Abstract
Histone acetylation levels in cells result from a dynamic equilibrium between competing histone acetylases and deacetylases. Changes in histone acetylation levels occur during both transcriptional activation and silencing. Cloning of the cDNA for a human histone deacetylase (HDAC1) has shown that it represents a human ortholog of the yeast transcriptional regulator RPD3. We have screened the expressed sequence tag database (National Center for Biotechnology Information) with the yeast RPD3 sequence and identified a human ortholog of RPD3, HDAC3. This cDNA encodes a protein of 428 amino acids with 58% sequence identity with HDAC1p. By using a specific polyclonal antiserum recognizing the C-terminal domain of HDAC3p and Western blotting, we detected a single ∼49-kDa band in several tumor cell lines. HDAC3p is expressed predominantly in the nuclear compartment. Immunoprecipitation experiments with either an antiserum against HDAC3p or an anti-FLAG antiserum and a flagged HDAC3 cDNA showed that HDAc3p exhibits deacetylase activity both on free histones and on purified nucleosomes. This deacetylase activity is inhibited by trichostatin, trapoxin, and butyrate in vitro to the same degree as the deacetylase activity associated to HDAC1p. These observations identify another member of a growing family of human HDAC genes.
Posttranslational modifications of the N-terminal domain of histones, which include acetylation, methylation, phosphorylation, and ubiquitination, are emerging as an important step in the transcriptional regulation of eukaryotic genes. In particular, acetylation of specific lysine residues in the N-terminal portion of all histones consists of the transfer of an acetyl group from acetyl CoA onto the ɛ-amino group of lysine residues. Regulation of acetylation of the core histones is a dynamic process under the control of competing enzymes: histone acetyltransferases and histone deacetylases (HDACs) (1, 2).
Although a correlation between core histone acetylation and gene activity has been known for many years, the identification of the first histone acetyltransferase (3) as a homologue of the yeast transcriptional coactivator GCN5 has brought considerable support to the paradigm that histone acetylation/deacetylation plays an active role in transcriptional regulation. More recent studies have shown that other proteins previously identified as transcriptional regulators also possess histone acetyltransferase activity. These include p300 and CBP, coactivators that interacts with DNA binding factors (4, 5); P/CAF, a human homologue of yeast GCN5 that interacts with p300/CBP (6); and TAFII250, a subunit of TFIID complex (7).
Independently, the target of action of the HDAC inhibitor trapoxin A was identified as a human ortholog of the yeast transcriptional regulator RPD3 (1). This gene, initially called HD1, is now called HDAC1, and its product exhibits HDAC activity (1). Another RPD3-related gene, initially called hRPD3 (8), now called HDAC2, encodes a protein interacting with YY1, a transcription factor that can act both as an activator or a repressor (9). HDAC1p and HDAC2p are part of two large repression complexes containing mSin3p, a mammalian homologue of the yeast corepressor SIN3 (9–11). Mammalian Sin3p was first identified as a protein interacting with Mad/Mxi proteins and required for their repressor function (12, 13). The HDAC1–Sin3 complex also plays a role of corepressor complex for N-Cor and SMRT, two silencing mediators for the unliganded retinoid and thyroid hormone receptors (14–16).
Herein, we report the identification and the characterization of a third human cDNA exhibiting homology to yeast RPD3. The protein encoded by this cDNA, HDAC3p, possesses intrinsic HDAC activity.
MATERIALS AND METHODS
Reagents and Cell Lines.
All cells were grown in Opti-MEM medium (GIBCO/BRL) supplemented with 2% fetal bovine serum (HyClone), penicillin (50 units/ml), streptomycin (50 μg/ml), and 2 mM glutamine at 37°C in a humidified 95% air/5% CO2 atmosphere. Trichostatin A (TSA) was obtained from Wako Pure Chemical Industries, and trapoxin (TPX) was obtained from M. Yoshida (University of Tokyo). Both drugs were stored in dimethyl sulfoxide at −20°C and diluted immediately before use. Sodium butyrate was purchased from Sigma.
Cloning of HDAC3 cDNA.
A clone containing a cDNA for a human RPD3 homologue from a human infant brain library was obtained from the I.M.A.G.E Consortium (Lawrence Livermore National Laboratory) (clone 32733) and fully sequenced on both strands. Because no ATG initiation codon was found in this cDNA, we performed rapid amplification of cDNA ends (CLONTECH) by using Marathon-Ready cDNA from spleen (CLONTECH) and an oligonucleotide corresponding to the 5′ region of the I.M.A.G.E cDNA clone (5′-AGAAAGCTTCTTGAACCCCGTCACCATGGTGGATGTC-3′). A 650-bp fragment was amplified by rapid amplification of cDNA ends, gel-purified, subcloned into the TA cloning vector (Invitrogen), and sequenced on both strands. This fragment contained an ATG in-frame with the rest of the ORF and at a position similar to the ATG of HDAC1. To reconstruct the full ORF of this cDNA, called HDAC3 hereafter, an EcoRI–HindIII fragment from the rapid amplification of cDNA ends was ligated to a HindIII–NotI fragment (the NotI site is derived from Lafmid BA vector) into the pcDNA3.1 (Invitrogen) vector digested with EcoRI and HindIII. The full-length cDNA for HDAC1 (clone 31480) was isolated from a human fetal lung library (17) and obtained from Y. Nakamura (Kami-Ikebukuro Toshima-ku, Tokyo). The complete cDNA was resequenced, digested with EcoRI and XhoI [these two sites are derived from the Bluescript vector (Stratagene)] and subcloned into the EcoRI/XhoI-digested pcDNA3.1 vector (Invitrogen). To generate a fusion protein between HDACp and the FLAG epitope, both cDNAs were amplified by PCR using primer oligonucleotides containing the sequence encoding the FLAG sequence and subcloned into the pcDNA3.1 vector (Invitrogen).
Generation of Specific Antisera Against HDAC1 and HDAC3.
HDAC1- and HDAC3-specific antisera were generated by immunizing rabbits with peptides corresponding to the predicted C-terminal domain of each protein coupled to keyhole limpet hemocyanin (amino acids 467–482, EEKPEAKGVKEEVKLA for HDAC1 and amino acids 413–428, FYDGDHDNDKESDVEI for HDAC3, respectively). An IgG fraction was affinity purified from serum on protein G-Sepharose 4 (Pharmacia). Specific anti-IκB-α rabbit polyclonal serum was purchased from Santa Cruz Biotechnology. The anti-FLAG M2 monoclonal antibody was purchased from Kodak.
Western Blots Analysis.
Total cellular extracts were prepared by incubating cells in RIPA buffer [1× PBS/1% Nonidet P-40/0.5% deoxycholate/0.1% SDS/0.6 mM phenylmethylsulfonyl fluoride/aprotinin (60 μg/ml)/1 mM sodium orthovanadate] for 1 h at 4°C. After centrifugation at 12,000 × g for 10 min at 4°C, protein concentration in the supernatant was quantified, and 20 μg of each extract was separated by electrophoresis on a 7.5% SDS/PAGE gel, transferred to nitrocellulose, and probed with a 1:4,000 dilution of the anti-HDAC1 IgG fraction (5.3 mg/ml) or 1:2,000 dilution of the anti-HDAC3 IgG fraction (7.8 mg/ml). Horseradish peroxidase-conjugated goat anti-mouse IgG (Amersham; 1:10,000 dilution) was used as a secondary antibody for ECL detection (Amersham).
Jurkat cells nuclear extract and cytoplasmic fraction (S-100) were prepared (18). Twenty-five micrograms of nuclear extract proteins and a proportional amount of cytoplasmic proteins (18.5 μg) were separated by electrophoresis on a 10% SDS/PAGE gel and analyzed by Western blotting as described above.
Immunoprecipitation–HDAC Assays.
Jurkat cells (5 × 107 cells) were washed twice with ice-cold PBS and resuspended in 1 ml of lysis buffer (50 mM Tris⋅HCl, pH 7.5/120 mM NaCl/5 mM EDTA/0.5% Nonidet P-40) in the presence of protease inhibitors [antipain (10 μg/ml)/aprotinin (2 μg/ml)/chymostatin (10 μg/ml)/leupeptin (1 μg/ml)/pepstatin (1 μg/ml)/0.5 mM phenylmethylsulfonyl fluoride]. The lysate was incubated for 1 h on ice and cleared by centrifugation at 12,000 × g for 10 min at 4°C. Supernatants were precleared with 30 μl of a 50% protein G-Sepharose slurry for 2 h at 4°C. Beads were pelleted by centrifugation and supernatants were incubated overnight at 4°C with 30 μg of IgG fraction from anti-HDAC1 or anti-HDAC3 polyclonal antiserum (preincubated 2 h at room temperature with either the homologous or heterologous immunizing peptide). As a control we used 10 μl of the corresponding preimmune serum. Thirty microliters of a 50% protein G-Sepharose slurry was added for 2 h at 4°C. Immune complexes were pelleted by centrifugation and washed three times with 1 ml of lysis buffer and three times with lysis buffer containing 0.5 M NaCl. Beads were resuspended in 200 μl of HDAC buffer (20 mM Tris⋅HCl, pH 8.0/150 mM NaCl/10% glycerol), and a HDAC assay was performed as described (1). Briefly, a synthetic peptide corresponding to amino acids 1–23 of histone H4 was synthesized and purified by reverse-phase HPLC. This peptide (0.5 mg) was acetylated in vitro by overnight incubation with [3H]acetate (5 mCi, 5.3 Ci/mmol; 1 Ci = 37 GBq; NEN) in 500 μl of EtOH in the presence of 0.24 M benzotriazole-1-yloxy-tris(dimethylamino)phosphonium hexafluorophosphate (Aldrich) and 0.2 M triethylamine. The reaction mixture was dried, resuspended in 10% methanol, and purified by reverse-phase HPLC. Fractions containing acetylated peptides were lyophilized and resuspended in water. For deacetylase assays, 20,000 cpm of acetylated peptide was incubated with the immunoprecipitate for 24 h at 16°C and the reaction was stopped by adding acetic acid (0.04 M, final concentration) and HCl (250 mM, final concentration). The mixture was extracted with ethyl acetate and the released [3H]acetic acid was quantified by scintillation counting. For inhibition studies, the immunoprecipitated complexes were preincubated with the various drugs for 30 min at 4°C.
Isolation of [3H]Acetyllysine-Labeled Nucleosomes.
Jurkat T cells (2 × 106 cells per ml) were incubated with [3H]acetic acid (250 μCi/ml) for 1 h at 37°C. Cells were washed three times in ice-cold 1× PBS/10 mM sodium butyrate, resuspended in buffer A (10 mM Tris⋅HCl, pH 7.4/10 mM NaCl/3 mM MgCl2/0.3 M sucrose/10 mM sodium butyrate/protease inhibitors) at 2.5 × 107 cells per ml and incubated for 10 min at 4°C. An equivalent volume of buffer A/0.2% Nonidet P-40 was added and cells were incubated for 10 min at 4°C. Nuclei were centrifuged at 600 × g for 10 min, resuspended at 106 cells per ml of buffer A/10 mM CaCl2, and digested with micrococcal nuclease (Sigma; 0.05 unit/ml) for 30 min at room temperature. The digestion was stopped by cooling and nuclei were centrifuged (500 × g, 10 min). The pellet was resuspended in 350 μl of 1 mM EDTA, pH 7/10 mM sodium butyrate for 10 min at 4°C. After centrifugation (10 min, 800 × g), the supernatant was adjusted to 150 mM sucrose and layered on top of a 1 M sucrose cushion (10 mM Tris⋅HCl, pH 7.5/10 mM sodium butyrate/protease inhibitors), and centrifuged at 50,000 × g for 48 h. The pellet was resuspended in 150 μl of 10 mM sodium cacodylate (pH 7) and extensively dialyzed against the same buffer. For the deacetylation assay, 5 μl of [3H]acetyllysine-labeled nucleosomes were incubated with HDAC1 or HDAC3 immunoprecipitated proteins, and the reaction products were resolved on a 15% SDS/PAGE gel. Gels were fixed and stained with Coomassie blue and enhanced with a fluorography enhancing solution (Amplify, Amersham) for 30 min. Gels were dried and exposed for autoradiography at −70°C for 3–15 days.
RESULTS
Cloning of cDNA for a Human RPD3 Ortholog, HDAC3.
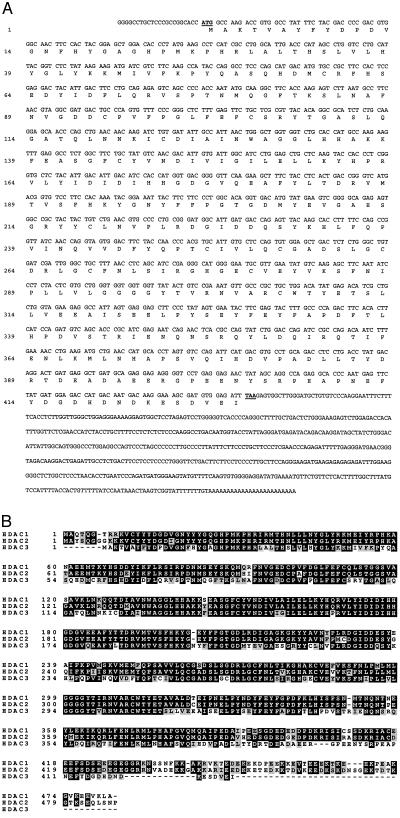
To identify RPD3 orthologs expressed in human cells, we screened the National Center for Biotechnology Information (National Institutes of Health) database of expressed sequence tags (ESTs) with the yeast RPD3 sequence and identified more than 40 EST clones. These sequences were compared with each other and with the published sequence of HDAC1. We focused our effort on a single clone from the I.M.A.G.E Consortium that appeared distinct from both HDAC1 and HDAC2 and was representative of several other ESTs. Analysis of the sequence of this cDNA, referred to as HDAC3 hereafter, revealed the existence of an ORF of 1,284 bp encoding for a putative protein of 428 amino acids (Fig. 1A). The predicted protein has a theoretical molecular mass of 48.8 kDa and a calculated isoelectric point of 4.8. The start codon is surrounded by a favorable translational initiation context (GCCACCATGG, where the initiation codon is in boldface type) (19) and is colinear with the ATG of HDAC1 and HDAC2. A potential adenylation signal (AATAAA) is located at nucleotide position 1,895, 20 nucleotides before the poly(A) tail.
Figure 1.
Nucleotide and predicted amino acid sequences of HDAC3. (A) The nucleotide and predicted amino acid sequences of HDAC3 are shown. (B) Protein sequence alignment of HDAC1p, -2p, and -3p. All three proteins were aligned by using clustal w (version 1.7) and printed by using boxshade (version 3.21). Identical residues are in black; conserved residues are in gray.
The deduced protein sequence of HDAC3 was aligned with HDAC1 and HDAC2 (Fig. 1B). HDAC3p exhibits extensive sequence homology with HDAC1p and HDAC2p. The similarity between HDAC1p and HDAC2p (84% identity and 90.5% homology) is greater than between HDAC3p and HDAC1p (58.4% identity and 73.7% homology) or HDAC3 and HDAC2 (58.5% identity and 74.8% homology), suggesting that HDAC3p might be functionally distinct from HDAC1p and HDAC2p.
HDAC3p Is a Ubiquitously Expressed and Predominantly Nuclear Protein.
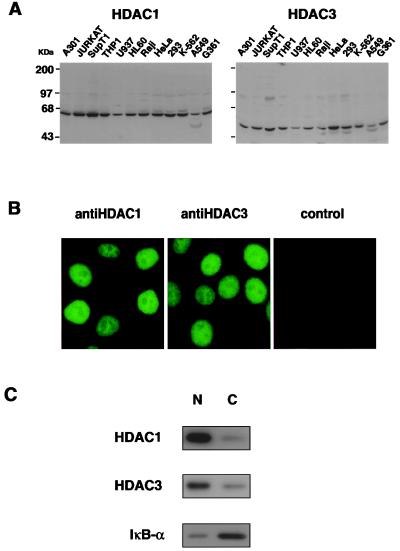
To obtain specific antisera against HDAC1p and HDAC3p, rabbits were immunized with specific peptides corresponding to the predicted C-terminal regions of HDAC1p and HDAC3p. Western blot analysis showed that the anti-HDAC3p antibody recognized a single major protein with a molecular mass of ∼50 kDa (Fig. 2A) in agreement with the predicted molecular mass (48.8 kDa). HDAC3p was detected in all human cell lines studied (Fig. 2A) and in several mouse and rat cell lines (data not shown). HDAC3p was expressed at the highest level in HeLa cells and at lower levels in the U937, Raji, and A549 cell lines (Fig. 2A). In comparison, the HDAC1p was expressed at more uniform levels in various cell lines (Fig. 2A). Next, we examined the cellular sublocalization of HDAC1p and HDAC3p by using indirect immunofluorescence and our specific anti-HDAC1 or anti-HDAC3 antiserum. Both proteins were detected as predominantly nuclear proteins in HeLa cells (Fig. 2B). To confirm this observation, we prepared nuclear and cytoplasmic extracts from Jurkat cells and analyzed proportional amounts of nuclear and cytoplasmic proteins by Western blotting with anti-HDAC1 or anti-HDAC3 antiserum. This analysis showed that both HDAC1p and HDAC3p are predominantly located in the cell nucleus (Fig. 2C). Western blot analysis of the same fractions with an anti-IκB-α antiserum showed that this protein is predominantly cytoplasmic, as predicted (Fig. 2C).
Figure 2.
HDAC1p and HDAC3p are ubiquitously expressed and predominantly nuclear proteins. (A) Western blot analysis. Western blots containing 20 μg of protein per lane from different cell lines were developed with a polyclonal serum raised against HDAC1p or HDAC3p. Coomassie blue staining of gels showed that similar amounts of cellular protein were loaded in different lanes (data not shown). Sizes of protein markers are indicated. (B) Immunofluorescence. HeLa cells grown on coverslips were fixed, stained with the anti-HDAC1 or anti-HDAC3 antiserum, and examined by immunofluorescence microscopy as described (20). (C) Subcellular localization of HDAC1p and HDAC3p. Jurkat cells nuclear (N) and cytoplasmic (C) fractions were prepared. Proportional amounts of each fraction were analyzed by Western blotting using specific antiserum directed against HDAC3, HDAC1, or IκB-α.
HDAC3p Is a HDAC.
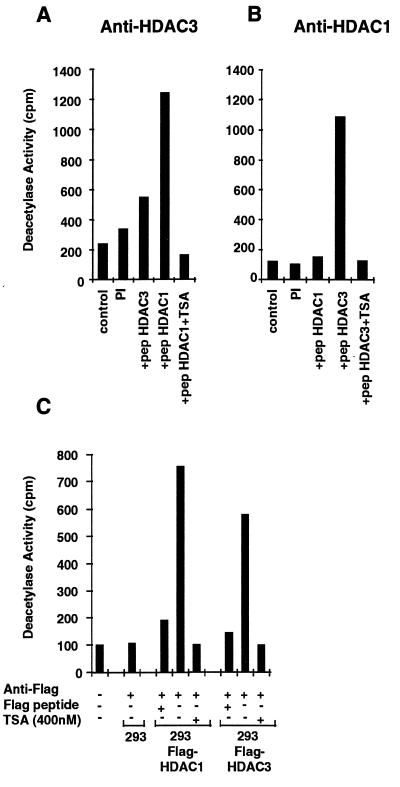
To determine whether HDAC3p exhibits HDAC activity, HDAC3p was immunoprecipitated from cellular extracts and its enzymatic activity was measured in vitro. A peptide corresponding to amino acids 1–24 of histone H4 was acetylated in vitro and used as a substrate for the deacetylase as described (1). Immunoprecipitates obtained with the anti-HDAC3 antiserum contained HDAC activity. Immunoprecipitation of this deacetylase activity was specifically blocked when the anti-HDAC3 antiserum was preincubated with an excess of the homologous HDAC3p immunizing peptide (Fig. 3A) but not with the heterologous immunizing peptide corresponding to HDAC1p (Fig. 3A). HDAC3 deacetylase activity was also inhibited in the presence of 400 nM TSA (Fig. 3A). Immunoprecipitates obtained with the anti-HDAC1 antiserum also contained HDAC activity (Fig. 3B). Immunoprecipitation of this activity was blocked by preincubating the antiserum with the homologous immunizing peptide from HDAC1p but not with the heterologous HDAC3p peptide (Fig. 3B).
Figure 3.
HDAC3 is a HDAC. (A) Cellular extracts from Jurkat cells (5 × 107 cells) were subjected to immunoprecipitation using a polyclonal antiserum against HDAC3p, in the presence of a 100-fold excess of HDAC3 peptide (residues 413–428) or HDAC1 peptide (residues 467–482). As a control, the corresponding preimmune serum was used. Immunoprecipitated complexes were tested for HDAC activity, by measuring the release of [3H]acetate from an acetylated N-terminal H4 peptide (in cpm), in the absence or presence of 400 nM TSA. (B) Same protocol as described for A, except that a polyclonal serum against HDAC1p was used. (C) Cellular extracts (2.5 × 107 cells) from 293 cells stably expressing the HDAC1-FLAG or HDAC3-FLAG fusion protein or control cells were subjected to immunoprecipitation using a monoclonal antibody against the FLAG epitope (10 μg) in the absence or presence of a 100-fold excess of the FLAG peptide. Immunoprecipitated complexes were tested for HDAC activity.
To confirm these observations, the FLAG-epitope-tagged proteins HDAC1-FLAG and HDAC3-FLAG were expressed in 293 cells after transfection of the corresponding cDNAs. HDAC activity was immunoprecipitated with an anti-FLAG monoclonal antibody from the 293/HDAC1-FLAG and the 293/HDAC3-FLAG cells (Fig. 3C). Both activities were inhibited by 400 nM TSA or when the antibody was preincubated with an excess of FLAG peptide (Fig. 3C). No HDAC activity was immunoprecipitated from control 293 cell extracts with the anti-FLAG antibody (Fig. 3C). Both HDAC1p and HDAC3p exhibit enzymatic activity when expressed in insert SF9 cells using baculovirus (W.F. and E.V., unpublished observations). These observations collectively demonstrate that a specific HDAC activity is associated with the HDAC3 protein.
HDAC1p and HDAC3p Deacetylate Nucleosomal Histones.
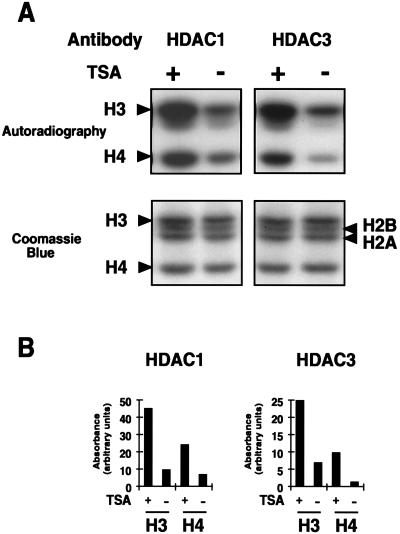
To examine the ability of HDAC1p and HDAC3p to deacetylate histones in their natural context (i.e., when incorporated into nucleosomes), we isolated nucleosomes after in vivo labeling with [3H]acetate, microccoccal nuclease digestion, and density centrifugation on sucrose cushions. Purified nucleosomes (mono-, di-, and trinucleosomes, predominantly) were incubated with immunoprecipitated HDAC1p or HDAC3p in the absence or presence of TSA. Histones were separated on 15% SDS/PAGE gels, stained with Coomassie blue, and autoradiographed. Immunoprecipitates containing HDAC1p or HDAC3p deacetylated both histone H3 and histone H4 (Fig. 4). In several experiments, we observed that histone HDAC3p more efficiently deacetylated histone H4 than HDAC1p. In the experiment shown, HDAC1 caused a 3.6-fold reduction in H4 acetylation, whereas HDAC3 caused a 8.2-fold reduction in H4 acetylation (Fig. 4). No significant deacetylation was observed with immunoprecipitates obtained with the preimmune antiserum (data not shown). In addition, preincubation of the anti-HDAC3 antiserum with the HDAC3 C-terminal peptide used to generate the antiserum completely abolished deacetylation, whereas preincubation with the unrelated HDAC1 peptide did not (data not shown). The opposite was observed with the anti-HDAC1 antiserum (data not shown). These observations therefore demonstrate that the anti-HDAC1 and anti-HDAC3 antisera specifically immunoprecipitate HDAC enzymes active on nucleosomal histones and suggest that these enzymes possess different substrate specificities.
Figure 4.
Acetylated nucleosomes are substrate for HDAC1p and HDAC3p. (A) [3H]Acetyllysine-labeled nucleosomes were incubated with anti-HDAC1 (Left) or anti-HDAC3 (Right) immunoprecipitated complexes in the absence or presence of TSA (400 nM). Reaction products were resolved on a 15% SDS/PAGE gel, and histones were visualized by staining with Coomassie blue (Lower) and by autoradiography after enhancement (Upper). The positions of the four histones H2A, H2B, H3, and H4 are indicated. (B) The autoradiogram was quantified after scanning by using the image analysis program (National Institutes of Health). Results are expressed as arbitrary absorbance values.
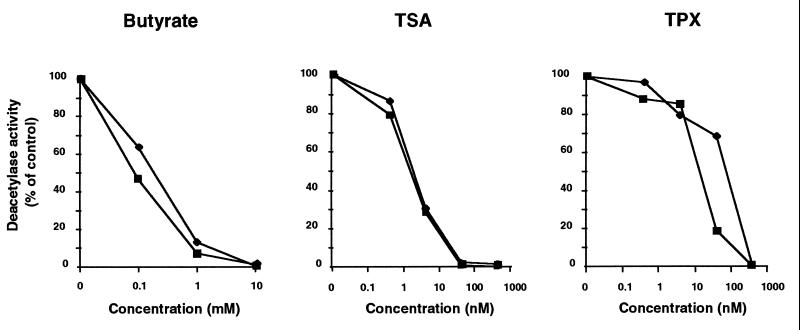
Specific Inhibition of HDAC1 and HDAC3 by Butyrate, TSA, and TPX.
To further characterize the properties of these proteins, HDAC1p and HDAC3p were used in deacetylase assays in the presence of various HDAC inhibitors (21, 22). Different concentrations of sodium butyrate, TSA, and TPX were incubated with immunoprecipitated HDAC3p and HDAC1p, and the enzymatic activity of the two proteins was measured by using in vitro-acetylated histone H4 peptide as a substrate. Both enzymes were inhibited to the same degree by butyrate and TSA and 50% inhibition was observed at 0.1 mM butyrate and 2 nM TSA for both enzymes (Fig. 5). In contrast, HDAC3 appeared more sensitive to TPX than HDAC1, with 50% inhibition at 10 nM TPX for HDAC1 and 50% inhibition at 100 nM for HDAC3 (Fig. 5).
Figure 5.
Inhibition of HDAC1p and HDAC3p deacetylase activity by butyrate, TSA, and TPX. Cellular extracts from Jurkat cells were subjected to immunoprecipitation using polyclonal antiserum against HDAC3 or HDAC1, and tested for HDAC activity, in the absence or the presence of increasing concentrations of the following inhibitors: sodium butyrate (0.1, 1, and 10 mM), TSA (0.4, 4, 40, and 400 nM), and TPX (0.4, 4, 40, and 400 nM). Results are expressed as a percentage of the HDAC activity obtained in the absence of inhibitors.
DISCUSSION
In this report, we describe the isolation and characterization of a human RPD3 ortholog, HDAC3. With a polyclonal serum directed against a C-terminal peptide of HDAC3p, we immunoprecipitated a specific HDAC activity, active both on a synthetic peptide derived from the N-terminal domain of histone H4 and on purified nucleosomal histones H3 and H4. This activity was inhibited by sodium butyrate, TPX, and TSA, indicating that these inhibitors can interact directly with the enzyme without prior cellular modifications. HDAC3p is present in all cell lines and tissues examined and is predominantly nuclear.
HDAC3p belongs to a family of proteins highly conserved from yeast to humans, highlighting the importance of the control of chromatin posttranslational modifications in eukaryotic cells. The fact that several HDAC enzymes coexist in cells (23, 24) suggests that these proteins have distinct functions. HDAC1p and HDAC2p associate in vivo with mSin3p, an integrator that targets HDACs to specific promoter regions by interacting with specific DNA binding proteins, such as Mad and Mxi (10, 11). mSin3 also interacts with the corepressors N-Cor and SMRT and, via this interaction, mediates the repressive activity of the unliganded retinoid and thyroid hormone receptors (14, 15). The interaction of HDAC1p and HDAC2p with mSin3p results in the specific targeting of a HDAC activity to specific promoter regions. Deacetylation of histone in a specific nucleosome in the context of such a promoter is thought to results in transcriptional activation or repression. Although HDAC1p and HDAC2p appear very similar at the sequence level and at the functional level, because they both interact with mSin3p, HDAC3p is more distantly related to both proteins at the amino acid sequence level. Preliminary experiments indicate that HDAC3p does not interact with mSin3p or RbAp48p (W.F., S.E., and E.V., unpublished observations) and low-stringency immunoprecipitation experiments using the anti-HDAC3 antiserum have identified several proteins distinct from those coimmunoprecipitated with the anti-HDAC1 antiserum (W.F. S.E., and E.V., unpublished observations). These experiments suggest that HDAC3p might be targeted to a subset of promoters distinct from those targeted by HDAC1p or HDAC2p via its interaction with DNA binding proteins. An additional level of specificity might also occur as a result of different substrate specificities of different HDACs. We have repeatedly observed that HDAC3p deacetylates histone H4 more completely than HDAC1 when examined in the context of nucleosomal histones (see Fig. 4). Future studies should examine the specific acetyllysine residues that are targeted in histone H3 and H4 by each of these enzymes. A difference in substrate specificity among HDAC1, -2, and -3 could result in markedly different biological effects because the acetylation/deacetylation of specific lysine residues in histones H3 and H4 has been associated with different biological functions (refs. 25–28; for review, see ref. 29).
Although several reports have shown a correlation between core histone hyperacetylation and transcriptional activation (30–35), other studies have suggested that HDACs play a significant role in both transcriptional silencing and transcriptional activation, depending on the promoter examined. First, disruption of either the RPD3 or SIN3 gene in yeast showed that both genes are required for full activation and/or or repression of specific promoters (36–38). In mammalian cells, inhibition of HDACs with TSA or TPX results in the activation or repression of a small fraction of cellular genes (39). Finally, mutations affecting the RPD3 gene in Drosophila and yeast reduced the transcriptional activity of genes near heterochromatin regions, indicating that rpd3p can counteract the genomic silencing imposed by these particular chromatin regions (40). Such differences might be explained by the local chromatin organization of different promoters. In the HIV and mouse mammary tumor virus promoters, precisely positioned nucleosomes are associated with transcriptional repression (41, 42). Inhibition of HDACs results in transcriptional activation of both promoters, consistent with a model in which the repressed state is associated with a hypoacetylated nucleosome (30, 43). In other promoters, positioned nucleosomes have been reported to facilitate transcription (44, 45). In such cases, stabilization of a nucleosome by deacetylation of core histones could result in the potentiation of transcriptional activation.
Although changes in histone acetylation have long been known to correlate with transcriptional activation and repression of specific genes, the recent identification of specific enzymes involved in these modifications, including HDAC3p in this report, will allow the direct investigation of the role of histone modifications in transcriptional regulation.
Acknowledgments
We thank David Spector (Cold Spring Harbor Laboratories) for performing the immunofluorescence microscopy analysis and Dr. Y. Nakamura (Kami-Ikebukuro Toshima-ku, Tokyo) and M.Yoshida (University of Tokyo, Tokyo) for reagents. We thank Chris Hassig (Harvard University) for discussions and for providing the protocol for in vitro acetylation of H4 peptide. C.V.L. is “Chercheur Qualifié” of the “Fonds National de la Recherche Scientifique” (Belgium). W.F. is supported by a fellowship from the Boehringer Ingelheim (Germany) and on a leave from the University of Tubingen (Germany). This work was supported in part by a grant from the National Institutes of Health of the United States Public Health Service (National Institute of General Medical Science/National Institute of Allergy and Infectious Diseases GM51671).
ABBREVIATIONS
- HDAC
histone deacetylase
- TSA
trichostatin A
- TPX
trapoxin
Note Added in Proof
While this manuscript was in preparation, Yang and coworkers (46) independently reported the identification of HDAC3.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF039703).
References
- 1.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 2.Wade P A, Pruss D, Wolffe A P. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 3.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 4.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 6.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 7.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, et al. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 8.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Y, Seto E, Chang L S, Shenk T. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 10.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 11.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 12.Ayer D E, Lawrence Q A, Eisenman R N. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, De Pinho R A. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 14.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, De Pinho R A. Nature (London) 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 15.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, et al. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 16.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 17.Sudo K, Chinen K, Nakamura Y. Genomics. 1994;24:276–279. doi: 10.1006/geno.1994.1616. [DOI] [PubMed] [Google Scholar]
- 18.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak M. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spector D L, Smith H C. Exp Cell Res. 1986;163:87–94. doi: 10.1016/0014-4827(86)90560-4. [DOI] [PubMed] [Google Scholar]
- 21.Kijima M, Yoshida M, Sugita K, Horinouchi S, Beppu T. J Biol Chem. 1993;268:22429–22435. [PubMed] [Google Scholar]
- 22.Yoshida M, Kijima M, Akita M, Beppu T. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 23.Carmen A A, Rundlett S E, Grunstein M. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 24.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durrin L K, Mann R K, Kayne P S, Grunstein M. Cell. 1991;65:1023–1031. doi: 10.1016/0092-8674(91)90554-c. [DOI] [PubMed] [Google Scholar]
- 26.Fischer-Adams G, Grunstein M. EMBO J. 1995;14:1468–1477. doi: 10.1002/j.1460-2075.1995.tb07133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner B M, Birley A J, Lavender J. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 28.Jeppesen P, Turner B M. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 29.Pazin M, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 30.Van Lint C, Emiliani S, Ott M, Verdin E. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 31.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hebbes T R, Thorne A W, Clayton A L, Crane-Robinson C. Nucleic Acids Res. 1992;20:1017–1022. doi: 10.1093/nar/20.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clayton A L, Hebbes T R, Thorne A W, Crane-Robinson C. FEBS Lett. 1993;336:23–26. doi: 10.1016/0014-5793(93)81601-u. [DOI] [PubMed] [Google Scholar]
- 34.Hebbes T R, Thorne A W, Crane-Robinson C. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe A P. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadosh D, Struhl K. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 37.Vidal M, Gaber R F. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vidal M, Strich R, Esposito R E, Gaber R F. Mol Cell Biol. 1991;11:6306–6316. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Lint C, Emiliani S, Verdin E. Gene Expr. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 40.De Rubertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P. Nature (London) 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- 41.Archer T K, Lefebvre P, Wolford R G, Hager G L. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 42.Verdin E, Paras P, Jr, Van Lint C. EMBO J. 1993;12:3249–3259. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartsch J, Truss M, Bode J, Beato M. Proc Natl Acad Sci USA. 1996;93:10741–10746. doi: 10.1073/pnas.93.20.10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cullen K E, Kladde M P, Seyfred M A. Science. 1993;261:203–206. doi: 10.1126/science.8327891. [DOI] [PubMed] [Google Scholar]
- 45.Schild C, Claret F X, Wahli W, Wolffe A P. EMBO J. 1993;12:423–433. doi: 10.1002/j.1460-2075.1993.tb05674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]