Abstract
Microorganisms use genetic circuits to integrate environmental information. We have constructed a synthetic AND gate in the bacterium Escherichia coli that integrates information from two promoters as inputs and activates a promoter output only when both input promoters are transcriptionally active. The integration occurs via an interaction between an mRNA and tRNA. The first promoter controls the transcription of a T7 RNA polymerase gene with two internal amber stop codons blocking translation. The second promoter controls the amber suppressor tRNA supD. When both components are transcribed, T7 RNA polymerase is synthesized and this in turn activates a T7 promoter. Because inputs and outputs are promoters, the design is modular; that is, it can be reconnected to integrate different input signals and the output can be used to drive different cellular responses. We demonstrate this modularity by wiring the gate to integrate natural promoters (responding to Mg2+ and AI-1) and using it to implement a phenotypic output (invasion of mammalian cells). A mathematical model of the transfer function is derived and parameterized using experimental data.
Keywords: genetic circuit, logic gate, signal integration, synthetic biology
Introduction
Genetically programming cells require sensors to receive information, circuits to process the inputs, and actuators to link the circuit output to a cellular response (Andrianantoandro et al, 2006; Chin, 2006; Voigt, 2006; Tan et al, 2007). In this paradigm, sensing, signal integration, and actuation are encoded by distinct ‘devices' comprised of genes and regulatory elements (Knight and Sussman, 1997; Endy, 2005). These devices communicate with one another through changes in gene expression and activity. For example, when a sensor is stimulated, this may lead to the activation of a promoter, which then acts as the input to a circuit. There has been a large effort to create and characterize different classes of devices and to make this information publicly available in the Registry of Standard Biological Parts (parts.mit.edu).
We have constructed a device that functions as an AND gate that can integrate two input signals and control a cellular response. An AND gate is a logical operation that integrates multiple input signals. The output of an AND gate is only ON when all of the inputs are ON. If any of the outputs are OFF, then the output is OFF. An AND gate forms the core of electronic computing and is a critical device to create different genetic programs. It is particularly useful to integrate signals from multiple sensors to identify an environment with high specificity.
Bacteria use a variety of mechanisms to sense their environment, including two-component systems, transcription factors, and small RNA molecules (Hoch and Silhavy, 1995). In some cases, an environment is defined by a single signal, such as the presence or absence of a small molecule (e.g., the lac, trp, and fur operons; Setty et al, 2003). In other cases, it is a complex array of signals that are integrated by the bacterium to identify an environment. Integrating multiple signals can increase the sensing specificity. Even signals that are too general to identify a specific environment (e.g., pH, temperature, and osmolarity) can achieve higher specificity together. A similar problem arises when programming cells to identify an environment that is not naturally encountered, and for which there is not a single dominant signal. In this case, genetic logic gates are required to integrate multiple signals to achieve sensing specificity.
Different logic gates have been built using biological components such as transcription factor genes and regulatory elements (Weiss et al, 1999; Guet et al, 2002; Kramer et al, 2004) or protein–protein interactions (Dueber et al, 2003). To date, the architecture of these circuits relies on the specific identity of a particular set of inputs and a particular output. In these examples, changing the identities of the inputs and outputs is not simple. In contrast, a circuit is modular if it can be rapidly connected to different inputs and used to drive different outputs. Modularity facilitates the incorporation of a circuit into different genetic programs.
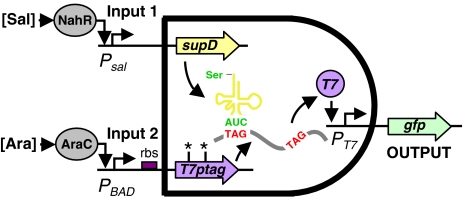
We have designed and constructed a modular genetic AND gate whose inputs and outputs are promoters (Figure 1). Only when both of the input promoters are active is the output promoter turned on. The architecture of the AND gate involves two parts, both of which are required to express T7 RNA polymerase. The first part is the T7 RNA polymerase gene, which has been modified to contain two amber stop codons that block translation. The second part is the nonsense suppressor tRNA supD, which enables the translation of polymerase. When both of these parts are transcribed from the input promoters, polymerase is expressed and this activates an output T7 promoter.
Figure 1.
A schematic representation of the genetic AND gate is shown. Two promoters are the inputs into the gate. The first promoter is linked to the transcription of the amber suppressor tRNA supD. The second promoter drives the transcription of T7 RNA polymerase. The polymerase gene has been modified to contain two amber stop codons (T7ptag). These stop codons are translated as serine when supD is also transcribed. Polymerase is expressed only when both SupD and T7ptag mRNA are present. To characterize the transfer function of the AND gate, two input promoters are used that respond to the small molecules salicylate and arabinose. In addition, the output is connected to the expression of fast-degrading green fluorescent protein.
Using two inducible systems as inputs (promoters that respond to arabinose and salicylate) and connecting the output to the expression of green fluorescent protein (gfp), we demonstrate that this circuit behaves as a near-digital AND gate. These data are used to parameterize a simple model of the steady-state input–output response (transfer function) of the circuit. This formalization will facilitate the integration of this circuit into larger genetic systems. To demonstrate the circuit is modular, two constructs are made that switch the input promoters and connect the output to a different response. First, new inputs are added that respond to the quorum signal AI-1 (luxR) and magnesium limitation (phoPQ). Second, the output is switched to the invasin gene, which enables the bacteria to invade mammalian cells. In both cases, the circuit behaves as an AND gate.
Results
Circuit design and construction
A two-input AND gate activates an output only when both inputs are on. If either or both of the inputs are off, then the output is off. Ideally, the circuit should be designed to be modular, such that the inputs and outputs can be rapidly rewired. In transcription-based systems, it is convenient that the connections between devices be promoters (Basu et al, 2004, 2005; Endy, 2005). For example, if a two-component system turns a promoter on, then this promoter can be used as the input into the next device. Similarly, if the output is a promoter, then this can either express a gene that produces a cellular phenotype or act as an input into the next device.
Our AND gate design uses two promoters as inputs and turns on an output promoter. Transcription occurs from the output promoter only when both input promoters are active. The circuit integrates the inputs via a translational interaction (Figure 1). The first input promoter drives the transcription of mRNA encoding T7 RNA polymerase that by itself cannot be translated due to the presence of amber stop codons. The second input promoter drives the transcription of an amber suppressor, which allows the activator to be expressed and activate an output promoter.
The suppression is based on the TAG amber stop codon and the SupD amber suppressor tRNA derived from Escherichia coli tRNA2Ser (Hoffman and Wilhelm, 1970). In wild-type bacteria, the TAG codon is decoded by release factor 1 resulting in translation termination. In the presence of SupD, TAG codons are decoded as serine, and translation resumes to generate the full-length protein. Because there are only 326 TAG codons in E. coli (Blattner et al, 1997), suppressed expression can be as efficient as 50% with no loss of viability (Anderson et al, 2002). We verified that the T7 RNA polymerase and supD parts do not affect the growth rate or morphology when expressed in E. coli (Supplementary information).
T7 RNA polymerase was chosen to be the activator in the circuit, although in principle any transcriptional regulator could be used in this design. The T7 gene was modified to contain amber codons at positions 8 and 14 (T7ptag). This causes premature translational termination, resulting in a non-functional polypeptide. This combination of mutations afforded the lowest basal T7 RNA polymerase activity and the highest gain in activity when coexpressed with an amber suppressor (Santoro et al, 2002). When both the polymerase and supD genes are expressed, full-length polymerase is synthesized, and the output T7 promoter becomes activated.
To characterize the circuit dynamics, two promoters that can be induced with small molecules were used as inputs (Figure 1, plasmid details in Supplementary information). The supD gene was placed under the control of a salicylate-activated promoter (Psal) (input 1). The T7ptag gene was placed under the control of an arabinose-inducible promoter (PBAD) (input 2). To monitor activation, a fast folding green fluorescent protein containing a degradation tag (GFPmut3_LAA), was placed under the control of the T7 promoter.
The first construct contained a strong ribosome binding site (rbs) (Table I) and did not function as an AND gate. The circuit was always inducible by arabinose, independent of the concentration of salicylate (not shown). Intuitively, this could result if the basal expression of T7ptag was high, where a sufficient amount of activator was produced even in the absence of arabinose. In other words, the range of the activity of the input promoter did not match the range required for the proper behavior of the circuit. This problem has been observed before in genetic circuit design (Yokobayashi et al, 2002). A successful approach to matching the range of the input to a downstream circuit has been to mutagenize the rbs, either using rational substitutions or random mutagenesis (Yokobayashi et al, 2002; Feng et al, 2004; Anderson et al, 2006).
Table 1.
Sequenced ribosome binding sites
| Parenta | GGAGGAATTAACCATG |
| Libraryb | NNNGGAATTAACCRTG |
| B9 | CTAGGAATTAACCGTG |
| F11 | AAAGGAATTAACCGTG |
| MgrB | AAAGGAATTAACCATG |
| Salc | TCAGGAGTCATCATTATTTATG |
Abbreviation: rbs; ribosome binding site.
aStart codons are in bold.
bN=A, T, C, or G; R=A and G.
cThe rbs used to measure fluorescence from the salicylate promoter.
To tune the range of the PBAD input, we designed a saturation mutagenic library of three positions in the rbs and the first base of the start codon (Table I). This library of 128 theoretical variants was plated on media containing arabinose and salicylate, and 50% of the colonies were visibly fluorescent green. Of these, 48 green colonies were subsequently grown in liquid media with no inducer, only salicylate, or both inducers and assayed by fluorimetry. Of the 48 assayed variants 44 showed at least five-fold gain in fluorescence when both inducers were added compared to values obtained when only one or no inducer was added (Supplementary information). Therefore, most variants displayed AND-gate behavior. Two variants, B9 and F11, were chosen for further characterization. The B9 clone has a weaker rbs and behaves as a functional AND gate. The F11 clone has a weaker rbs than the initial sequence, but it produces a similar salicylate-independent response.
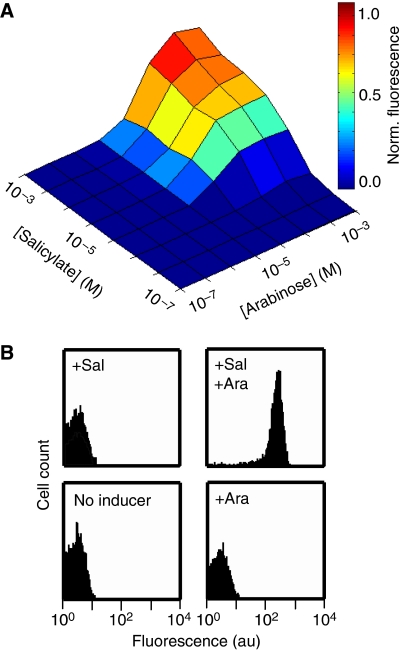
The B9 clone was used to further characterize the function of the AND gate circuit. The output of the circuit was measured by growing cells to mid-log phase in different combinations of the two inducers (Materials and methods). The output of the circuit was measured using fluorimetry (Figure 2). The transitions between the on and off states were very steep, thus producing a near-digital AND gate. Flow cytometry was used to measure the population heterogeneity (Figure 2B and Supplementary information). There was no detectable expression in the absence of either inducer and a 1000-fold induction when both inducers are present.
Figure 2.
Integration of two inducible promoters by the AND gate. (A) The fluorescence was measured for 64 combinations of inducer in a fluorimeter. The data are shown for (left to right) 0, 3.2 × 10−7, 1.3 × 10−6, 5.2 × 10−6, 2.1 × 10−5, 8.3 × 10−5, 3.3 × 10−4, and 1.3 × 10−3 M arabinose, and (bottom to top) 0, 1.5 × 10−7, 6.1 × 10−7, 2.4 × 10−6, 9.8 × 10−6, 3.9 × 10−5, 1.6 × 10−4, and 6.2 × 10−4 M salicylate. (B) The fluorescence was measured in individual cells using a flow cytometer to determine the population level behavior. The entire population of cells is turned on in the presence of both arabinose and salicylate (1.3 × 10−3 and 6.2 × 10−4 M, respectively). When either inducer is not added, the entire population is turned off. There is a 1000-fold induction between the ON and OFF states. The data for this figure were obtained using plasmids pAC-SalSer914, pBACr-AraT7940, and pBR939b (Supplementary information).
Transfer function model
The transfer function of a genetic circuit describes the steady-state response as a function of the activity of the input promoters. For a logic gate, this is a two-dimensional function, where two inputs are being integrated into a single output. An analytical form for the transfer function was derived on the basis of biochemical interaction underlying the circuit architecture and a simple model of translation control (Gilchrist and Wagner, 2006). The model relates the normalized output of the AND gate (G/Gmax) to the individual transfer functions of the two input promoters, I1 and I2.
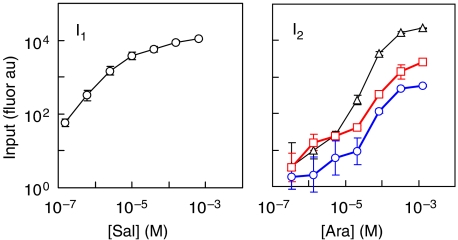
The transfer function was derived in order to understand how the range of the input promoters affects the function of the circuits. To characterize the circuit, two promoters are used that can be induced by small molecules (salicylate and arabinose). However, to generalize the model, I1 and I2 should be the activity of the PBAD and Psal promoters and not dependent on the concentrations of the small molecules. The activity of these promoters can be measured independently by fusing gfp to the promoter and measuring the output in response to the small-molecule inducer, thus producing a one-dimensional function (Figure 3). The individual responses of the two promoters are then used to parameterize the transfer function.
Figure 3.
The individual transfer functions for the input promoters are shown. The transfer functions were measured by fusing the promoter to gfp and measuring the fluorescence as a function of the concentration of small molecule inducer (salicylate or arabinose). The transfer functions were used to parameterize the AND gate model. The left panel shows the activation of Psal in response to salicylate. This promoter is leaky even in the complete absence of inducer. The right panel shows the transfer functions for the PBAD parent rbs (△), F11 (red ▪), and B9 (blue ○) clones (Table I). The average and standard deviation of four fluorimetry experiments are shown (the error is often smaller than the size of the data point). The data shown in this figure were obtained using plasmids pBACr899 (Psal), pBAC872s (PBAD, parent rbs), pBAC978 (PBAD, F11 rbs), pBAC987 (PBAD, B9 rbs) (Supplementary information).
The full derivation of the transfer function is described in the Supplementary information. The form of transfer function relating fluorescence measurements is as follows:
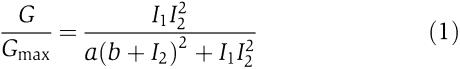 |
where Gmax is the maximum fluorescence observed for the output. Once a and b are calculated, these parameters could be used in conjunction with any two input promoters, provided that their one-dimensional transfer function was determined under the same standard growth conditions and plasmids.
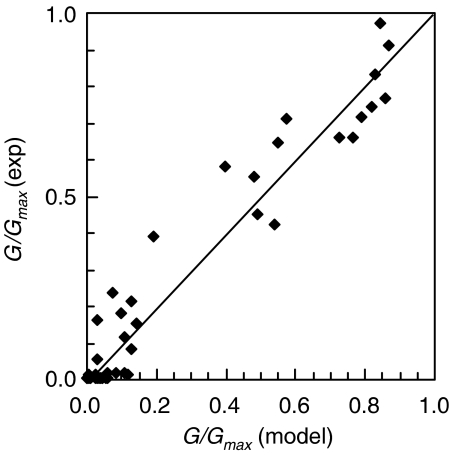
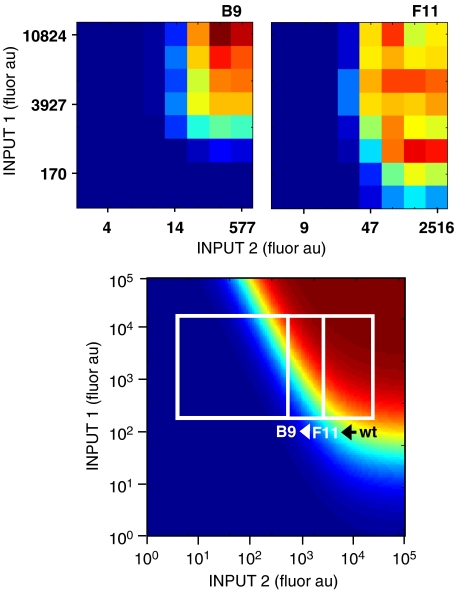
Equation (1) was parameterized using the full set of experimental data for the B9 clone when both inducers were systematically varied (Figure 2A). To calculate the one-dimensional transfer functions for I1 and I2, the gene for a fast-degrading green fluorescent protein was fused to the salicylate and arabinose promoters (Supplementary information). For the salicylate promoter, a strong rbs was used as a standard measure of promoter activity (Table I). For the arabinose reporters, the original, B9, and F11 rbss were inserted upstream of GFP. The fusions were cloned into a plasmid and the fluorescence was measured as a function of inducer concentration (Figure 3). For each pair of salicylate and arabinose concentrations, the one-dimensional fluorescence data were used to obtain I1 and I2, respectively. The two-dimensional data were used to obtain the value of G/Gmax. Fitting these data to equation (1) yielded the following: a=50±20 and b=3000±1000 (Figure 4; Supplementary Figure S1).
Figure 4.
The AND gate model was parameterized using fluorescence data. The fit to the AND gate transfer function is shown for the B9 clone. Each point represents one experimental data point from the two-dimensional array of inducer combinations (Figure 2). This was compared to the value of G/Gmax calculated using equation (1) and using the values of I1 and I2 from the one-dimensional data (Figure 3). The fit was performed using a non-linear regression algorithm to yield a=50 and b=3000. The Pearson correlation coefficient for the fit is 0.971. The full fit to all of the data is shown in the Supplementary information.
The parameterized transfer function captures the behavior of the circuit when input promoters with different transfer functions are connected to the AND gate (Figure 5). When the model is extrapolated outside of the B9 data used for parameterization, it can be seen that different ranges of I2 lead to different responses. In particular, a stronger rbs (as for the original and F11 clones) extends the upper limit of the range. This leads to a loss of the AND gate function, as now the circuit is inducible independent of I1. Note that for any range of I1, it is possible to generate a functional gate by altering the strength of the rbs of I2. This suggests that it is better that leaky promoters (such as Psal) be used as I1, as this can be compensated for by adjusting the rbs of I2. Here, we have relied on random mutagenesis and screening to identify functional rbss. Our ultimate goal is to use the transfer function and lists of standardized functional rbss (e.g., Registry of Standard Biology Parts entries BBa_J61100-39) to predict the rbs required to make the circuit functional for two input promoters.
Figure 5.
The activity range of the input promoters affects the function of the AND gate circuit. In top panel, the experimental fluorescence of the B9 and F11 gates is shown as a function of I1 and I2. In the bottom panel, the theoretical transfer function (bottom) is calculated using equation (1) and the fit values for the parameters a and b. The white boxes show the ranges for the wild type as well as the F11 and B9 mutants, which have progressively weaker rbss. These boxes are drawn on the basis of range of the one-dimensional transfer functions (Figure 3). It is only the B9 clone that behaves like an AND gate, requiring the maximal activation of both promoters before the output is turned on. In contrast, the F11 clone always shows some activity at high levels of I2, independent of I1.
Circuit modularity
The AND gate is designed to be modular, where the inputs and outputs can be changed by swapping promoters and directing mutagenesis to the rbs. Using this device, any two environmental signals can be connected to the AND gate, as long as they lead to the activation of a promoter. Similarly, the T7 promoter can be used to drive a cellular response or act as the input to another downstream circuit.
To demonstrate the modularity of the circuit, two constructs containing different inputs and outputs were designed and analyzed. First, the inputs are swapped for two natural promoters (quorum sensing and Mg2+ responsive). Next, the output is swapped from gfp to the invasin gene, which enables the bacteria to invade mammalian cells. Both of these circuits demonstrated AND-gate behavior, thus demonstrating the modularity of the circuit and the capability to integrate natural inputs and control cellular behavior as an output.
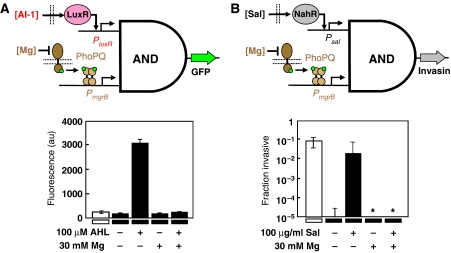
To replace the inputs, PBAD and Psal were replaced with PmgrB and Plux, which respond to magnesium limitation and the AI-1 quorum signal, respectively (Supplementary Figure S7). Two-component systems are a ubiquitous sensing motif in bacteria, consisting of a membrane-bound sensor and a cytoplasmic response regulator (Hoch and Silhavy, 1995). When stimulated by an environmental signal, the sensor phosphorylates the response regulator, which can then modulate gene expression. The E. coli PhoPQ two-component system responds to the external magnesium concentration. The PhoQ regulator activates the mgrB promoter in the absence of exogenous magnesium (Kato et al, 1999; Minagawa et al, 2003). Quorum sensing systems are used by bacteria to communicate (Bassler and Losick, 2006). These systems have been used extensively as communication devices in synthetic genetic systems to program cells to form patterns (Basu et al, 2005), regulate their density (You et al, 2004), and kill malignant cells in response to bacterial density (Anderson et al, 2006). The lux promoter and luxR gene, derived from the Vibrio fischeri quorum sensing circuit, is induced in response to exogenous N-3-oxohexanoyl-L-homoserine lactone (AI-1) (Sitnikov et al, 1995).
The inputs were connected to the circuit one at a time. First, the mgrB promoter was placed in front of the T7ptag gene with the B9 rbs. The circuit function was then assayed using the salicylate input to drive supD. This construct did not generate an AND-gate as cells showed no fluorescence when induced with salicylate. Presumably, this is because insufficient transcript was produced from PmgrB. As before, this problem was overcome by retuning the rbs. A clone was identified with a stronger rbs (Table I) that yielded a functional AND-gate. Next, Plux and the luxR gene were used to transcribe the supD gene. The circuit shows 15-fold fluorescence over background when fully induced but undetectable fluorescence in the presence of Mg2+ or the absence of AI-1 (Figure 6).
Figure 6.
The modularity of the AND gate was demonstrated by swapping the inputs and output of the circuit. (A) The inputs of the circuit were exchanged with the lux and mgrB promoters. The lux promoter responds to the quorum signal input AI-1 and the mgrB promoter responds to the absence of exogenous magnesium via the PhoPQ two-component system. Only when both promoters are active (in the presence of AI-1 and under magnesium limitation) is the output on. The white bar shows the background fluorescence of E. coli DH10B cells with no plasmids. The plasmids used to obtain these data are pBACr-Mgr940, pSupDLuxR, and pBR939b (Supplementary information). (B) Replacing the output gfp gene with the inv gene results in the invasion of mammalian cells only when both input promoters are on. The invasiveness of the bacteria is equivalent to the expression of inv from a constitutive promoter (white bar). The stars indicate no invasion being detected. In both panels, the error bars show the standard deviation of four replicates. The plasmids used to obtain these data are pSalSer914 and pBACr-Mgr951 (Supplementary information).
We next examined whether the output of the AND gate could control a cellular behavior. The expression of the invasin gene (inv) of Yersinia pseudotuberculosis in E. coli confers the ability to invade mammalian cells expressing β1-integrin (Isberg et al, 1987). We have previously shown that singular environmentally-responsive promoters controlling inv confer environment-dependent invasion (Anderson et al, 2006). Here, the inv gene is substituted for gfp and the AND gate is tested using the salicylate and Mg2+ inputs.
In a previous study, we created an rbs variant of inv that conferred arabinose-dependent invasion under a PBAD promoter (Anderson et al, 2006). This construct was placed under the control of the T7 promoter and combined with the AND gate plasmids that have PmgrB and Psal as inputs. After being grown in different combinations of the inputs, the bacteria were assayed for invasion of HeLa cells (Materials and methods). Bacteria grown in the presence of Mg2+ or the absence of salicylate show no detectable invasion (Figure 6B). The bacteria only invade when both of the input promoters are on. These experiments demonstrate the ability of a modular AND gate to integrate multiple environmental signals and respond with cellular behavior.
Discussion
We have constructed a modular AND gate based on the amber suppression of T7 RNA polymerase. Only when two input promoters are active is an output turned on. Because the inputs and outputs of this gate are transcriptional signals, they can be easily replaced. This modularity is demonstrated by swapping the inputs and outputs of the circuit while preserving the AND-gate behavior. In this paradigm, changes in transcription become a common currency allowing the modular integration of individual devices (Weiss et al, 1999; Endy, 2005).
The modular nature of this type of transcriptional logic gate will facilitate its use in a variety of engineering applications. In particular, AND logic would be useful in obtaining gene expression in a specific microenvironment. A set of promoters—identified rationally or with a microarray—could be used as inputs to the AND gate. Rather than directly detecting the presence of a new environment with a single engineered promoter or sensing system, independent promoters sense different aspects of the environment. The AND gate activates only when all conditions are present to induce cellular responses. Often microenvironments are defined by multiple nonspecific signals such as oxygen, pH, cell density, lactate, and glucose. It is only when several of these inputs are integrated that specificity can be achieved.
A two-dimensional transfer function of a simple mathematical form captures the input–output behavior of the AND gate. This model could be used to predict the genetic changes required to connect two inputs to the circuit to produce a functional AND gate. To be able to use the model, the inputs have to be characterized using the plasmid and fluorescent reporter system used in this study. In this sense, this work represents a step toward standardization, where circuits are characterized quantitatively to understand their collective function when connected in series. This will be a critical approach in the design of large integrated systems consisting of multiple genetic circuits.
The transfer function we derived relies on a steady-state assumption and is based on a deterministic model. However, the response of the circuit may have dynamic or stochastic aspects that are not predicted from the model. For some applications where the induction of inputs is transient, the dynamics of the circuit could be critical to the successful implementation of the output process. A further obstacle to standardization is that the circuit may show different response characteristics in different environments or stages of growth. For example, this AND gate produces a lower gain at low cell densities (not shown). This change in the output range could impact its connection to a downstream circuit.
The ultimate goal in genetic circuit design is to incorporate them into more complex systems consisting of multiple circuits, sensors, and actuators. Unlike electronic circuits, where the spatial wiring of a circuit determines the flow of information through the circuit, intracellular circuits prevent cross-communication through specificity of biochemical interactions. Once a part—such as T7 or SupD—is used, it cannot be used in any of the other devices. Therefore, the use of the T7 RNA polymerase-based gate makes this valuable gene unavailable for protein overexpression within the same system. An advantage of our design is that any particular transcriptional activator, including engineered sequence-specific transcription factors (Mandell and Barbas, 2006), could be used in place of the polymerase gene. Similarly, the use of amber suppression precludes its use in other systems. For example, translational recoding with unnatural amino acids using amber suppression could not coexist with this logic gate (Wang et al, 2006). Other translational regulators that could be used in this gate include nonsense, missense, and frameshift suppressors, or riboregulators (Anderson et al, 2002; Isaacs et al, 2004).
Pushing the boundary of genetic engineering will require a toolbox of genetic circuits that perform prescribed functions and are designed to be incorporated into larger systems. Toward this end, we have described the construction and analysis of a genetic AND gate. We have demonstrated that this gate is modular, so that it can be connected to different promoter-based inputs and used to drive different outputs. Further, in a step toward standardization, we developed a model that could be empirically parameterized, and used to predict how new promoters will connect to the gate. Circuits like this will find broad application in genetic engineering of systems in which multiple transcriptional signals must be combined to produce a specific cellular response.
Materials and methods
Strains and growth conditions
All manipulations were performed in E. coli strain MC1061, DH10B, or EC100D™ pir-116 (Epicentre, Madison, WI) growing in 2YT liquid media or LB agar plates supplemented with antibiotics at 25 μg/ml at 37°C. HeLa cells were obtained from the UCSF Cell Culture Facility (San Francisco, CA) and grown in DMEM media supplemented with 10% FCS and 1% streptomycin/penicillin solution. DNA-modifying enzymes were purchased from New England Biolabs. Oligonucleotides were synthesized by Sigma-Genosys (The Woodlands, TX) and used unpurified. PCR was performed with the Roche High-Fidelity PCR kit. Plasmid pLC113 containing the salicylate promoter was a gift from Sandy Parkinson (Ames et al, 2002). Plasmid pAC-SupD and the double amber mutant of T7 RNA polymerase were described previously (Anderson et al, 2002; Santoro et al, 2002). N-3-oxohexanoyl-L-homoserine lactone, L-arabinose, and sodium salicylate were from Sigma.
Plasmid design
Sequences of the plasmids constructed for this study are available through the Registry of Standard Biological Parts (http://parts.mit.edu). Reporter plasmid pBR939B is a pBR322 derivative containing the pMB1 origin of replication, an ampicillin resistance gene, and a GFPmut3_LAA gene under the control of a T7 promoter and a rrnB terminator. Plasmid pAC-SalSer914 is a pACYC184 derivative containing the p15A origin of replication, a chloramphenicol resistance gene, and the supD gene under the control of a salicylate operon and an rrnB terminator. To construct pAC-SalSer914, the salicylate operon was PCR-amplified from plasmid pLC113 with oligonucleotides ca899F and ca899R and inserted into the NotI and BamHI sites of plasmid pAC581 (Anderson et al, 2006) to obtain plasmid pAC899. Subsequently, the supD gene was PCR-amplified from plasmid pAC-SupD with oligonucleotides ca914F and ca914R and inserted into the BamHI and EcoRI sites of plasmid pAC899. T7 RNA polymerase-expressing plasmids were constructed in plasmid pBAC872s (Anderson et al, 2006) containing the F plasmid origin of replication and par genes, an R6K origin of replication, a kanamycin resistance gene, and a BamHI/EcoRI cassette flanked by an arabinose promoter and a TrrnB terminator. In strain EC100D™ pir-116, the R6K origin confers high copy number. In strain MC1061, pBAC872s derivatives are single-copy plasmids. Arabinose-inducible GFP reporter plasmids pBAC987 and pBAC978 containing the B9 and F11-derived rbss were constructed from plasmid pBAC872s. The GFP cassette present in pBAC872s was PCR-amplified with oligonucleotides ca978F or ca987F and ca606R and inserted into the BamHI and EcoRI sites of pBAC872s. The GFP sequence contains a degradation tag that confers a half-life of 40 min (Miller et al, 2000).
To construct Plasmid pSupDLuxR, a variant of pAC581 (pAC-SupDb) was first constructed with BamHI and EcoRI sites upstream of the supD gene. The entire lux operon was PCR-amplified with oligonucleotides ca742F and ca721R from plasmid pAC-LuxGfp (Anderson et al, 2006) and inserted into the BamHI and EcoRI sites of pAC-SupDb to obtain plasmid pSupDLux. The luxI gene in pSupDLux was then excised by inverse PCR with oligonucleotides ca752F and ca747R and recircularization with BglII to obtain pSupDLuxR.
Plasmid pBACr-Mgr940 was constructed from plasmid pBAC874t, a variant of pBAC872s with a Ptet promoter (Anderson et al, 2006). The mgrB promoter was PCR-amplified from E. coli strain MG1655 genomic DNA with oligonucleotides ca901F and ca901R and inserted into the NotI and BamHI sites of pBAC874t to obtain plasmid pBACr-Mgr901. Subsequently, T7ptag variants were inserted into the BamHI and EcoRI sites of pBACr-Mgr901.
Plasmid pBACr-Mgr951 was constructed from plasmids pBACr-Mgr940 and pBACr-Inv939. Plasmid pBACr-Inv939 is a pBAC874t derivative containing a T7-GFP cassette derived from pBR939b within the NotI and BamHI sites upstream of the BamHI/EcoRI cassette from plasmid pBACr-AraInv (Anderson et al, 2006) containing the rbs and the invasin gene. The T7-GFP-Invasin fragment of pBACr-Inv939 was PCR-amplified with oligonucleotides ca279 and ca951R and inserted into the NotI site of pBACr-Mgr940 to yield plasmid pBACr-Mgr951.
Saturation mutagenesis and library screening
To construct the T7 RNA polymerase rbs library, the T7 RNA polymerase gene with two amber stop codons and a short 5′ linker sequence was PCR-amplified with oligonucleotides ca940F and ca564R from plasmid pREP2-HLAA02 (Santoro et al, 2002) and inserted into the BamHI and EcoRI sites of pBAC872s or pBACr-Mgr901 under the control of the arabinose promoter. In this manner, the 5′ flanking bases of the rbs and the first base of the start codon were replaced with random sequence. The library of 5′UTR variants was inserted into MC1061 cells harboring pBR939b and pAC-SalSer914 and plated on LB media supplemented with the appropriate antibiotics, 100 μg/ml salicylate and arabinose. Individual green-fluorescing colonies were grown in 500 μl aliquots in a 96-well block in the presence of 100 μg/ml salicylate, 100 μg/ml arabinose, 10 μM N-3-oxohexanoyl-L-homoserine lactone and/or 50 mM MgCl2, or no additive, and then assayed for fluorescence in a Tecan Safire fluorescence plate reader (Tecan). From this screen, individual variants B9, F11, and pBACr-Mgr940 were characterized further. A third variant was sequenced and found to introduce a hairpin occluding the rbs, which may result in aberrant translation (Isaacs et al, 2004). Thus, this variant is not considered further.
Induction experiments
To observe AND gate induction by cytometry, 50 ml cultures of MC1061 cells (salicylate and arabinose gate) or DH10B cells (AHL and no Mg2+ gate) were grown in 2YT media in the presence or absence of inducers and the appropriate antibiotics for 3 h at 37°C to OD600=1 in a baffled flask with aeration. Cytometry analysis was performed with a Becton Dickinson FACSCalibur™ on bacteria diluted in PBS buffer. Counts were gated by side and forward scatter. Data for 30 000 cells were collected for each experiment. Fluorescence was tuned relative to bacteria without a GFP gene centered within the first decade of fluorescence. For fluorimetry measurements, bacteria were grown in 400 μl cultures in 96-well blocks with shaking for 8 h at 37°C to OD600=1. Aliquots of 100 μl each were transferred to 96-well plates and assayed for fluorescence in a Tecan.
Invasion assays
Bacterial cultures were diluted 100-fold in 2YT media, and 50 μl was added to 1 ml of DMEM media in 24-well plates containing a confluent culture of HeLa cells (MOI=5). After 1 h incubation at 37°C, the wells were washed once in DMEM media and then incubated for 1 h with 1 ml of DMEM supplemented with 100 μg/ml gentamicin. Subsequently the wells were washed three times with DMEM, lysed with 1 ml of 1% Triton X-100, and then spread on LB agar plates and grown 24 h. Percent invasion was determined as the ratio of recovered bacteria divided by the number of CFU present in the original diluted culture as determined by titer. Values were averaged over four repetitions of the experiment
Supplementary Material
Supplementary Information
Acknowledgments
We thank Sandy Parkinson for supplying plasmid pLC113. JCA is supported by a Damon Runyon Cancer Research Foundation Postdoctoral Fellowship. APA was supported by the Howard Hughes Medical Institute. CAV was supported by the Sloan Foundation, Pew Fellowship, ONR, Packard Fellowship, NIH EY016546, NIH AI067699, NSF BES-0547637, UC-Discovery, and a Sandler Family Opportunity Award. APA, CAV, and JCA are supported by the SynBERC NSF ERC (www.synberc.org).
The authors declare that they have no competing financial interests.
References
- Ames P, Studdert CA, Reiser RH, Parkinson JS (2002) Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci USA 99: 7060–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Clarke EJ, Arkin AP, Voigt CA (2006) Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol 355: 619–627 [DOI] [PubMed] [Google Scholar]
- Anderson JC, Magliery TJ, Schultz PG (2002) Exploring the limits of codon and anticodon size. Chem Biol 9: 237–244 [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E, Basu S, Karig DK, Weiss R (2006) Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol 2, 2006.0028 10.1038/msb4100073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler BL, Losick R (2006) Bacterially speaking. Cell 125: 237–246 [DOI] [PubMed] [Google Scholar]
- Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R (2005) A synthetic multicellular system for programmed pattern formation. Nature 434: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Basu S, Mehreja R, Thiberge S, Chen MT, Weiss R (2004) Spatiotemporal control of gene expression with pulse-generating networks. Proc Natl Acad Sci USA 101: 6355–6360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett Gr, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y (1997) The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1474 [DOI] [PubMed] [Google Scholar]
- Chin JW (2006) Programming and engineering biological networks. Curr Opin Struct Biol 16: 551–556 [DOI] [PubMed] [Google Scholar]
- Dueber JE, Yeh BJ, Chak K, Lim WA (2003) Reprogramming control of an allosteric signaling switch through modular recombination. Science 301: 1904–1908 [DOI] [PubMed] [Google Scholar]
- Endy D (2005) Foundations for engineering biology. Nature 438: 449–453 [DOI] [PubMed] [Google Scholar]
- Feng XJ, Hooshangi S, Chen D, Li GY, Weiss R, Rabitz H (2004) Optimizing genetic circuits by global sensitivity analysis. Biophys J 87: 2195–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist MA, Wagner A (2006) A model of protein translation including codon bias, nonsense errors, and ribosome recycling. J Theor Biol 239: 417–434 [DOI] [PubMed] [Google Scholar]
- Guet CC, Elowitz MB, Hsing W, Leibler S (2002) Combinatorial synthesis of genetic networks. Science 296: 1466–1470 [DOI] [PubMed] [Google Scholar]
- Hoch JA, Silhavy TJ (1995) Two-Component Signal Transduction. ASM Press: Washington DC [Google Scholar]
- Hoffman EP, Wilhelm RC (1970) Genetic mapping and dominance of the amber suppressor, Su1 (supD), in Escherichia coli K-12. J Bacteriol 103: 32–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ (2004) Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol 22: 841–847 [DOI] [PubMed] [Google Scholar]
- Isberg RR, Voorhis DL, Falkow S (1987) Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50: 769–778 [DOI] [PubMed] [Google Scholar]
- Kato A, Tanabe H, Utsumi R (1999) Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J Bacteriol 181: 5516–5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight TK, Sussman GJ (1997) Cellular gate technology. Unconventional Models of Computation 257–272 [Google Scholar]
- Kramer BP, Fischer C, Fussenegger M (2004) BioLogic gates enable logical transcription control in mammalian cells. Biotechnol Bioeng 87: 478–484 [DOI] [PubMed] [Google Scholar]
- Mandell JG, Barbas CFr (2006) Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res 34: W516–W523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WG, Leveau JH, Lindow SE (2000) Improved gfp and inaZ broad-host-range promoter–probe vectors. Mol Plant Microbe Interact 13: 1243–1250 [DOI] [PubMed] [Google Scholar]
- Minagawa S, Ogasawara H, Kato A, Yamamoto K, Eguchi Y, Oshima T, Mori H, Ishihama A, Utsumi R (2003) Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J Bacteriol 185: 3696–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Wang L, Herberich B, King DS, Schultz PG (2002) An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat Biotechnol 20: 1044–1048 [DOI] [PubMed] [Google Scholar]
- Setty Y, Mayo AE, Surette MG, Alon U (2003) Detailed map of a cis-regulatory input function. Proc Natl Acad Sci USA 100: 7702–7707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnikov DM, Schineller JB, Baldwin TO (1995) Transcriptional regulation of bioluminesence genes from Vibrio fischeri. Mol Microbiol 17: 801–812 [DOI] [PubMed] [Google Scholar]
- Tan C, Song H, Niemi J, You L (2007) A synthetic biology challenge: making cells compute. Mol Biosyst 3: 343–353 [DOI] [PubMed] [Google Scholar]
- Voigt CA (2006) Genetic devices to program bacteria. Curr Opin Biotech 17: 548–557 [DOI] [PubMed] [Google Scholar]
- Wang L, Xie J, Schultz PG (2006) Expanding the genetic code. Annu Rev Biophys Biomol Struct 35: 225–249 [DOI] [PubMed] [Google Scholar]
- Weiss R, Homsy GE, Knight TF (1999) Toward in vivo digital circuits. In: DIMACS Workshop on Evolution as Computation. Princeton University [Google Scholar]
- Yokobayashi Y, Weiss R, Arnold FH (2002) Directed evolution of a genetic circuit. Proc Natl Acad Sci USA 99: 16587–16591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, Cox R Sr, Weiss R, Arnold FH (2004) Programmed population control by cell–cell communication and regulated killing. Nature 428: 868–871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information