Fig. 3). One gene of interest lost in CD2–C3 DN3s but not Rag2 null DN3s was Hes-1 which is induced by Notch receptor-ligand interactions. Notch signalling is required throughout DN to DP stage of thymocyte development to support T cell metabolism and survival [39–42]. A RhoA requirement for Notch signalling has not been described but it is known that RhoA is necessary for integrin mediated cell adhesion and thymocyte cell migration [19]. Accordingly, decreased Hes-1 expression could reflect that pre-T cells lacking Rho function cannot make normal contacts with stromal cells that express the Notch ligands [43,42,44,45].
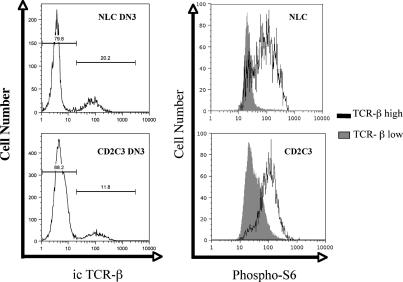
There were also unique transcriptional changes seen in Rag2−/− cells that were not found in the CD2–C3 DN3s (Supplementary Fig. 4) which indicates that the pre-TCR can regulate a program of gene transcription via RhoA independent pathways The unique changes in Rag2−/− cells not surprisingly included loss of expression of rearranged TCR-β subunits. One other gene downregulated in Rag2−/− DN3s but not CD2–C3 DN3s was RPS6, which encodes the ribosomal S6 subunit. In this respect, we recently described high basal levels of S6 phosphorylation in ex vivo β selected DN3s and no detectable S6 phosphorylation in ex vivo Rag2−/− DN3s [13]. The data in Fig. 4 quantify S6 phosphorylation in CD2–C3 DN3s compared to wild type cells. Normal DN3s are heterogeneous for phosphoS6 with the majority of cells being phosphoS6low but a significant percentage of cells are phosphoS6high. The DN3 thymocyte subpopulation can be subdivided into cells that have not yet completed TCR-β locus rearrangements and those that express a functional TCR-β subunit that allows surface expression and signalling of the pre-TCR complex. Analysis of TCRβ expression by intracellular staining revealed that DN3s that express icTCR-β chains are generally phosphoS6high whereas DN3s that are icTCR-β null are uniformly phosphoS6low. In CD2–C3 DN3s there were also high levels of phosphoS6 in TCR-β high cells. The presence of normal S6 phosphorylation in β selected DN3s lacking Rho function is a further indication that Rho is necessary for a subset of responses in DN3 thymocytes but does not globally block signalling.
Fig. 4.
Endogenous RhoA function is not required for pre-TCR induced phosphorylation of S6. FACS histograms shows intracellular TCR-β staining profiles (left panel) for wild type and CD2–C3 DN3 cells and subsequent levels of phospho-S6 (right panel) in TCR-β low or high DN3 cells.
One important question is whether the transcriptional changes in CD2–C3 DN3s explain why loss of RhoA function results in failed thymocyte differentiation? In this respect, microarray analysis of Rag2−/− pre-T cells revealed decreased expression of TCR-β subunits as a major defect (Supplementary Fig. 4) and failed expression of the TCR-β subunit is indeed responsible for the DN3 developmental block in Rag2−/− mice. In CD2–C3 DN3s there were several gene defects that would explain why RhoA function is essential for pre-T cell development. For example, RhoA is required for expression of Egr3 and Ets1 and loss of either of these transcription factors inhibits pre-T cell proliferation and is suboptimal for transition through early pre-TCR-dependent stages of thymocyte development [46,36]. Loss of individual Egr family or AP-1 transcription factors does not completely abrogate thymocyte differentiation but this reflects that there is considerable redundancy in AP-1 complexes due to the ability of different family members to pair as dimers [47–49]. The simultaneous elimination of Egr1, Egr3, Ets1 and multiple Fos/Jun family members in CD2–C3 DN3s would circumvent the possibility of redundancy [50,51]. This comprehensive loss of AP-1 activity in pre-T cell lacking RhoA function plus the decreased expression of other transcription factors would rapidly cause global changes in the transcriptional program of a cell. Moreover, the transcription factor defects would be exacerbated by the decreased expression of chromatin modifying enzymes such as SATB1.
In summary, loss of Rho function or failed expression of the pre-TCR complex in Rag2−/− null mice block T cell development at a common stage. The present data show that the genetic consequences of loss of Rho function versus loss of pre-TCR expression for T cell progenitors are not identical. The present results also show that loss of Rho function in pre-T cells results in downregulation of genes encoding members of the Fos/Jun and Early growth response (Egr) family of transcription factors. The collective loss of these transcription factors and the resultant secondary genetic changes explains why loss of RhoA function prevents pre-T cell development.
Acknowledgements
We thank the transgenic animal facility for animal maintenance, Dr Simon Arthur for supplying Real time PCR primers and use of the BioRad iCycler. Members of the Cantrell lab for helpful discussions. This work was supported by a Wellcome Trust Principle Research Fellowship Grant. GR065975. The Academy of Finland, National Technology Agency of Finland and the Turku University Research Fund (R. Lahesmaa).
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.febslet.2007.07.077.
Appendix A. Supplementary data
List of total genes downregulated or upregulated in CD2–C3 DN 3 thymocytes.
List of genes that are commonly downregulated or upregulated in CD2–C3 and RAG2−/− DN3 thymocytes.
List of genes that are uniquely downregulated in CD2–C3 DN3 thymocytes.
List of genes that are uniquely downregulated or upregulated in RAG2−/−DN3 thymocytes.
References
- 1.von Boehmer H., Aifantis I., Feinberg J., Lechner O., Saint-Ruf C., Walter U., Buer J., Azogui O. Pleiotropic changes controlled by the pre-T-cell receptor. Curr. Opin. Immunol. 1999;11:135–142. doi: 10.1016/s0952-7915(99)80024-7. [DOI] [PubMed] [Google Scholar]
- 2.von Boehmer H., Aifantis I., Azogui O., Saint-Ruf C., Grassi F. The impact of pre-T-cell receptor signals on gene expression in developing T cells. Cold Spring Harb. Symp. Quant. Biol. 1999;64:283–289. doi: 10.1101/sqb.1999.64.283. [DOI] [PubMed] [Google Scholar]
- 3.Michie A.M., Zuniga-Pflucker J.C. Regulation of thymocyte differentiation: pre-TCR signals and beta-selection. Semin. Immunol. 2002;14:311–323. doi: 10.1016/s1044-5323(02)00064-7. [DOI] [PubMed] [Google Scholar]
- 4.von Boehmer H. Unique features of the pre-T-cell receptor alpha-chain: not just a surrogate. Nat. Rev. Immunol. 2005;5:571–577. doi: 10.1038/nri1636. [DOI] [PubMed] [Google Scholar]
- 5.Aifantis I., Mandal M., Sawai K., Ferrando A., Vilimas T. Regulation of T-cell progenitor survival and cell-cycle entry by the pre-T-cell receptor. Immunol. Rev. 2006;209:159–169. doi: 10.1111/j.0105-2896.2006.00343.x. [DOI] [PubMed] [Google Scholar]
- 6.Mariathasan S., Jones R.G., Ohashi P.S. Signals involved in thymocyte positive and negative selection. Semin. Immunol. 1999;11:263–272. doi: 10.1006/smim.1999.0182. [DOI] [PubMed] [Google Scholar]
- 7.Germain R.N. T-cell development and the CD4–CD8 lineage decision. Nat. Rev. Immunol. 2002;2:309–322. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 8.Singer A., Bosselut R. CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: analysis of the CD4/CD8 lineage decision. Adv. Immunol. 2004;83:91–131. doi: 10.1016/S0065-2776(04)83003-7. [DOI] [PubMed] [Google Scholar]
- 9.Ciofani M., Zuniga-Pflucker J.C. A survival guide to early T cell development. Immunol. Res. 2006;34:117–132. doi: 10.1385/IR:34:2:117. [DOI] [PubMed] [Google Scholar]
- 10.Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka Y., Ardouin L., Gillet A., Lin S.Y., Magnan A., Malissen B., Malissen M. Early T-cell development in CD3-deficient mice. Immunol. Rev. 1995;148:171–199. doi: 10.1111/j.1600-065x.1995.tb00098.x. [DOI] [PubMed] [Google Scholar]
- 12.Matthews S.A., Cantrell D.A. The role of serine/threonine kinases in T-cell activation. Curr. Opin. Immunol. 2006;18:314–320. doi: 10.1016/j.coi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Hinton H.J., Clarke R.G., Cantrell D.A. Antigen receptor regulation of phosphoinositide-dependent kinase 1 pathways during thymocyte development. FEBS Lett. 2006;580:5845–5850. doi: 10.1016/j.febslet.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 14.Na S., Li B., Grewal I.S., Enslen H., Davis R.J., Hanke J.H., Flavell R.A. Expression of activated CDC42 induces T cell apoptosis in thymus and peripheral lymph organs via different pathways. Oncogene. 1999;18:7966–7974. doi: 10.1038/sj.onc.1203122. [DOI] [PubMed] [Google Scholar]
- 15.Galandrini R., Henning S.W., Cantrell D.A. Different functions of the GTPase Rho in prothymocytes and late pre-T cells. Immunity. 1997;7:163–174. doi: 10.1016/s1074-7613(00)80519-1. [DOI] [PubMed] [Google Scholar]
- 16.Gomez M., Kioussis D., Cantrell D.A. The GTPase Rac-1 controls cell fate in the thymus by diverting thymocytes from positive to negative selection. Immunity. 2001;15:703–713. doi: 10.1016/s1074-7613(01)00235-7. [DOI] [PubMed] [Google Scholar]
- 17.Henning S.W., Galandrini R., Hall A., Cantrell D.A. The GTPase Rho has a critical regulatory role in thymus development. Embo. J. 1997;16:2397–2407. doi: 10.1093/emboj/16.9.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cleverley S., Henning S., Cantrell D. Inhibition of Rho at different stages of thymocyte development gives different perspectives on Rho function. Curr. Biol. 1999;9:657–660. doi: 10.1016/s0960-9822(99)80289-9. [DOI] [PubMed] [Google Scholar]
- 19.Vielkind S., Gallagher-Gambarelli M., Gomez M., Hinton H.J., Cantrell D.A. Integrin regulation by RhoA in thymocytes. J. Immunol. 2005;175:350–357. doi: 10.4049/jimmunol.175.1.350. [DOI] [PubMed] [Google Scholar]
- 20.Tybulewicz V.L. Vav-family proteins in T-cell signalling. Curr. Opin. Immunol. 2005;17:267–274. doi: 10.1016/j.coi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds L.F., de Bettignies C., Norton T., Beeser A., Chernoff J., Tybulewicz V.L. Vav1 transduces T cell receptor signals to the activation of the Ras/ERK pathway via LAT, Sos, and RasGRP1. J. Biol. Chem. 2004;279:18239–18246. doi: 10.1074/jbc.M400257200. [DOI] [PubMed] [Google Scholar]
- 22.Teramoto H., Malek R.L., Behbahani B., Castellone M.D., Lee N.H., Gutkind J.S. Identification of H-Ras, RhoA, Rac1 and Cdc42 responsive genes. Oncogene. 2003;22:2689–2697. doi: 10.1038/sj.onc.1206364. [DOI] [PubMed] [Google Scholar]
- 23.Marinissen M.J., Chiariello M., Tanos T., Bernard O., Narumiya S., Gutkind J.S. The small GTP-binding protein RhoA regulates c-jun by a ROCK-JNK signaling axis. Mol. Cell. 2004;14:29–41. doi: 10.1016/s1097-2765(04)00153-4. [DOI] [PubMed] [Google Scholar]
- 24.Berenjeno I.M., Nunez F., Bustelo X.R. Transcriptomal profiling of the cellular transformation induced by Rho subfamily GTPases. Oncogene. 2007 doi: 10.1038/sj.onc.1210194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haks M.C., Oosterwegel M.A., Blom B., Spits H.M., Kruisbeek A.M. Cell-fate decisions in early T cell development: regulation by cytokine receptors and the pre-TCR. Semin. Immunol. 1999;11:23–37. doi: 10.1006/smim.1998.0153. [DOI] [PubMed] [Google Scholar]
- 26.Zuniga-Pflucker J.C. T-cell development made simple. Nat. Rev. Immunol. 2004;4:67–72. doi: 10.1038/nri1257. [DOI] [PubMed] [Google Scholar]
- 27.Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat. Rev. Immunol. 2006;6:127–135. doi: 10.1038/nri1781. [DOI] [PubMed] [Google Scholar]
- 28.Kohwi-Shigematsu T., Maass K., Bode J. A thymocyte factor SATB1 suppresses transcription of stably integrated matrix-attachment region-linked reporter genes. Biochemistry. 1997;36:12005–12010. doi: 10.1021/bi971444j. [DOI] [PubMed] [Google Scholar]
- 29.Nakayama Y., Mian I.S., Kohwi-Shigematsu T., Ogawa T. A nuclear targeting determinant for SATB1, a genome organizer in the T cell lineage. Cell Cycle. 2005;4:1099–1106. [PubMed] [Google Scholar]
- 30.Pavan Kumar P., Purbey P.K., Sinha C.K., Notani D., Limaye A., Jayani R.S., Galande S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol. Cell. 2006;22:231–243. doi: 10.1016/j.molcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 31.Zhang T., Wolfe M.W., Roberson M.S. An early growth response protein (Egr) 1 cis-element is required for gonadotropin-releasing hormone-induced mitogen-activated protein kinase phosphatase 2 gene expression. J. Biol. Chem. 2001;276:45604–45613. doi: 10.1074/jbc.M107075200. [DOI] [PubMed] [Google Scholar]
- 32.Tanzola M.B., Kersh G.J. The dual specificity phosphatase transcriptome of the murine thymus. Mol. Immunol. 2006;43:754–762. doi: 10.1016/j.molimm.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Li C., Scott D.A., Hatch E., Tian X., Mansour S.L. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development. 2007;134:167–176. doi: 10.1242/dev.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bettini M., Xi H., Milbrandt J., Kersh G.J. Thymocyte development in early growth response gene 1-deficient mice. J. Immunol. 2002;169:1713–1720. doi: 10.4049/jimmunol.169.4.1713. [DOI] [PubMed] [Google Scholar]
- 35.Carleton M., Haks M.C., Smeele S.A., Jones A., Belkowski S.M., Berger M.A., Linsley P., Kruisbeek A.M., Wiest D.L. Early growth response transcription factors are required for development of CD4(−)CD8(−) thymocytes to the CD4(+)CD8(+) stage. J. Immunol. 2002;168:1649–1658. doi: 10.4049/jimmunol.168.4.1649. [DOI] [PubMed] [Google Scholar]
- 36.Xi H., Kersh G.J. Early growth response gene 03 regulates thymocyte proliferation during the transition from CD4-CD8- to CD4+CD8+ J. Immunol. 2004;172:964–971. doi: 10.4049/jimmunol.172.2.964. [DOI] [PubMed] [Google Scholar]
- 37.Safford M., Collins S., Lutz M.A., Allen A., Huang C.T., Kowalski J., Blackford A., Horton M.R., Drake C., Schwartz R.H., Powell J.D. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat. Immunol. 2005;6:472–480. doi: 10.1038/ni1193. [DOI] [PubMed] [Google Scholar]
- 38.Xi H., Schwartz R., Engel I., Murre C., Kersh G.J. Interplay between RORgammat, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006;24:813–826. doi: 10.1016/j.immuni.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 39.Tomita K., Hattori M., Nakamura E., Nakanishi S., Minato N., Kageyama R. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 1999;13:1203–1210. doi: 10.1101/gad.13.9.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kageyama R., Ohtsuka T., Tomita K. The bHLH gene Hes1 regulates differentiation of multiple cell types. Mol. Cells. 2000;10:1–7. doi: 10.1007/s10059-000-0001-0. [DOI] [PubMed] [Google Scholar]
- 41.Ciofani M., Schmitt T.M., Ciofani A., Michie A.M., Cuburu N., Aublin A., Maryanski J.L., Zuniga-Pflucker J.C. Obligatory role for cooperative signaling by pre-TCR and notch during thymocyte differentiation. J. Immunol. 2004;172:5230–5239. doi: 10.4049/jimmunol.172.9.5230. [DOI] [PubMed] [Google Scholar]
- 42.Huang J., Garrett K.P., Pelayo R., Zuniga-Pflucker J.C., Petrie H.T., Kincade P.W. Propensity of adult lymphoid progenitors to progress to DN2/3 stage thymocytes with notch receptor ligation. J. Immunol. 2005;175:4858–4865. doi: 10.4049/jimmunol.175.8.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deftos M.L., Bevan M.J. Notch signaling in T cell development. Curr. Opin. Immunol. 2000;12:166–172. doi: 10.1016/s0952-7915(99)00067-9. [DOI] [PubMed] [Google Scholar]
- 44.Hill C.S., Wynne J., Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 45.Lehar S.M., Dooley J., Farr A.G., Bevan M.J. Notch ligands delta 1 and jagged1 transmit distinct signals to T-cell precursors. Blood. 2005;105:1440–1447. doi: 10.1182/blood-2004-08-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eyquem S., Chemin K., Fasseu M., Bories J.C. The Ets-1 transcription factor is required for complete pre-T cell receptor function and allelic exclusion at the T cell receptor beta locus. Proc. Natl. Acad. Sci. USA. 2004;101:15712–15717. doi: 10.1073/pnas.0405546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Dam H., Castellazzi M. Distinct roles of Jun:Fos and Jun:ATF dimers in oncogenesis. Oncogene. 2001;20:2453–2464. doi: 10.1038/sj.onc.1204239. [DOI] [PubMed] [Google Scholar]
- 48.Mechta-Grigoriou F., Gerald D., Yaniv M. The mammalian Jun proteins: redundancy and specificity. Oncogene. 2001;20:2378–2389. doi: 10.1038/sj.onc.1204381. [DOI] [PubMed] [Google Scholar]
- 49.Hess J., Angel P., Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 50.Shaulian E., Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 51.Shaulian E., Karin M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of total genes downregulated or upregulated in CD2–C3 DN 3 thymocytes.
List of genes that are commonly downregulated or upregulated in CD2–C3 and RAG2−/− DN3 thymocytes.
List of genes that are uniquely downregulated in CD2–C3 DN3 thymocytes.
List of genes that are uniquely downregulated or upregulated in RAG2−/−DN3 thymocytes.